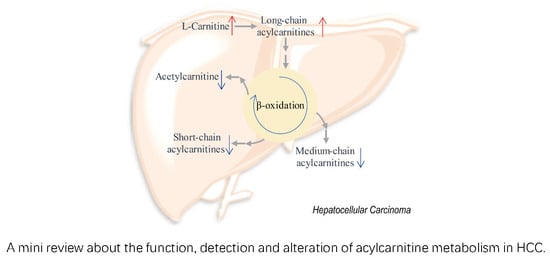
Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma
Abstract
1. Introduction
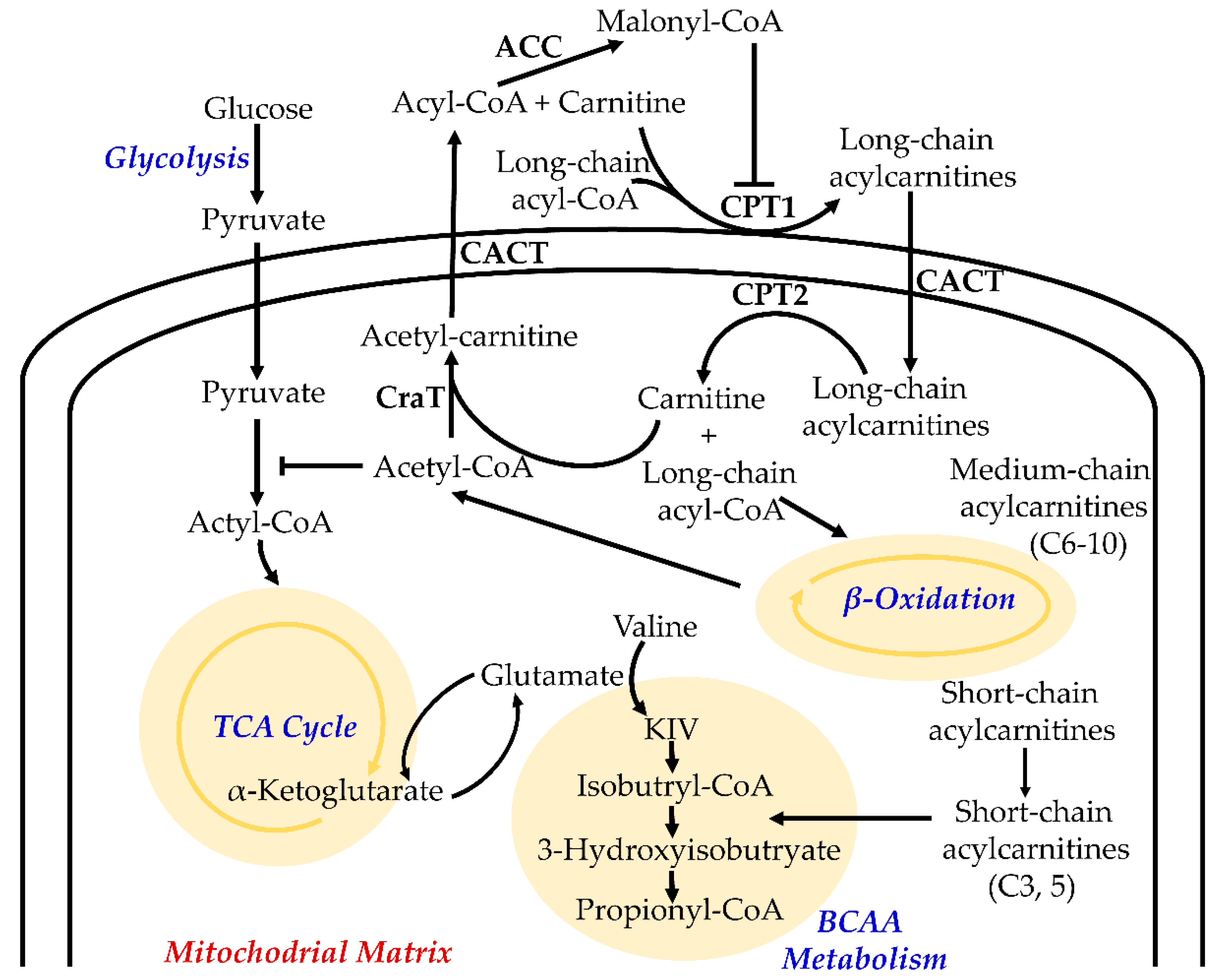
2. Function of Acylcarnitines in Cellular Metabolism
3. Detection of Acylcarnitines in Biological Samples
4. Alteration of Acylcarnitine Metabolism in HCC
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gingold, J.A.; Zhu, D.; Lee, D.F.; Kaseb, A.; Chen, J. Genomic profiling and metabolic homeostasis in primary liver cancers. Trends Mol. Med. 2018, 24, 395–411. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Janjua, N.Z.; Chong, M.; Grebely, J.; Aspinall, E.J.; Innes, H.; Valerio, H.; Hajarizadeh, B.; Hayes, P.C.; Krajden, M.; et al. Trends in hepatocellular carcinoma incidence and survival among people with hepatitis C: An international study. J. Viral Hepat. 2018, 25, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human fatty liver disease: Old questions and new insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Ling, Y.; Wang, H.Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis. Oncol. 2018, 2, 6. [Google Scholar] [CrossRef]
- Charrez, B.; Qiao, L.; Hebbard, L. Hepatocellular carcinoma and non-alcoholic steatohepatitis: The state of play. World J. Gastroenterol. 2016, 22, 2494–2502. [Google Scholar] [CrossRef]
- Agosti, P.; Sabba, C.; Mazzocca, A. Emerging metabolic risk factors in hepatocellular carcinoma and their influence on the liver microenvironment. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 607–617. [Google Scholar] [CrossRef]
- Starley, B.Q.; Calcagno, C.J.; Harrison, S.A. Non-alcoholic fatty liver disease and hepatocellular carcinoma: A weighty connection. Hepatology 2010, 51, 1820–1832. [Google Scholar] [CrossRef]
- Khan, F.Z.; Perumpail, R.B.; Wong, R.J.; Ahmed, A. Advances in hepatocellular carcinoma: Non-alcoholic steatohepatitis-related hepatocellular carcinoma. World J. Hepatol. 2015, 7, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M.; Matsuda, A.; Diederichs, S.; Patel, T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017, 67, 603–618. [Google Scholar] [CrossRef] [PubMed]
- Umeda, S.; Kanda, M.; Kodera, Y. Emerging evidence of molecular biomarkers in hepatocellular carcinoma. Histol. Histopathol. 2018, 33, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Kimhofer, T.; Fye, H.; Taylor-Robinson, S.; Thursz, M.; Holmes, E. Proteomic and metabonomic biomarkers for hepatocellular carcinoma: A comprehensive review. Br. J. Cancer 2015, 112, 1141–1156. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Solis, K.; McAlister, V. Wading through the noise of “multi-omics” to identify prognostic biomarkers in hepatocellular carcinoma. Hepatobil. Surg. Nutr. 2015, 4, 293–294. [Google Scholar] [CrossRef]
- De Stefano, F.; Chacon, E.; Turcios, L.; Marti, F.; Gedaly, R. Novel biomarkers in hepatocellular carcinoma. Dig. Liver Dis. 2018, 50, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tan, H.Y.; Wang, N.; Wang, X.; Feng, Y. Deciphering hepatocellular carcinoma through metabolomics: from biomarker discovery to therapy evaluation. Cancer Manag. Res. 2018, 10, 715–734. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, A.; Sun, H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology 2013, 57, 2072–2077. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Baniasadi, H.; Gowda, G.A.; Gu, H.; Zeng, A.; Zhuang, S.; Skill, N.; Maluccio, M.; Raftery, D. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis 2013, 34, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhu, B.; Zheng, R.; Zhao, X.; Yin, P.; Lu, X.; Jiao, B.; Xu, G.; Yao, Z. Development of urinary pseudotargeted LC-MS-based metabolomics method and its application in hepatocellular carcinoma biomarker discovery. J. Proteome Res. 2015, 14, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Tan, Y.; Yin, P.; Ye, G.; Gao, P.; Lu, X.; Wang, H.; Xu, G. Metabolic characterization of hepatocellular carcinoma using non-targeted tissue metabolomics. Cancer Res. 2013, 73, 4992–5002. