Propolis Exerts an Anti-Inflammatory Effect on PMA-Differentiated THP-1 Cells via Inhibition of Purine Nucleoside Phosphorylase
Abstract
:1. Introduction
2. Results
2.1. Cytotoxicity of Propolis Extracts against PMA-Differentiated THP-1 Cells
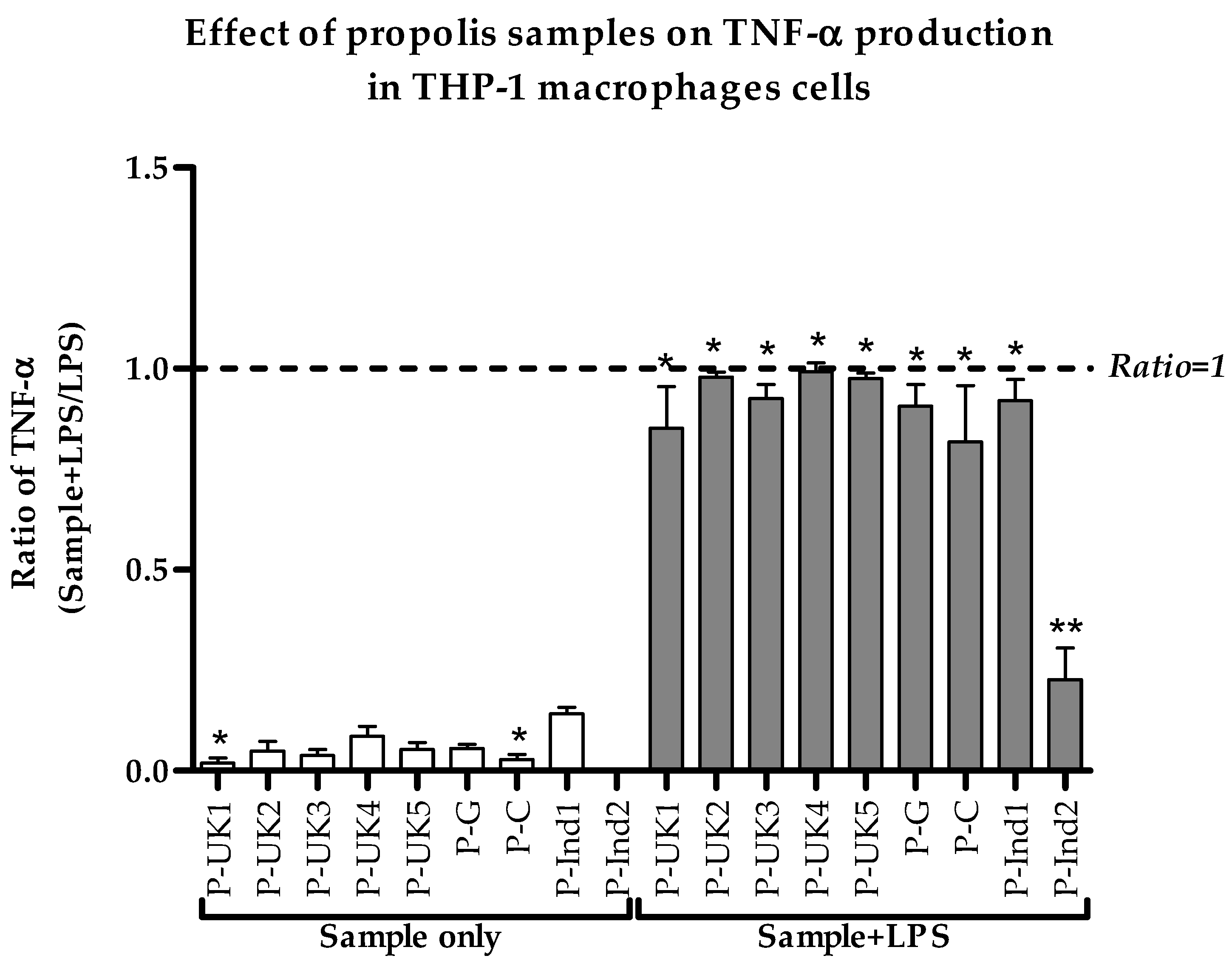
2.2. Effect of Propolis Extracts on Pro-Inflammatory TNF-α Cytokine Production
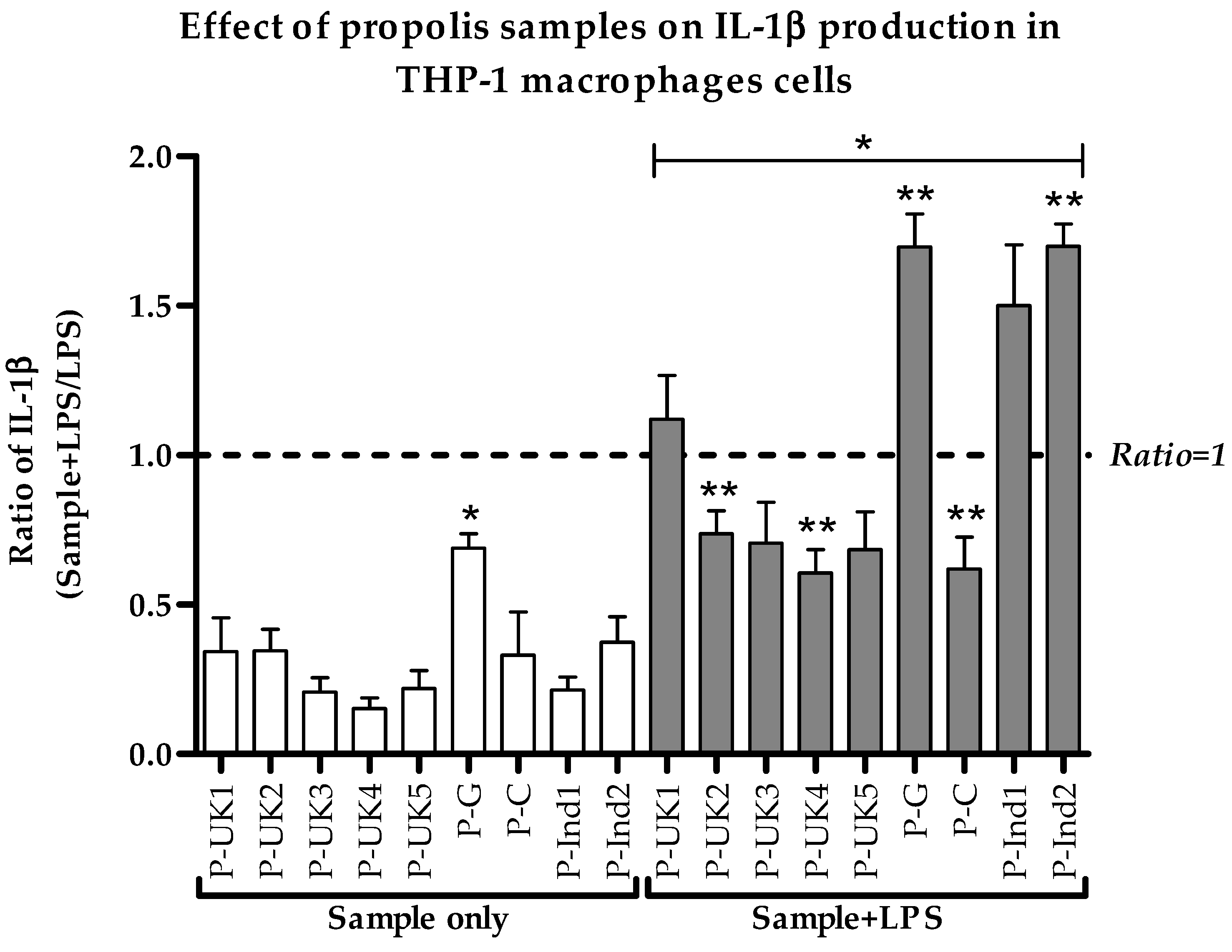
2.3. Effect of Propolis Extracts on Pro-Inflammatory IL-1β Cytokine Production
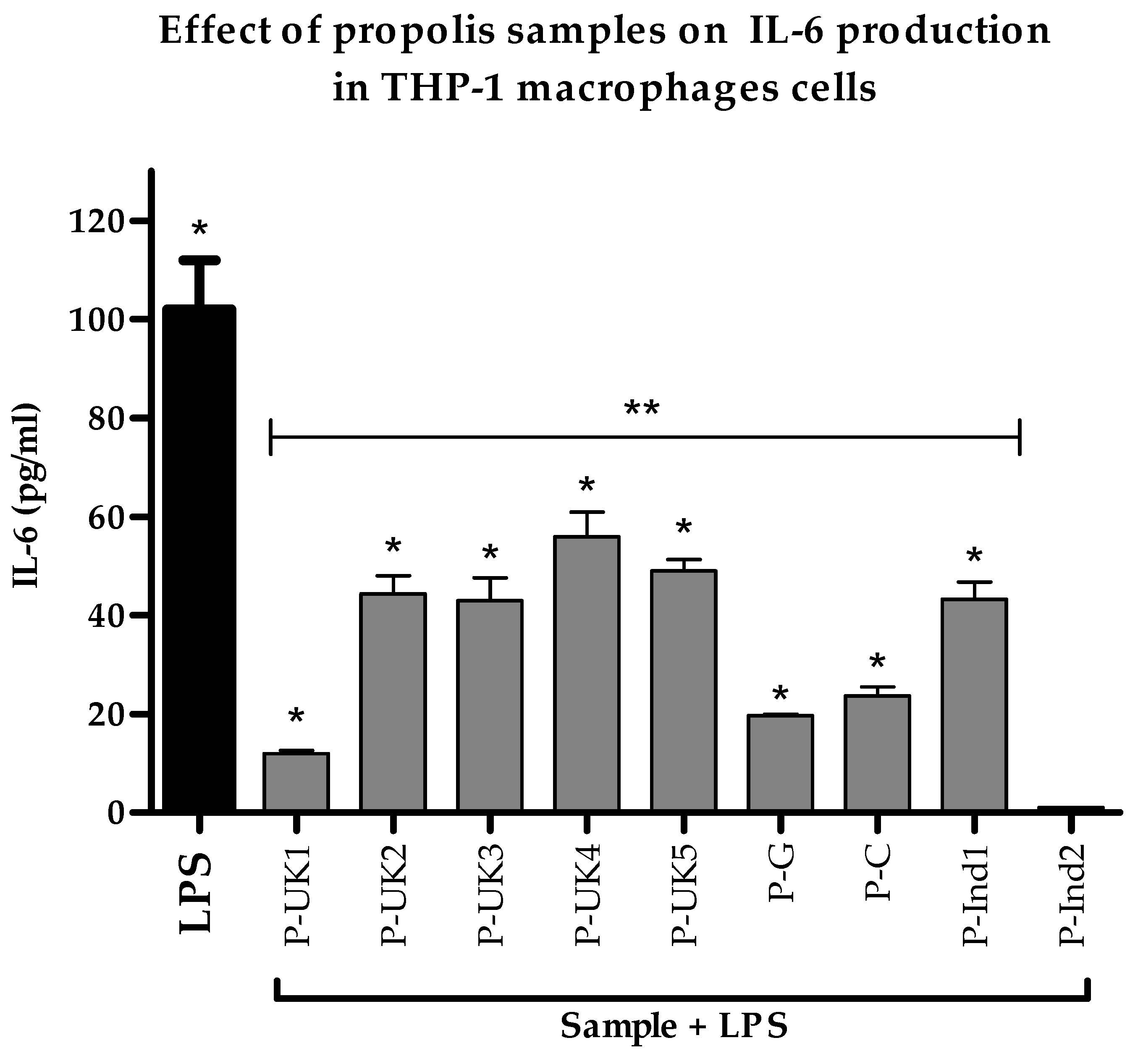
2.4. Effect of Propolis Extracts on Pro-Inflammatory IL-6 Cytokine Production
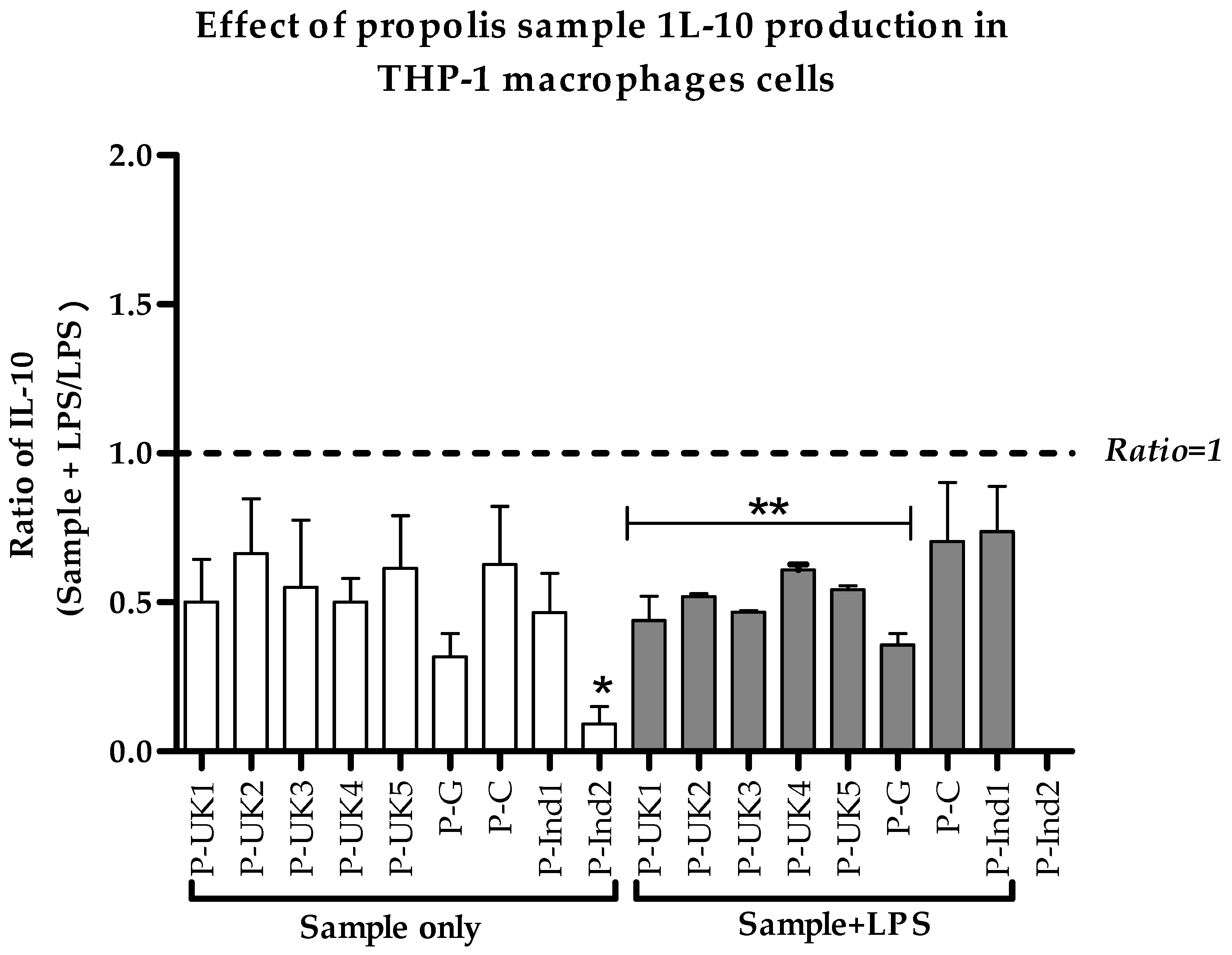
2.5. Effect of Propolis Extracts on Anti-Inflammatory IL-10 Cytokine Production
2.6. Effect of Cameroonian Propolis (P-C) on the Cell Metabolome
3. Discussion
4. Materials and Methods
4.1. Extract Preparation
4.2. Cell Culture and Differentiation
4.3. Cell Viability Assay
4.4. Cytokine Production
4.5. Enzyme-Linked Immunosorbent Assay (ELISA)
4.6. Metabolite Extraction
4.7. LC-MS Conditions
4.8. Data Extraction and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fiala, M.; Liu, Q.N.; Sayre, J.; Pop, V.; Brahmandam, V.; Graves, M.C.; Vinters, H.V. Cyclooxygenase- 2- positive macrophages infiltrate the Alzheimer’s disease brain and damage the blood-brain barrier. Eur. J. Clin. Investig. 2002, 32, 360–371. [Google Scholar] [CrossRef]
- Ohishi, S. Evaluation of time course and inter-relationship of inflammatory mediators in experimental inflammatory reaction. Yakugaku Zasshi J. Pharm. Soc. Jpn. 2000, 120, 455–462. [Google Scholar] [CrossRef]
- Marcio, A.R.A.; Silvana, A.L.; Rosane, N.M.G.; Maria Nilce, S.R.; Flávia, R.F.N. Mechanisms of action underlying the anti-inflammatory and immunomodulatory effects of propolis: A brief review. Rev. Bras. Farmacogn. 2011, 22, 208–219. [Google Scholar]
- Ramos, A.; Miranda, J. Propolis: A review of its anti- inflammatory and healing actions. J. Venom. Anim. Toxins Incl. Trop. Dis. 2007, 13, 697–710. [Google Scholar] [CrossRef]
- Majno, G. Chronic Inflammation. Am. J. Pathol. 1998, 153, 1035–1039. [Google Scholar] [CrossRef]
- Verhoeckx, K.C.; Bijlsma, S.; Jespersen, S.; Ramaker, R.; Verheij, E.R.; Witkamp, R.F.; van der Greef, J.; Rodenburg, R.J. Characterization of anti-inflammatory compounds using transcriptomics, proteomics, and metabolomics in combination with multivariate data analysis. Int. Immunopharmacol. 2004, 4, 1499–1514. [Google Scholar] [CrossRef]
- Silva, B.B.; Rosalen, P.L.; Cury, J.A.; Ikegaki, M.; Souza, V.C.; Esteves, A.; Alencar, S.M. Chemical composition and botanical origin of red propolis, a new type of Brazilian propolis. Evid. Based Complement. Altern. Med. 2008, 5, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Tiveron, A.P.; Rosalen, P.L.; Franchin, M.; Lacerda, R.C.C.; Bueno-Silva, B.; Benso, B.; Denny, C.; Ikegaki, M.; de Alencar, S.M. Chemical Characterization and Antioxidant, Antimicrobial, and Anti- Inflammatory Activities of South Brazilian Organic Propolis.(Research Article). PLoS ONE 2016, 11, e0165588. [Google Scholar] [CrossRef]
- Zancanela, D.; Herculano, R.; Funari, C.; Marcos, C.; Almeida, A.; Guastaldi, A. Physical, chemical and antimicrobial implications of the association of propolis with a natural rubber latex membrane. Mater. Lett. 2017, 209, 39–42. [Google Scholar] [CrossRef]
- Andrade, J.K.S.; Denadai, M.; de Oliveira, C.S.; Nunes, M.L.; Narain, N. Evaluation of bioactive compounds potential and antioxidant activity of brown, green and red propolis from Brazilian northeast region. Food Res. Int. 2017, 101, 129–138. [Google Scholar] [CrossRef] [PubMed]
- López, B.G.-C.; Schmidt, E.M.; Eberlin, M.N.; Sawaya, A.C.H.F. Phytochemical markers of different types of red propolis. Food Chem. 2014, 146, 174–180. [Google Scholar] [CrossRef]