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Batchuluun, B.; Al Rijjal, D.; Prentice, K.J.; Eversley, J.A.; Burdett, E.; Mohan, H.; Bhattacharjee, A.; Gunderson, E.P.; Liu, Y.; Wheeler, M.B. Elevated medium-chain acylcarnitines are associated with gestational diabetes mellitus and early progression to type 2 diabetes and induce pancreatic beta-cell dysfunction. Diabetes 2018, 67, 885–897. [Google Scholar] [CrossRef]
- Tarasenko, T.N.; Cusmano-Ozog, K.; McGuire, P.J. Tissue acylcarnitine status in a mouse model of mitochondrial beta-oxidation deficiency during metabolic decompensation due to influenza virus infection. Mol. Genet. Metab. 2018, 125, 144–152. [Google Scholar] [CrossRef]
- McCoin, C.S.; Knotts, T.A.; Adams, S.H. Acylcarnitines—old actors auditioning for new roles in metabolic physiology. Nat. Rev. Endocrinol. 2015, 11, 617–625. [Google Scholar] [CrossRef]
- Hinder, L.M.; Figueroa-Romero, C.; Pacut, C.; Hong, Y.; Vivekanandan-Giri, A.; Pennathur, S.; Feldman, E.L. Long-chain acyl coenzyme A synthetase 1 overexpression in primary cultured Schwann cells prevents long chain fatty acid-induced oxidative stress and mitochondrial dysfunction. Antioxid. Redox Signal. 2014, 21, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Murthy, M.S.R.; Pande, S.V. Mechanism of carnitine acylcarnitine translocase-catalyzed import of acylcarnitines into mitochondria. J. Biol. Chem. 1984, 259, 9082–9089. [Google Scholar] [PubMed]
- Pande, S.V.; Murthy, M.S. Carnitine-acylcarnitine translocase deficiency: Implications in human pathology. Biochim. Biophys. Acta Mol. Basis Dis. 1994, 1226, 269–276. [Google Scholar] [CrossRef]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zheng, X.; Yang, M.; Zhao, A.; Li, M.; Chen, T.; Panee, J.; Jia, W.; Ji, G. Serum lipid alterations identified in chronic hepatitis B, hepatitis B virus-associated cirrhosis and carcinoma patients. Sci. Rep. 2017, 7, 42710. [Google Scholar] [CrossRef]
- Qu, Q.; Zeng, F.; Liu, X.; Wang, Q.J.; Deng, F. Fatty acid oxidation and carnitine palmitoyltransferase I: Emerging therapeutic targets in cancer. Cell Death Dis. 2016, 7, e2226. [Google Scholar] [CrossRef]
- Xiang, L.; Wei, J.; Tian, X.Y.; Wang, B.; Chan, W.; Li, S.; Tang, Z.; Zhang, H.; Cheang, W.S.; Zhao, Q.; et al. Comprehensive analysis of acylcarnitine species in db/db mouse using a novel method of high-resolution parallel reaction monitoring reveals widespread metabolic dysfunction induced by diabetes. Anal. Chem. 2017, 89, 10368–10375. [Google Scholar] [CrossRef]
- Seiler, S.E.; Koves, T.R.; Gooding, J.R.; Wong, K.E.; Stevens, R.D.; Ilkayeva, O.R.; Wittmann, A.H.; DeBalsi, K.L.; Davies, M.N.; Lindeboom, L.; et al. Carnitine acetyltransferase mitigates metabolic inertia and muscle fatigue during exercise. Cell Metab. 2015, 22, 65–76. [Google Scholar] [CrossRef]
- Casals, N.; Zammit, V.; Herrero, L.; Fado, R.; Rodriguez-Rodriguez, R.; Serra, D. Carnitine palmitoyltransferase 1C: From cognition to cancer. Prog. Lipid Res. 2016, 61, 134–148. [Google Scholar] [CrossRef]
- Kim, H.I.; Raffler, J.; Lu, W.; Lee, J.J.; Abbey, D.; Saleheen, D.; Rabinowitz, J.D.; Bennett, M.J.; Hand, N.J.; Brown, C.; et al. Fine mapping and functional analysis reveal a role of SLC22A1 in acylcarnitine transport. Am. J. Hum. Genet. 2017, 101, 489–502. [Google Scholar] [CrossRef]
- Hagenbuchner, J.; Scholl-Buergi, S.; Karall, D.; Ausserlechner, M.J. Very long-/ and long Chain-3-Hydroxy Acyl CoA Dehydrogenase Deficiency correlates with deregulation of the mitochondrial fusion/fission machinery. Sci. Rep. 2018, 8, 3254. [Google Scholar] [CrossRef] [PubMed]
- Vargas, C.R.; Ribas, G.S.; da Silva, J.M.; Sitta, A.; Deon, M.; de Moura Coelho, D.; Wajner, M. Selective Screening of fatty acids oxidation defects and organic acidemias by liquid chromatography/tandem mass spectrometry acylcarnitine analysis in Brazilian patients. Arch. Med. Res. 2018, 49, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Diekman, E.F.; Visser, G.; Schmitz, J.P.J.; Nievelstein, R.A.J.; de Sain-van der Velden, M.; Wardrop, M.; Van der Pol, W.L.; Houten, S.M.; van Riel, N.A.W.; Takken, T.; et al. Altered energetics of exercise explain risk of rhabdomyolysis in very long-chain acyl-coa dehydrogenase deficiency. PLoS ONE 2016, 11, e0147818. [Google Scholar] [CrossRef] [PubMed]
- Makrecka-Kuka, M.; Sevostjanovs, E.; Vilks, K.; Volska, K.; Antone, U.; Kuka, J.; Makarova, E.; Pugovics, O.; Dambrova, M.; Liepinsh, E. Plasma acylcarnitine concentrations reflect the acylcarnitine profile in cardiac tissues. Sci. Rep. 2017, 7, 17528. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Boenzi, S.; Inglese, R.; la Marca, G.; Muraca, M.; Martinez, T.B.; Johnson, D.W.; Zelli, E.; Dionisi-Vici, C. Measurement of succinyl-carnitine and methylmalonyl-carnitine on dried blood spot by liquid chromatography-tandem mass spectrometry. Clin. Chim. Acta 2014, 429, 30–33. [Google Scholar] [CrossRef] [PubMed]
- DiBattista, A.; McIntosh, N.; Lamoureux, M.; Al-Dirbashi, O.Y.; Chakraborty, P.; Britz-McKibbin, P. Temporal signal pattern recognition in mass spectrometry: a method for rapid identification and accurate quantification of biomarkers for inborn errors of metabolism with quality assurance. Anal. Chem. 2017, 89, 8112–8121. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liang, L.; Gao, X.; Zhang, H.; Yao, P.; Hu, Y.; Ma, Y.; Wang, F.; Jin, Q.; Li, H.; et al. Early prediction of developing type 2 diabetes by plasma acylcarnitines: A population-based study. Diabetes Care 2016, 39, 1563–1570. [Google Scholar] [CrossRef]
- Burkhardt, R.; Kirsten, H.; Beutner, F.; Holdt, L.M.; Gross, A.; Teren, A.; Tonjes, A.; Becker, S.; Krohn, K.; Kovacs, P.; et al. Integration of genome-wide snp data and gene-expression profiles reveals six novel loci and regulatory mechanisms for amino acids and acylcarnitines in whole blood. PLoS Genet 2015, 11, e1005510. [Google Scholar] [CrossRef]
- Guasch-Ferre, M.; Zheng, Y.; Ruiz-Canela, M.; Hruby, A.; Martinez-Gonzalez, M.A.; Clish, C.B.; Corella, D.; Estruch, R.; Ros, E.; Fito, M.; et al. Plasma acylcarnitines and risk of cardiovascular disease: Effect of Mediterranean diet interventions. Am. J. Clin. Nutr. 2016, 103, 1408–1416. [Google Scholar] [CrossRef]
- Ahmad, T.; Kelly, J.P.; McGarrah, R.W.; Hellkamp, A.S.; Fiuzat, M.; Testani, J.M.; Wang, T.S.; Verma, A.; Samsky, M.D.; Donahue, M.P.; et al. Prognostic implications of long-chain acylcarnitines in heart failure and reversibility with mechanical circulatory support. J. Am. Coll. Cardiol. 2016, 67, 291–299. [Google Scholar] [CrossRef]
- Virmani, A.; Pinto, L.; Bauermann, O.; Zerelli, S.; Diedenhofen, A.; Binienda, Z.K.; Ali, S.F.; van der Leij, F.R. The carnitine palmitoyl transferase (CPT) system and possible relevance for neuropsychiatric and neurological conditions. Mol. Neurobiol. 2015, 52, 826–836. [Google Scholar] [CrossRef]