- Akyol, S.; Ozturk, G.; Ginis, Z.; Armutcu, F.; Yigitoglu, M.R.; Akyol, O. In vivo and in vitro antıneoplastic actions of caffeic acid phenethyl ester (CAPE): Therapeutic perspectives. Nutr. Cancer 2013, 65, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Sahinler, N.; Kaftanoglu, O. Natural product propolis: Chemical composition. Nat. Prod. Res. 2005, 19, 183–188. [Google Scholar] [CrossRef]
- Marcucci, M.C. Propolis: Chemical composition, biological properties and therapeutic activity. Apidologie 1995, 26, 83–99. [Google Scholar] [CrossRef]
- Falcão, S.I.; Vilas-Boas, M.; Estevinho, L.M.; Barros, C.; Domingues, M.R.; Cardoso, S.M. Phenolic characterization of Northeast Portuguese propolis: Usual and unusual compounds. Anal. Bioanal. Chem. 2010, 396, 887–897. [Google Scholar] [CrossRef]
- Raghukumar, R.; Vali, L.; Watson, D.; Fearnley, J.; Seidel, V. Antimethicillin-resistant Staphylococcus aureus (MRSA) activity of ‘pacific propolis’ and isolated prenylflavanones. Phytother. Res. 2010, 24, 1181–1187. [Google Scholar] [PubMed]
- Cuesta-Rubio, O.; Frontana-Uribe, B.A.; Ramírez-Apan, T.; Cárdenas, J. Polyisoprenylated Benzophenones In Cuban Propolis; Biological Activity Of Nemorosone §. Z. Für Nat. C 2002, 57, 372–378. [Google Scholar] [CrossRef]
- Marcucci, M. Chemical composition, plant origin and biological activity of Brazilian propolis. Curr. Top. Phytochem. 1999, 2, 115–123. [Google Scholar]
- Kumar, H.; Kawai, T.; Akira, S. Toll- like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef]
- Pasare, C.; Medzhitov, R. Toll- like receptors and acquired immunity. Semin. Immunol. 2004, 16, 23–26. [Google Scholar] [CrossRef]
- Bueno-Silva, B.; Rosalen, P.L.; Alencar, S.M.; Mayer, M.P.A. Anti- inflammatory mechanisms of neovestitol from Brazilian red propolis in LPS-activated macrophages. J. Funct. Foods 2017, 36, 440–447. [Google Scholar] [CrossRef]
- Ansorge, S.; Reinhold, D.; Lendeckel, U. Propolis and some of its constituents down-regulate DNA synthesis and inflammatory cytokine production but induce TGF-β1 production of human immune cells. Z. Für Nat. C 2003, 58, 580–589. [Google Scholar] [CrossRef]
- Park, Y.K.; Alencar, S.M.; Aguiar, C.L. Botanical origin and chemical composition of Brazilian propolis. J. Agric. Food Chem. 2002, 50, 2502–2506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, G.; Gurley, E.; Zhou, H. Flavonoid Apigenin Inhibits Lipopolysaccharide- Induced Inflammatory Response through Multiple Mechanisms in Macrophages. PLoS ONE 2014, 9, e107072. [Google Scholar] [CrossRef] [PubMed]
- Soromou, L.W.; Chu, X.; Jiang, L.; Wei, M.; Huo, M.; Chen, N.; Guan, S.; Yang, X.; Chen, C.; Feng, H.; et al. In vitro and in vivo protection provided by pinocembrin against lipopolysaccharide- induced inflammatory responses. Int. Immunopharmacol. 2012, 14, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.N.; Shin, S.A.; Choo, G.S.; Kim, H.J.; Park, Y.S.; Kim, B.S.; Kim, S.K.; Cho, S.D.; Nam, J.S.; Choi, C.S.; et al. Anti-inflammatory effect of quercetin and galangin in LPS-stimulated RAW264.7 macrophages and DNCB-induced atopic dermatitis animal models.(lipopolysaccharides)(Report). Int. J. Mol. Med. 2018, 41, 888–898. [Google Scholar] [PubMed]
- Missima, F.; Pagliarone, A.; Orsatti, C.; Araújo, J., Jr.; Sforcin, J. The Effect of propolis on Th1/Th2 cytokine expression and production by melanoma-bearing mice submitted to stress. Phytother. Res. 2010, 24, 1501–1507. [Google Scholar] [CrossRef]
- Tanaka, M.; Okamoto, Y.; Fukui, T.; Masuzawa, T. Suppression of interleukin 17 production by Brazilian propolis in mice with collagen-induced arthritis. Inflammopharmacology 2012, 20, 19–26. [Google Scholar] [CrossRef]
- Almutairi, S.; Eapen, B.; Chundi, S.M.; Akhalil, A.; Siheri, W.; Clements, C.; Fearnley, J.; Watson, D.G.; Edrada-Ebel, R. New anti-trypanosomal active prenylated compounds from African propolis. Phytochem. Lett. 2014, 10, 35–39. [Google Scholar] [CrossRef]
- Mbawala, A.; Fohouo, F.T.; Roger, D.; Milliere, J. Spectra of antibacterial activity of propolis (Promax-C) samples from two localities of Adamaoua Province (Cameroon). Res. J. Microbiol. 2009, 4, 150–157. [Google Scholar] [CrossRef]
- Njintang, Y.; Tatsadjieu, N.; Ngakou, A.; Danra, D.; Tchuenguem-Fohouo, F. Antiradical activity and polyphenol content of ethanolic extracts of Propolis. Int. J. Biosci. 2012, 2, 56–63. [Google Scholar]
- Kardar, M.N.; Zhang, T.; Coxon, G.D.; Watson, D.G.; Fearnley, J.; Seidel, V. Characterisation of triterpenes and new phenolic lipids in Cameroonian propolis. Phytochemistry 2014, 106, 156–163. [Google Scholar] [CrossRef]
- Ngenge, T.A.; Carol, M.; Emmanuel, T.; Vernyuy, T.P.; Joseph, M.T.; Popova, M.; Bankova, V. Chemical constituents and anti-ulcer activity of propolis from the North-West region of Cameroon. Res. J. Phytochem. 2016, 10, 45–57. [Google Scholar]
- Papachroni, D.; Graikou, K.; Kosalec, I.; Damianakos, H.; Ingram, V.; Chinou, I. Phytochemical analysis and biological evaluation of selected African propolis samples from Cameroon and Congo. Nat. Prod. Commun. 2015, 10, 67–70. [Google Scholar] [CrossRef]
- Rochfort, S. Metabolomics Reviewed: A New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef]
- Krivov, S.V.; Fenton, H.; Goldsmith, P.J.; Prasad, R.K.; Fisher, J.; Paci, E. Optimal reaction coordinate as a biomarker for the dynamics of recovery from kidney transplant. PLoS Comput. Biol. 2014, 10, e1003685. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.; Willing, B.; Lucio, M.; Fekete, A.; Dicksved, J.; Halfvarson, J.; Tysk, C.; Schmitt-Kopplin, P. Metabolomics Reveals Metabolic Biomarkers of Crohn’s Disease. PLoS ONE 2009, 4, e6386. [Google Scholar] [CrossRef]
- Sugimoto, M.; Sakagami, H.; Yokote, Y.; Onuma, H.; Kaneko, M.; Mori, M.; Sakaguchi, Y.; Soga, T.; Tomita, M. Non-targeted metabolite profiling in activated macrophage secretion. Metabolomics 2012, 8, 624–633. [Google Scholar] [CrossRef]
- Li, P.; Yin, Y.-L.; Li, D.; Woo Kim, S.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef]
- Rodríguez-Prados, J.C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragone, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010, 185, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Young, S.P.; Kapoor, S.R.; Viant, M.R.; Byrne, J.J.; Filer, A.; Buckley, C.D.; Kitas, G.D.; Raza, K. The Impact of Inflammation on Metabolomic Profiles in Patients With Arthritis. Arthritis Rheum. 2013, 65, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Bingle, L.; Brown, N.J.; Lewis, C.E. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J. Pathol. J. Pathol. Soc. G. B. Irel. 2002, 196, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-W.; Karin, M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Investig. 2007, 117, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Ngenge, T.A.; Emmanuel, T.; Maurice, T.F.; Joseph, M.T. A New Spinastane-type Triterpenoid from a Cameroonian Propolis sample and Evaluation of Antibacterial and Anti-inflammatory Potential of Extracts. J. Chem. Chem. Sci. 2017, 7, 763–770. [Google Scholar]
- Zingue, S.; Nde, C.B.M.; Michel, T.; Ndinteh, D.T.; Tchatchou, J.; Adamou, M.; Fernandez, X.; Fohouo, F.-N.T.; Clyne, C.; Njamen, D. Ethanol-extracted Cameroonian propolis exerts estrogenic effects and alleviates hot flushes in ovariectomized Wistar rats. BMC Complement. Altern. Med. 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Singh, S.; Burke, T.R.; Grunberger, D.; Aggarwal, B.B. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proc. Natl. Acad. Sci. USA 1996, 93, 9090–9095. [Google Scholar] [CrossRef] [PubMed]