- Nakagawa, H.; Hayata, Y.; Kawamura, S.; Yamada, T.; Fujiwara, N.; Koike, K. Lipid metabolic reprogramming in hepatocellular carcinoma. Cancers (Basel) 2018, 10, 447. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Chen, Y.X.; Guan, L.H.; Zhang, H.Z.; Huang, Y.Y.; Johnson, C.H.; Wu, Z.M.; Gonzalez, F.J.; Yu, A.M.; Huang, P.; et al. Carnitine palmitoyltransferase 1C regulates cancer cell senescence through mitochondria- associated metabolic reprograming. Cell Death Differ. 2018, 25, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Nakagawa, H.; Enooku, K.; Kudo, Y.; Hayata, Y.; Nakatsuka, T.; Tanaka, Y.; Tateishi, R.; Hikiba, Y.; Misumi, K.; et al. CPT2 downregulation adapts HCC to lipid-rich environment and promotes carcinogenesis via acylcarnitine accumulation in obesity. Gut 2018, 67, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Wettersten, H.I.; Hakimi, A.A.; Morin, D.; Bianchi, C.; Johnstone, M.E.; Donohoe, D.R.; Trott, J.F.; Aboud, O.A.; Stirdivant, S.; Neri, B.; et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015, 75, 2541–2552. [Google Scholar] [CrossRef] [PubMed]
- Valentino, A.; Calarco, A.; Di Salle, A.; Finicelli, M.; Crispi, S.; Calogero, R.A.; Riccardo, F.; Sciarra, A.; Gentilucci, A.; Galderisi, U.; et al. Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: Molecular basis and pathophysiological consequences. Oncogene 2017, 36, 6030–6040. [Google Scholar] [CrossRef] [PubMed]
- Ganti, S.; Taylor, S.L.; Kim, K.; Hoppel, C.L.; Guo, L.; Yang, J.; Evans, C.; Weiss, R.H. Urinary acylcarnitines are altered in human kidney cancer. Int. J. Cancer 2012, 130, 2791–2800. [Google Scholar] [CrossRef]
- Chughtai, K.; Jiang, L.; Greenwood, T.R.; Glunde, K.; Heeren, R.M. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumour models. J. Lipid Res. 2013, 54, 333–344. [Google Scholar] [CrossRef]
- Yu, D.; Zhou, L.; Xuan, Q.; Wang, L.; Zhao, X.; Lu, X.; Xu, G. Strategy for comprehensive identification of acylcarnitines based on liquid chromatography-high-resolution mass spectrometry. Anal. Chem. 2018, 90, 5712–5718. [Google Scholar] [CrossRef]
- Mansour, F.R.; Wei, W.; Danielson, N.D. Separation of carnitine and acylcarnitines in biological samples: A review. Biomed. Chromatogr. 2013, 27, 1339–1353. [Google Scholar] [CrossRef]
- Horvath, T.D.; Stratton, S.L.; Bogusiewicz, A.; Owen, S.N.; Mock, D.M.; Moran, J.H. Quantitative measurement of urinary excretion of 3-hydroxyisovaleryl carnitine by LC-MS/MS as an indicator of biotin status in humans. Anal. Chem. 2010, 82, 9543–9548. [Google Scholar] [CrossRef] [PubMed]
- Heinig, K.; Henion, J. Determination of carnitine and acylcarnitines in biological samples by capillary electrophoresis-mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 1999, 735, 171–188. [Google Scholar] [CrossRef]
- Vernez, L.; Wenk, M.; Krahenbuhl, S. Determination of carnitine and acylcarnitines in plasma by high-performance liquid chromatography/electrospray ionization ion trap tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Pasquali, M. Acidified acetonitrile and methanol extractions for quantitative analysis of acylcarnitines in plasma by stable isotope dilution tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 827, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Pormsila, W.; Morand, R.; Krahenbuhl, S.; Hauser, P.C. Capillary electrophoresis with contactless conductivity detection for the determination of carnitine and acylcarnitines in clinical samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011, 879, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.G.; Struys, E.A.; Bootsma, A.; tenBrink, H.J.; Dorland, L.; deAlmeida, I.T.; Duran, M.; Jakobs, C. Quantitative analysis of plasma acylcarnitines using gas chromatography chemical ionization mass fragmentography. J. Lipid Res. 1997, 38, 173–182. [Google Scholar] [PubMed]
- Sun, D.; Cree, M.G.; Zhang, X.J.; Boersheim, E.; Wolfe, R.R. Measurement of stable isotopic enrichment and concentration of long-chain fatty acyl-carnitines in tissue by HPLC-MS. J. Lipid Res. 2006, 47, 431–439. [Google Scholar] [CrossRef]
- Schoonen, J.W.; van Duinen, V.; Oedit, A.; Vulto, P.; Hankemeier, T.; Lindenburg, P.W. Continuous-flow microelectroextraction for enrichment of low abundant compounds. Anal. Chem. 2014, 86, 8048–8056. [Google Scholar] [CrossRef]
- Qiu, C.L.; Raynie, D.E. The use of extraction technologies in food safety studies. LC GC Eur. 2017, 30, 662–669. [Google Scholar]
- Isaguirre, A.C.; Olsina, R.A.; Martinez, L.D.; Lapierre, A.V.; Cerutti, S. Development of solid phase extraction strategies to minimize the effect of human urine matrix effect on the response of carnitine by UPLC-MS/MS. Microchem. J. 2016, 129, 362–367. [Google Scholar] [CrossRef]
- Morand, R.; Donzelli, M.; Haschke, M.; Krahenbuhl, S. Quantification of plasma carnitine and acylcarnitines by high-performance liquid chromatography-tandem mass spectrometry using online solid-phase extraction. Anal. Bioanal. Chem. 2013, 405, 8829–8836. [Google Scholar] [CrossRef] [PubMed]
- Vernez, L.; Thormann, W.; Krahenbuhl, S. Analysis of carnitine and acylcarnitines in urine by capillary electrophoresis. J. Chromatogr. A 2000, 895, 309–316. [Google Scholar] [CrossRef]
- Kivilompolo, M.; Ohrnberg, L.; Oresic, M.; Hyotylainen, T. Rapid quantitative analysis of carnitine and acylcarnitines by ultra-high performance-hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2013, 1292, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Magiera, S.; Baranowski, J. Determination of carnitine and acylcarnitines in human urine by means of microextraction in packed sorbent and hydrophilic interaction chromatography-ultra-high-performance liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2015, 109, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Minkler, P.E.; Stoll, M.S.K.; Ingalls, S.T.; Hoppel, C.L. Selective and accurate C5 acylcarnitine quantitation by UHPLC-MS/MS: Distinguishing true isovaleric acidemia from pivalate derived interference. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1061-1062, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Minkler, P.E.; Stoll, M.S.; Ingalls, S.T.; Kerner, J.; Hoppel, C.L. Validated method for the quantification of free and total carnitine, butyrobetaine, and acylcarnitines in biological samples. Anal. Chem. 2015, 87, 8994–9001. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Ito, T.; Suzuki, A.; Kurono, Y.; Ueta, A.; Yokoi, K.; Sumi, S.; Togari, H.; Sugiyama, N. Simultaneous quantification of acylcarnitine isomers containing dicarboxylic acylcarnitines in human serum and urine by high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 799–806. [Google Scholar] [CrossRef]
- Maeda, Y.; Nakajima, Y.; Gotoh, K.; Hotta, Y.; Kataoka, T.; Sugiyama, N.; Shirai, N.; Ito, T.; Kimura, K. Kinetic and molecular orbital analyses of dicarboxylic acylcarnitine methylesterification show that derivatization may affect the screening of newborns by tandem mass spectrometry. Bioorg. Med. Chem. Lett. 2016, 26, 121–125. [Google Scholar] [CrossRef]
- Zuniga, A.; Li, L. Ultra-high performance liquid chromatography tandem mass spectrometry for comprehensive analysis of urinary acylcarnitines. Anal. Chim. Acta 2011, 689, 77–84. [Google Scholar] [CrossRef]
- Peng, M.; Liu, L.; Jiang, M.; Liang, C.; Zhao, X.; Cai, Y.; Sheng, H.; Ou, Z.; Luo, H. Measurement of free carnitine and acylcarnitines in plasma by HILIC-ESI-MS/MS without derivatization. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 932, 12–18. [Google Scholar] [CrossRef]
- Li, K.; Sun, Q. Simultaneous determination of free and total carnitine in human serum by HPLC with UV detection. J. Chromatogr. Sci. 2010, 48, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.R.; Ren, S.; Park, M.J.; Choi, Y.J.; Lee, B.J. Determination of highly soluble l-carnitine in biological samples by reverse phase high performance liquid chromatography with fluorescent derivatization. Arch. Pharm. Res. 2007, 30, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Xu, L.; Li, W.; Wu, L. Simultaneous determination of thirteen kinds of amino acid and eight kinds of acylcarnitine in human serum by LC-MS/MS and its application to measure the serum concentration of lung cancer patients. Biomed. Chromatogr. 2016, 30, 1796–1806. [Google Scholar] [CrossRef] [PubMed]