- Franchin, M.; Freires, I.A.; Lazarini, J.G.; Nani, B.D.; Da Cunha, M.G.; Colón, D.F.; de Alencar, S.M.; Rosalen, P.L. The use of Brazilian propolis for discovery and development of novel anti- inflammatory drugs. Eur. J. Med. Chem. 2018, 153, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Wen-Chien, H.; Hsin-Chi, T.; Young-Fa, C.; Tsui-Kang, H.; Cheng-Chun, W.; Jyh-Larng, C.; Feng-Cheng, S.; Yi-Chou, C. The effects of propolis to anti-inflammatory in tumor necrosis factor-o-stimulated human periodontal. Res. J. Biotechnol. 2016, 11, 9. [Google Scholar]
- Liu, K.; Pi, F.; Zhang, H.; Ji, J.; Xia, S.; Cui, F.; Sun, J.; Sun, X. Metabolomics Analysis to Evaluate the Anti- Inflammatory Effects of Polyphenols: Glabridin Reversed Metabolism Change Caused by LPS in RAW 264.7 Cells. J. Agric. Food Chem. 2017, 65, 6070–6079. [Google Scholar] [CrossRef]
- Infantino, V.; Convertini, P.; Di Noia, M.A.; Calvello, R.; Palmieri, F.; Iacobazzi, V.; Cucci, L.; Panaro, M.A. The mitochondrial citrate carrier: A new player in inflammation. Biochem. J. 2011, 438, 433–436. [Google Scholar] [CrossRef]
- Möller, B.; Villiger, P.M. Inhibition of IL-1, IL-6, and TNF-α in immune-mediated inflammatory diseases. Springer Semin. Immunopathol. 2006, 27, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Chan, G.C.-F.; Cheung, K.-W.; Sze, D.M.-Y. The Immunomodulatory and Anticancer Properties of Propolis. Clin. Rev. Allergy Immunol. 2012, 44, 1–12. [Google Scholar] [CrossRef]
- Ewelina, S.; Alicja, Z.K.; Anna, S.-Ł.; Anna, M.; Zenon, P.C.; Wojciech, K. Chemical Composition and Anti- Inflammatory Effect of Ethanolic Extract of Brazilian Green Propolis on Activated J774A. 1 Macrophages. Evid. Based Complement. Altern. Med. 2013, 2013, 976415. [Google Scholar]
- Krawczyk, C.M.; Holowka, T.; Sun, J.; Blagih, J.; Amiel, E.; DeBerardinis, R.J.; Cross, J.R.; Jung, E.; Thompson, C.B.; Jones, R.G. Toll-like receptor induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010, 115, 4742–4749. [Google Scholar] [CrossRef]
- Alqarni, A.; Ferro, V.; Parkinson, J.; Dufton, M.; Watson, D. Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells. Vaccines 2018, 6, 72. [Google Scholar] [CrossRef]
- Kelly, B.; Neill, L.A.J. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef]
- Peña-Altamira, L.E.; Polazzi, E.; Giuliani, P.; Beraudi, A.; Massenzio, F.; Mengoni, I.; Poli, A.; Zuccarini, M.; Ciccarelli, R.; Di Iorio, P. Release of soluble and vesicular purine nucleoside phosphorylase from rat astrocytes and microglia induced by pro-inflammatory stimulation with extracellular ATP via P2X7 receptors. Neurochem. Int. 2018, 115, 37–49. [Google Scholar] [CrossRef]
- Kazmers, I.S.; Mitchell, B.S.; Dadonna, P.E.; Wotring, L.L.; Townsend, L.B.; Kelley, W.N. Inhibition of purine nucleoside phosphorylase by 8-aminoguanosine: Selective toxicity for T lymphoblasts. Science 1981, 214, 1137–1139. [Google Scholar] [CrossRef] [PubMed]
- Markert, M.L. Purine nucleoside phosphorylase deficiency. Immunodefic. Rev. 1991, 3, 45–81. [Google Scholar] [PubMed]
- Savarese, T.M.; Crabtree, G.W.; Parks, R.E., Jr. 5′-methylthioadenosine phosphorylase—I: Substrate activity of 5′-deoxyadenosine with the enzyme from Sarcoma 180 cells. Biochem. Pharmacol. 1981, 30, 189–199. [Google Scholar] [CrossRef]
- Arpaia, E.; Benveniste, P.; Di Cristofano, A.; Gu, Y.; Dalal, I.; Kelly, S.; Hershfield, M.; Pandolfi, P.P.; Roifman, C.M.; Cohen, A. Mitochondrial Basis for Immune Deficiency: Evidence from Purine Nucleoside Phosphorylase–deficient Mice. J. Exp. Med. 2000, 191, 2197–2208. [Google Scholar] [CrossRef]