- Minkler, P.E.; Stoll, M.S.K.; Ingalls, S.T.; Hoppel, C.L. Correcting false positive medium-chain acyl-CoA dehydrogenase deficiency results from newborn screening; synthesis, purification, and standardization of branched-chain C8 acylcarnitines for use in their selective and accurate absolute quantitation by UHPLC-MS/MS. Mol. Genet. Metab. 2017, 120, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Higgins, R.; Lim, M.D.; Atkinson, K.; Yang, J.; Lin, K.; Borchers, C.H. Isotope-labelling derivatization with 3-nitrophenylhydrazine for LC/multiple-reaction monitoring-mass-spectrometry-based quantitation of carnitines in dried blood spots. Anal. Chim. Acta 2018, 1037, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, C.; Magera, M.J.; Allard, P.; Tortorelli, S.; Gavrilov, D.; Oglesbee, D.; Raymond, K.; Rinaldo, P.; Matern, D. Combined newborn screening for succinylacetone, amino acids, and acylcarnitines in dried blood spots. Clin. Chem. 2008, 54, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.W. Inaccurate measurement of free carnitine by the electrospray tandem mass spectrometry screening method for blood spots. J. Inherit. Metab. Dis. 1999, 22, 201–202. [Google Scholar] [CrossRef]
- Cyr, D.; Giguere, R.; Giguere, Y.; Lemieux, B. Determination of urinary acylcarnitines: A complementary aid for the high-risk screening of several organic acidurias using a simple and reliable GC/MS-based method. Clin. Biochem. 2000, 33, 151–155. [Google Scholar] [CrossRef]
- Moreira, V.; Brasili, E.; Fiamoncini, J.; Marini, F.; Miccheli, A.; Daniel, H.; Lee, J.J.H.; Hassimotto, N.M.A.; Lajolo, F.M. Orange juice affects acylcarnitine metabolism in healthy volunteers as revealed by a mass-spectrometry based metabolomics approach. Food Res. Int. 2018, 107, 346–352. [Google Scholar] [CrossRef]
- Kriisa, K.; Leppik, L.; Balotsev, R.; Ottas, A.; Soomets, U.; Koido, K.; Volke, V.; Innos, J.; Haring, L.; Vasar, E.; et al. Profiling of acylcarnitines in first episode psychosis before and after antipsychotic treatment. J. Proteome Res. 2017, 16, 3558–3566. [Google Scholar] [CrossRef]
- Simcox, J.; Geoghegan, G.; Maschek, J.A.; Bensard, C.L.; Pasquali, M.; Miao, R.; Lee, S.; Jiang, L.; Huck, I.; Kershaw, E.E.; et al. Global analysis of plasma lipids identifies liver-derived acylcarnitines as a fuel source for brown fat thermogenesis. Cell Metab. 2017, 26, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Strand, E.; Pedersen, E.R.; Svingen, G.F.T.; Olsen, T.; Bjorndal, B.; Karlsson, T.; Dierkes, J.; Njolstad, P.R.; Mellgren, G.; Tell, G.S.; et al. Serum acylcarnitines and risk of cardiovascular death and acute myocardial infarction in patients with stable angina pectoris. J. Am. Heart Assoc. 2017, 6, e003620. [Google Scholar] [CrossRef] [PubMed]
- Schooneman, M.G.; Houtkooper, R.H.; Hollak, C.E.; Wanders, R.J.; Vaz, F.M.; Soeters, M.R.; Houten, S.M. The impact of altered carnitine availability on acylcarnitine metabolism, energy expenditure and glucose tolerance in diet-induced obese mice. Biochim. Biophys. Acta Mol. Basis Dis. 2016, 1862, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.K.; Islam, M.T.; Bhuyan, G.S.; Sultana, N.; Begum, N.; Al Mahmud-Un-Nabi, M.; Howladar, M.A.A.N.; Dipta, T.F.; Muraduzzaman, A.K.M.; Qadri, S.K.; et al. Impaired acylcarnitine profile in transfusion-dependent beta-thalassemia major patients in Bangladesh. J. Adv. Res. 2018, 12, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Sun, L.; Sun, Q.; Liang, L.; Gao, X.; Li, R.; Pan, A.; Li, H.; Deng, Y.; Hu, F.B.; et al. Associations of plasma amino acid and acylcarnitine profiles with incident reduced glomerular filtration rate. Clin. J. Am. Soc. Nephrol. 2018, 13, 560–568. [Google Scholar] [CrossRef]