- Herken, H.; Gurel, A.; Selek, S.; Armutcu, F.; Ozen, M.E.; Bulut, M.; Kap, O.; Yumru, M.; Savas, H.A.; Akyol, O. Adenosine deaminase, nitric oxide, superoxide dismutase, and xanthine oxidase in patients with major depression: Impact of antidepressant treatment. Arch. Med Res. 2007, 38, 247–252. [Google Scholar] [CrossRef]
- Tritsch, G.; Niswander, P. Positive correlation between superoxide release and intracellular adenosine deaminase activity during macrophage membrane perturbation regardless of nature or magnitude of stimulus. Mol. Cell. Biochem. 1982, 49, 49–51. [Google Scholar] [CrossRef]
- Sono, M. The roles of superoxide anion and methylene blue in the reductive activation of indoleamine 2, 3-dioxygenase by ascorbic acid or by xanthine oxidase-hypoxanthine. J. Biol. Chem. 1989, 264, 1616–1622. [Google Scholar]
- Munn, D.H.; Mellor, A.L. Indoleamine 2, 3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013, 34, 137–143. [Google Scholar] [CrossRef]
- Tomek, P.; Palmer, B.D.; Flanagan, J.U.; Sun, C.; Raven, E.L.; Ching, L.-M. Discovery and evaluation of inhibitors to the immunosuppressive enzyme indoleamine 2, 3-dioxygenase 1 (IDO1): Probing the active site-inhibitor interactions. Eur. J. Med. Chem. 2017, 126, 983–996. [Google Scholar] [CrossRef]
- Esser, C.; Rannug, A.; Stockinger, B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009, 30, 447–454. [Google Scholar] [CrossRef]
- Gutierrez-Vazquez, C.; Quintana, F.J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef]
- Fukuda, K. Etiological classification of depression based on the enzymes of tryptophan metabolism. BMC Psychiatry 2014, 14, 372. [Google Scholar] [CrossRef]
- Regan, T.; Gill, A.C.; Clohisey, S.M.; Barnett, M.W.; Pariante, C.M.; Harrison, N.A.; Consortium, M.I.; Hume, D.A.; Bullmore, E.T.; Freeman, T.C. Effects of anti-inflammatory drugs on the expression of tryptophan-metabolism genes by human macrophages. J. Leukoc. Biol. 2018, 103, 681–692. [Google Scholar] [CrossRef]
- Hellsten-Westing, Y.; Norman, B.; Balsom, P.D.; Sjodin, B. Decreased resting levels of adenine nucleotides in human skeletal muscle after high-intensity training. J. Appl. Physiol. 1993, 74, 2523–2528. [Google Scholar] [CrossRef] [PubMed]
- Ipata, P.L.; Pesi, R. Metabolic interaction between purine nucleotide cycle and oxypurine cycle during skeletal muscle contraction of different intensities: A biochemical reappraisal. Metabolomics 2018, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Neill, L.A.J. A critical role for citrate metabolism in LPS signalling. Biochem. J. 2011, 438, e5–e6. [Google Scholar] [CrossRef] [PubMed]
- Puchalska, P.; Huang, X.; Martin, S.E.; Han, X.; Patti, G.J.; Crawford, P.A. Isotope Tracing Untargeted Metabolomics Reveals Macrophage Polarization-State-Specific Metabolic Coordination across Intracellular Compartments. iScience 2018, 9, 298–313. [Google Scholar] [CrossRef]
- Mills, E.L.; O’Neill, L.A. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur. J. Immunol. 2016, 46, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Rattigan, K.M.; Pountain, A.W.; Regnault, C.; Achcar, F.; Vincent, I.M.; Goodyear, C.S.; Barrett, M.P. Metabolomic profiling of macrophages determines the discrete metabolomic signature and metabolomic interactome triggered by polarising immune stimuli. PLoS ONE 2018, 13, e0194126. [Google Scholar] [CrossRef]
- Qualls, J.E.; Subramanian, C.; Rafi, W.; Smith, A.M.; Balouzian, L.; DeFreitas, A.A.; Shirey, K.A.; Reutterer, B.; Kernbauer, E.; Stockinger, S. Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host Microbe 2012, 12, 313–323. [Google Scholar] [CrossRef] [PubMed]
- El Kasmi, K.C.; Qualls, J.E.; Pesce, J.T.; Smith, A.M.; Thompson, R.W.; Henao-Tamayo, M.; Basaraba, R.J.; König, T.; Schleicher, U.; Koo, M.-S. Toll-like receptor–induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 2008, 9, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Barber, K.D.; Barber, D.L.; Shenderov, K.; White, S.D.; Wilson, M.S.; Cheever, A.; Kugler, D.; Hieny, S.; Caspar, P.; Núñez, G. Cutting edge: Caspase-1 independent IL-1β production is critical for host resistance to Mycobacterium tuberculosis and does not require TLR signaling in vivo. J. Immunol. 2010, 184, 3326–3330. [Google Scholar] [CrossRef]
- Mishra, B.B.; Rathinam, V.A.; Martens, G.W.; Martinot, A.J.; Kornfeld, H.; Fitzgerald, K.A.; Sassetti, C.M. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat. Immunol. 2013, 14, 52–60. [Google Scholar] [CrossRef]
- El Kasmi, K.C.; Stenmark, K.R. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin. Immunol. 2015, 27, 267–275. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-Mcdermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Jha, A.K.; Huang, S.C.-C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef]
- Dröse, S. Differential effects of complex II on mitochondrial ROS production and their relation to cardioprotective pre- and postconditioning. Biochim. Biophys. Acta Bioenergy 2012, 1827, 578–587. [Google Scholar] [CrossRef]
- Blagih, J.; Jones, R.G. Polarizing Macrophages through Reprogramming of Glucose Metabolism. Cell Metab. 2012, 15, 793–795. [Google Scholar] [CrossRef]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The Sedoheptulose Kinase CARKL Directs Macrophage Polarization through Control of Glucose Metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef]
- Zhang, R.; Watson, D.G.; Wang, L.; Westrop, G.D.; Coombs, G.H.; Zhang, T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography- high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A 2014, 1362, 168–179. [Google Scholar] [CrossRef]
- Stoop, J.; Zegers, B.; Hendrickx, G.; Siegenbeek van Heukelom, L.; Staal, G.; De Bree, P.; Wadman, S.; Ballieux, R. Purine nucleoside phosphorylase deficiency associated with selective cellular immunodeficiency. N. Engl. J. Med. 1977, 296, 651–655. [Google Scholar] [CrossRef]
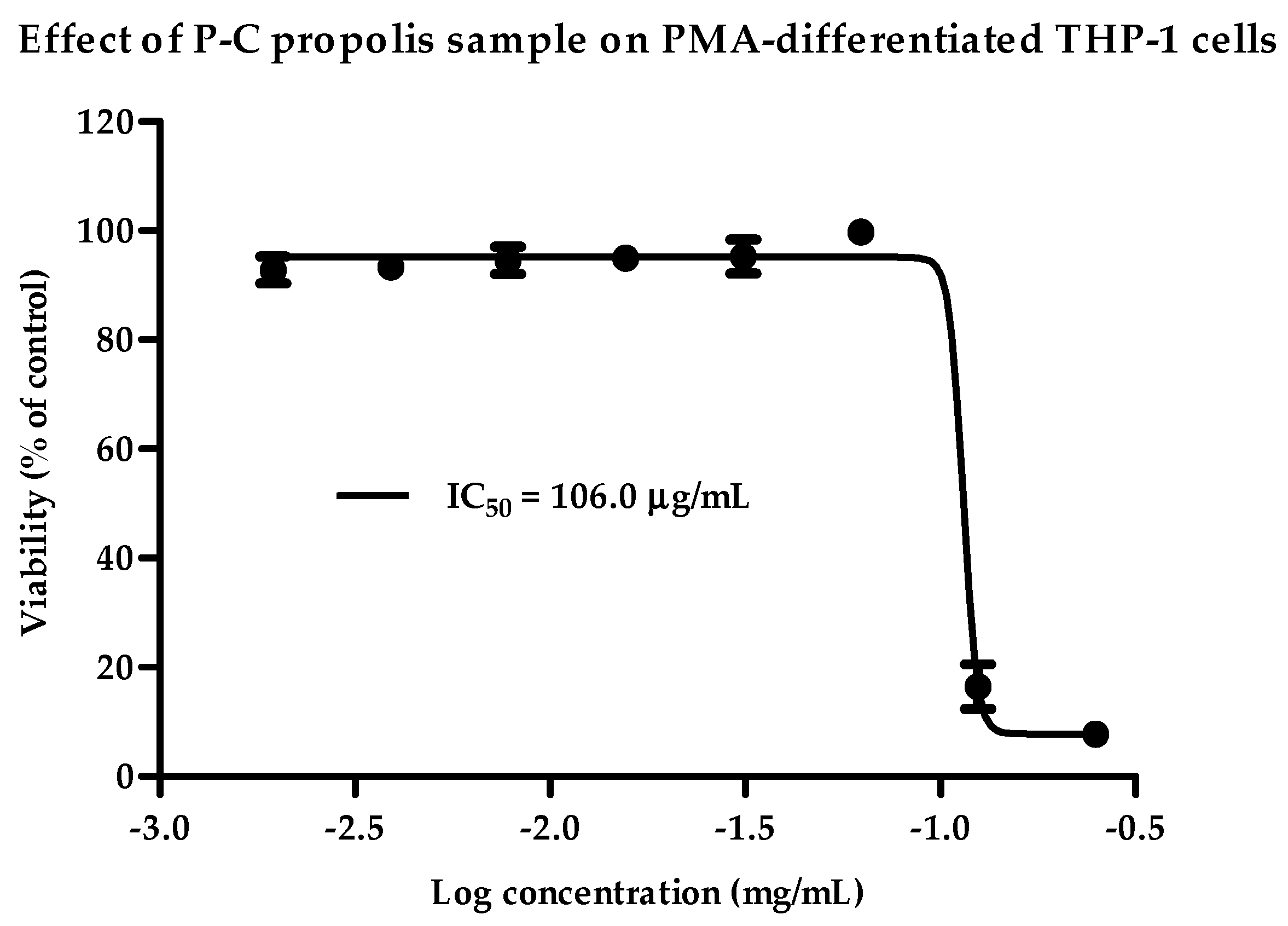

| Propolis Samples | IC50 (µg/mL) | Selected Final Concentration (µg/mL) | |
|---|---|---|---|
| Region | Extract Code | ||
| United Kingdom | P-UK1 | 57.95 | 30 |
| P-UK2 | 48.99 | 15 | |
| P-UK3 | 53.55 | 20 | |
| P-UK4 | 48.08 | 15 | |
| P-UK5 | 46.28 | 15 | |
| Ghana | P-G | 86.95 | 15 |
| Cameroon | P-C | 106 | 70 |
| Indonesia | P-Ind1 | 25.73 | 15 |
| P-Ind2 | >250 | 250 | |
| Mass | Rt | Putative Metabolite | LPS/C | P-C/C | (P-C+LPS)/LPS | |||
|---|---|---|---|---|---|---|---|---|
| Ratio | p-Value | Ratio | p-Value | Ratio | p-Value | |||
| Arginine and Proline Metabolism | - | - | - | - | - | - | ||
| 145.08 | 15.39 | 4-Guanidinobutanoate | 1.991 | <0.001 | 1.549 | 0.002 | 1.419 | 0.002 |
| 115.06 | 13.01 | l-Proline * | 1.137 | 0.046 | 1.467 | <0.001 | 1.303 | 0.001 |
| 113.06 | 11.14 | Creatinine | 0.884 | ns | 1.112 | ns | 1.454 | 0.030 |
| 175.10 | 16.04 | l-Citrulline * | 1.790 | <0.001 | 2.148 | <0.001 | 1.090 | ns |
| 129.09 | 16.02 | 4-Guanidinobutanal | 1.494 | 0.032 | 1.723 | <0.001 | 1.062 | ns |
| 290.12 | 16.78 | N-(l-Arginino)succinate | 0.556 | 0.001 | 0.919 | ns | 1.278 | 0.020 |
| 189.06 | 13.95 | N-Acetyl-l-glutamate * | 0.662 | <0.001 | 3.983 | <0.001 | 4.955 | <0.001 |
| 145.16 | 26.37 | Spermidine * | 1.198 | ns | 0.169 | <0.001 | 0.078 | <0.001 |
| Glycolysis/TCA cycle | ||||||||
| 260.03 | 16.83 | d-Glucose 1-phosphate * | 0.996 | ns | 0.531 | <0.001 | 0.476 | <0.001 |