- Ruiz, M.; Labarthe, F.; Fortier, A.; Bouchard, B.; Thompson Legault, J.; Bolduc, V.; Rigal, O.; Chen, J.; Ducharme, A.; Crawford, P.A.; et al. Circulating acylcarnitine profile in human heart failure: A surrogate of fatty acid metabolic dysregulation in mitochondria and beyond. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H768–H781. [Google Scholar] [CrossRef]
- Giesbertz, P.; Ecker, J.; Haag, A.; Spanier, B.; Daniel, H. An LC-MS/MS method to quantify acylcarnitine species including isomeric and odd-numbered forms in plasma and tissues. J. Lipid Res. 2015, 56, 2029–2039. [Google Scholar] [CrossRef]
- Hassan, A.; Tsuda, Y.; Asai, A.; Yokohama, K.; Nakamura, K.; Sujishi, T.; Ohama, H.; Tsuchimoto, Y.; Fukunishi, S.; Abdelaal, U.M.; et al. Effects of oral l-carnitine on liver functions after transarterial chemoembolization in intermediate-stage HCC patients. Mediat. Inflamm. 2015, 2015, 608216. [Google Scholar] [CrossRef]
- Flanagan, J.L.; Simmons, P.A.; Vehige, J.; Willcox, M.D.; Garrett, Q. Role of carnitine in disease. Nutr. Metab. (Lond.) 2010, 7, 30. [Google Scholar] [CrossRef]
- Baffy, G.; Brunt, E.M.; Caldwell, S.H. Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J. Hepatol. 2012, 56, 1384–1391. [Google Scholar] [CrossRef]
- Ding, H.-r.; Wang, J.-l.; Ren, H.-z.; Shi, X.-l.J.B.R.I. Lipometabolism and Glycometabolism in Liver Diseases. Biomed Res. Int. 2018, 2018, 1287127. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in non-alcoholic fatty liver disease. Metabolism 2011, 60, 404–413. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, Y.; Ye, G.; Li, X.; Yin, P.; Ruan, Q.; Xu, G. Classification and differential metabolite discovery of liver diseases based on plasma metabolic profiling and support vector machines. J. Sep. Sci. 2011, 34, 3029–3036. [Google Scholar] [CrossRef]
- Krahenbuhl, S.; Reichen, J. Carnitine metabolism in patients with chronic liver disease. Hepatology 1997, 25, 148–153. [Google Scholar] [CrossRef]
- Cheng, M.L.; Shiao, M.S.; Chiu, D.T.; Weng, S.F.; Tang, H.Y.; Ho, H.Y. Biochemical disorders associated with antiproliferative effect of dehydroepiandrosterone in hepatoma cells as revealed by LC-based metabolomics. Biochem. Pharmacol. 2011, 82, 1549–1561. [Google Scholar] [CrossRef]
- Yaligar, J.; Teoh, W.W.; Othman, R.; Verma, S.K.; Phang, B.H.; Lee, S.S.; Wang, W.W.; Toh, H.C.; Gopalan, V.; Sabapathy, K.; et al. Longitudinal metabolic imaging of hepatocellular carcinoma in transgenic mouse models identifies acylcarnitine as a potential biomarker for early detection. Sci. Rep. 2016, 6, 20299. [Google Scholar] [CrossRef]
- Ishikawa, H.; Takaki, A.; Tsuzaki, R.; Yasunaka, T.; Koike, K.; Shimomura, Y.; Seki, H.; Matsushita, H.; Miyake, Y.; Ikeda, F.; et al. l-carnitine prevents progression of non-alcoholic steatohepatitis in a mouse model with upregulation of mitochondrial pathway. PLoS ONE 2014, 9, e100627. [Google Scholar] [CrossRef]
- Tan, Y.; Yin, P.; Tang, L.; Xing, W.; Huang, Q.; Cao, D.; Zhao, X.; Wang, W.; Lu, X.; Xu, Z.; et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: Potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol. Cell. Proteom. 2012, 11, M111 010694. [Google Scholar] [CrossRef]
- Hu, J.; Lin, Y.Y.; Chen, P.J.; Watashi, K.; Wakita, T. Cell and animal models for studying hepatitis B virus infection and drug development. Gastroenterology 2019, 156, 338–354. [Google Scholar] [CrossRef]