| 260.03 | 15.95 | d-Fructose 6-phosphate * | 1.424 | 0.001 | 1.486 | 0.001 | 1.229 | 0.046 |
| 340.00 | 18.05 | d-Fructose 1,6-bisphosphate * | 2.242 | <0.001 | 1.163 | 0.006 | 0.754 | 0.003 |
| 170.00 | 16.00 | d-Glyceraldehyde 3-phosphate * | 0.717 | 0.001 | 1.899 | <0.001 | 1.622 | <0.001 |
| 260.02 | 17.72 | d-Glucose 6-sulfate | 1.339 | 0.004 | 1.461 | <0.001 | 1.118 | ns |
| 167.98 | 17.45 | Phosphoenolpyruvate | 1.254 | ns | 2.503 | 0.001 | 2.062 | 0.004 |
| 88.02 | 7.67 | Pyruvate * | 1.609 | 0.007 | 0.916 | ns | 0.483 | <0.001 |
| 809.13 | 12.28 | Acetyl-CoA | 0.681 | 0.002 | 1.678 | <0.001 | 1.912 | 0.001 |
| 190.01 | 15.80 | Oxalosuccinate | 0.588 | 0.044 | 5.385 | ns | 1.691 | ns |
| 132.01 | 15.74 | Oxaloacetate * | 0.700 | 0.041 | 1.064 | ns | 1.566 | 0.016 |
| 192.03 | 18.08 | Citrate * | 1.453 | 0.002 | 1.636 | <0.001 | 1.214 | 0.040 |
| 118.03 | 14.94 | Succinate * | 1.323 | 0.005 | 1.534 | <0.001 | 1.182 | ns |
| 116.01 | 14.92 | Fumarate | 0.894 | ns | 0.752 | 0.001 | 0.842 | ns |
| 131.07 | 14.89 | Creatine * | 0.647 | <0.001 | 1.179 | 0.011 | 1.544 | <0.001 |
| 427.03 | 15.19 | ADP * | 0.814 | 0.041 | 3.140 | <0.001 | 2.425 | <0.001 |
| 443.02 | 18.08 | GDP * | 0.727 | 0.003 | 1.917 | <0.001 | 2.158 | <0.001 |
| 507.00 | 16.55 | ATP * | 0.776 | 0.019 | 1.341 | 0.022 | 1.303 | 0.004 |
| 522.99 | 19.50 | GTP * | 0.923 | ns | 1.309 | 0.002 | 1.237 | 0.016 |
| 665.12 | 13.29 | NADH * | 0.757 | 0.004 | 1.623 | <0.001 | 1.620 | 0.001 |
| 663.11 | 14.24 | NAD+ * | 0.363 | <0.001 | 0.877 | ns | 1.893 | <0.001 |
| Oxidative Stress/ Pentose Phosphate Pathway | ||||||||
| 276.02 | 17.61 | 6-Phospho-d-gluconate * | 1.355 | 0.001 | 0.521 | <0.001 | 0.273 | <0.001 |
| 196.06 | 13.15 | d-Gluconic acid * | 0.889 | 0.006 | 1.539 | <0.001 | 1.310 | <0.001 |
| 177.94 | 15.81 | Pyrophosphate | 1.271 | 0.009 | 1.406 | <0.001 | 1.189 | 0.024 |
| 290.04 | 16.09 | d-Sedoheptulose 7-phosphate | 1.367 | <0.001 | 1.970 | <0.001 | 1.570 | <0.001 |
| 370.01 | 18.21 | d-Sedoheptulose 1,7-bisphosphate | 1.900 | <0.001 | 1.757 | <0.001 | 0.893 | ns |
| 210.07 | 14.03 | Sedoheptulose | 1.741 | 0.002 | 1.202 | ns | 0.931 | ns |
| 230.02 | 15.68 | d-Ribose 5-phosphate * | 1.572 | 0.036 | 1.018 | ns | 0.736 | ns |
| 307.08 | 14.22 | Glutathione | 0.579 | <0.001 | 1.508 | <0.001 | 1.856 | <0.001 |
| 612.15 | 17.27 | Glutathione disulphide * | 0.838 | 0.017 | 3.157 | <0.001 | 3.791 | <0.001 |
| 745.09 | 16.87 | NADPH | 0.494 | <0.001 | 00.00 | <0.001 | 00.00 | <0.001 |
| 743.08 | 16.68 | NADP+ * | 0.896 | ns | 2.267 | <0.001 | 2.063 | <0.001 |
| Purine Metabolism | ||||||||
| 347.06 | 15.61 | dGMP | 0.461 | <0.001 | 1.575 | 0.001 | 1.937 | <0.001 |
| 267.10 | 9.35 | Adenosine * | 1.276 | ns | 3.435 | <0.001 | 2.445 | <0.001 |
| 283.09 | 12.80 | Guanosine * | 1.677 | ns | 4.070 | <0.001 | 3.202 | 0.004 |
| 268.08 | 11.06 | Inosine * | 2.967 | <0.001 | 10.511 | <0.001 | 13.160 | <0.001 |
| 284.08 | 10.51 | Xanthosine | 2.064 | ns | 2.378 | 0.005 | 1.002 | ns |
| 251.10 | 7.93 | Deoxyadenosine | 2.912 | <0.001 | 0.399 | <0.001 | 0.292 | <0.001 |
| 152.04 | 11.30 | Xanthine * | 1.766 | 0.002 | 1.772 | <0.001 | 1.013 | ns |
| 168.03 | 12.34 | Urate * | 1.752 | 0.008 | 1.897 | 0.000 | 1.135 | ns |
| 136.04 | 10.41 | Hypoxanthine * | 14.695 | <0.001 | 2.277 | 0.009 | 0.714 | 0.013 |
| 398.14 | 16.78 | S-Adenosyl-L-methionine * | 1.237 | ns | 2.578 | <0.001 | 1.695 | <0.001 |
| 297.09 | 16.78 | 5’-Methylthioadenosine * | 0.700 | 0.001 | 2.727 | <0.001 | 1.570 | <0.001 |
| 348.05 | 15.39 | IMP * | 2.815 | <0.001 | 3.341 | <0.001 | 1.168 | ns |
| 347.06 | 13.75 | AMP * | 0.558 | <0.001 | 2.164 | <0.001 | 2.080 | <0.001 |
| Pyrimidine Metabolism/Glycan Chain Formation | ||||||||
| 125.06 | 10.94 | 5-Methylcytosine | 1.486 | 0.004 | 2.124 | <0.001 | 1.396 | 0.003 |
| 482.98 | 18.68 | CTP * | 0.603 | <0.001 | 0.894 | ns | 1.403 | <0.001 |
| 244.07 | 12.14 | Pseudouridine | 1.199 | 0.038 | 1.914 | <0.001 | 1.668 | <0.001 |
| 323.05 | 15.42 | CMP * | 0.742 | 0.002 | 1.915 | <0.001 | 1.760 | <0.001 |
| 111.04 | 11.13 | Cytosine* | 0.250 | <0.001 | 0.359 | 0.003 | 1.142 | ns |
| 114.04 | 14.89 | 5,6-Dihydrouracil | 0.631 | <0.001 | 1.150 | ns | 1.604 | <0.001 |
| 259.05 | 11.93 | Glucosamine 1-phosphate | 1.202 | ns | 1.711 | 0.001 | 1.360 | 0.020 |
| 324.04 | 15.06 | UMP * | 0.630 | <0.001 | 2.223 | <0.001 | 1.878 | <0.001 |
| 404.00 | 16.48 | UDP * | 0.433 | <0.001 | 2.482 | <0.001 | 2.412 | <0.001 |
| 483.97 | 17.79 | UTP * | 0.591 | 0.000 | 0.669 | 0.002 | 0.954 | ns |
| 566.05 | 16.04 | UDP-glucose * | 0.503 | <0.001 | 0.955 | ns | 1.711 | <0.001 |
| 536.04 | 16.07 | UDP-d-xylose | 0.666 | 0.002 | 2.042 | <0.001 | 2.321 | <0.001 |
| 580.03 | 18.83 | UDP-glucuronate | 0.583 | <0.001 | 1.426 | 0.001 | 1.706 | <0.001 |
| 607.08 | 14.91 | UDP-N-acetyl-d-glucosamine * | 0.579 | <0.001 | 0.984 | ns | 1.539 | 0.001 |
| Tryptophan Metabolism | ||||||||
| 220.09 | 9.74 | 5-Hydroxy-l-tryptophan isomer | 8.133 | <0.001 | 1.285 | 0.014 | 0.228 | <0.001 |
| 191.06 | 10.29 | 5-Hydroxyindoleacetate * | 1.527 | <0.001 | 0.869 | 0.035 | 0.503 | <0.001 |
| 219.05 | 10.31 | 5-Hydroxyindolepyruvate | 1.515 | <0.001 | 0.872 | 0.015 | 0.523 | <0.001 |
| 117.06 | 11.07 | Indole * | 1.723 | 0.011 | 1.565 | 0.002 | 0.987 | ns |
| 204.09 | 11.85 | l-Tryptophan * | 1.182 | ns | 1.849 | <0.001 | 1.531 | 0.013 |
| 208.09 | 11.09 | l-Kynurenine * | 1.835 | 0.001 | 1.591 | 0.001 | 0.810 | ns |
| Miscellaneous | ||||||||
| 131.06 | 14.61 | N-Acetyl-beta-alanine | 1.306 | <0.001 | 1.521 | <0.001 | 1.263 | 0.011 |
| 210.04 | 17.19 | d-Glucarate | 0.611 | 0.005 | 1.385 | 0.020 | 2.367 | <0.001 |
| 161.11 | 13.50 | l-Carnitine * | 0.899 | ns | 1.267 | <0.001 | 1.504 | 0.002 |
| 149.05 | 11.72 | l-Methionine * | 1.380 | 0.011 | 1.527 | <0.001 | 1.230 | 0.037 |
| 175.05 | 14.17 | N-Acetyl-l-aspartate * | 0.625 | <0.001 | 1.117 | ns | 1.721 | <0.001 |
| 203.12 | 11.26 | O-Acetylcarnitine * | 0.945 | ns | 3.291 | <0.001 | 3.114 | <0.001 |
| 226.11 | 15.85 | Carnosine * | 1.166 | ns | 1.558 | <0.001 | 1.279 | 0.025 |
| 119.06 | 14.52 | l-Threonine | 1.432 | <0.001 | 1.558 | <0.001 | 1.140 | ns |
| 105.04 | 15.85 | l-Serine * | 1.432 | 0.008 | 1.625 | <0.001 | 1.194 | ns |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alqarni, A.M.; Niwasabutra, K.; Sahlan, M.; Fearnley, H.; Fearnley, J.; Ferro, V.A.; Watson, D.G. Propolis Exerts an Anti-Inflammatory Effect on PMA-Differentiated THP-1 Cells via Inhibition of Purine Nucleoside Phosphorylase. Metabolites 2019, 9, 75. https://doi.org/10.3390/metabo9040075
Alqarni AM, Niwasabutra K, Sahlan M, Fearnley H, Fearnley J, Ferro VA, Watson DG. Propolis Exerts an Anti-Inflammatory Effect on PMA-Differentiated THP-1 Cells via Inhibition of Purine Nucleoside Phosphorylase. Metabolites. 2019; 9(4):75. https://doi.org/10.3390/metabo9040075
Chicago/Turabian StyleAlqarni, Abdulmalik M., Kanidta Niwasabutra, Muhamad Sahlan, Hugo Fearnley, James Fearnley, Valerie A. Ferro, and David G. Watson. 2019. "Propolis Exerts an Anti-Inflammatory Effect on PMA-Differentiated THP-1 Cells via Inhibition of Purine Nucleoside Phosphorylase" Metabolites 9, no. 4: 75. https://doi.org/10.3390/metabo9040075
APA StyleAlqarni, A. M., Niwasabutra, K., Sahlan, M., Fearnley, H., Fearnley, J., Ferro, V. A., & Watson, D. G. (2019). Propolis Exerts an Anti-Inflammatory Effect on PMA-Differentiated THP-1 Cells via Inhibition of Purine Nucleoside Phosphorylase. Metabolites, 9(4), 75. https://doi.org/10.3390/metabo9040075