- Bukh, J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology 2012, 142, 1279–1287. [Google Scholar] [CrossRef]
- Lerat, H.; Higgs, M.; Pawlotsky, J.M. Animal models in the study of hepatitis C virus-associated liver pathologies. Expert Rev. Gastroenterol. Hepatol. 2011, 5, 341–352. [Google Scholar] [CrossRef]
- Degos, F.; Christidis, C.; Ganne-Carrie, N.; Farmachidi, J.P.; Degott, C.; Guettier, C.; Trinchet, J.C.; Beaugrand, M.; Chevret, S. Hepatitis C virus related cirrhosis: Time to occurrence of hepatocellular carcinoma and death. Gut 2000, 47, 131–136. [Google Scholar] [CrossRef]
- Ieluzzi, D.; Covolo, L.; Donato, F.; Fattovich, G. Progression to cirrhosis, hepatocellular carcinoma and liver-related mortality in chronic hepatitis B patients in Italy. Dig. Liver Dis. 2014, 46, 427–432. [Google Scholar] [CrossRef]
- Zhou, T.C.; Lai, X.; Feng, M.H.; Tang, Y.; Zhang, L.; Wei, J. Systematic review and meta-analysis: Development of hepatocellular carcinoma in chronic hepatitis B patients with hepatitis e antigen seroconversion. J. Viral Hepat. 2018, 25, 1172–1179. [Google Scholar] [CrossRef]
- Liu, S.Y.; Zhang, R.L.; Kang, H.; Fan, Z.J.; Du, Z. Human liver tissue metabolic profiling research on hepatitis B virus-related hepatocellular carcinoma. World J. Gastroenterol. 2013, 19, 3423–3432. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Q.; Yin, P.; Xing, W.; Wu, Z.; Chen, S.; Lu, X.; Zhang, Y.; Lin, X.; Xu, G. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal. Bioanal. Chem. 2012, 403, 203–213. [Google Scholar] [CrossRef]
- Lin, X.; Yang, F.; Zhou, L.; Yin, P.; Kong, H.; Xing, W.; Lu, X.; Jia, L.; Wang, Q.; Xu, G. A support vector machine-recursive feature elimination feature selection method based on artificial contrast variables and mutual information. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 910, 149–155. [Google Scholar] [CrossRef]
- Chen, S.L.; Kong, H.W.; Lu, X.; Li, Y.; Yin, P.Y.; Zeng, Z.D.; Xu, G.W. Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometry. Anal. Chem. 2013, 85, 8326–8333. [Google Scholar] [CrossRef]
- Lu, Y.; Li, N.; Gao, L.; Xu, Y.J.; Huang, C.; Yu, K.; Ling, Q.; Cheng, Q.; Chen, S.; Zhu, M.; et al. Acetylcarnitine is a candidate diagnostic and prognostic biomarker of hepatocellular carcinoma. Cancer Res. 2016, 76, 2912–2920. [Google Scholar] [CrossRef]
- Shariff, M.I.; Ladep, N.G.; Cox, I.J.; Williams, H.R.; Okeke, E.; Malu, A.; Thillainayagam, A.V.; Crossey, M.M.; Khan, S.A.; Thomas, H.C.; et al. Characterization of urinary biomarkers of hepatocellular carcinoma using magnetic resonance spectroscopy in a Nigerian population. J. Proteome Res. 2010, 9, 1096–1103. [Google Scholar] [CrossRef]
- Shariff, M.I.; Gomaa, A.I.; Cox, I.J.; Patel, M.; Williams, H.R.; Crossey, M.M.; Thillainayagam, A.V.; Thomas, H.C.; Waked, I.; Khan, S.A.; et al. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: A validation study. J. Proteome Res. 2011, 10, 1828–1836. [Google Scholar] [CrossRef]
- Cox, I.J.; Aliev, A.E.; Crossey, M.M.; Dawood, M.; Al-Mahtab, M.; Akbar, S.M.; Rahman, S.; Riva, A.; Williams, R.; Taylor-Robinson, S.D. Urinary nuclear magnetic resonance spectroscopy of a Bangladeshi cohort with hepatitis-B hepatocellular carcinoma: A biomarker corroboration study. World J. Gastroenterol. 2016, 22, 4191–4200. [Google Scholar] [CrossRef]
- Xiao, J.F.; Varghese, R.S.; Zhou, B.; Nezami Ranjbar, M.R.; Zhao, Y.; Tsai, T.H.; Di Poto, C.; Wang, J.; Goerlitz, D.; Luo, Y.; et al. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J. Proteome Res. 2012, 11, 5914–5923. [Google Scholar] [CrossRef]
- Liu, Y.; Hong, Z.; Tan, G.; Dong, X.; Yang, G.; Zhao, L.; Chen, X.; Zhu, Z.; Lou, Z.; Qian, B.; et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int. J. Cancer 2014, 135, 658–668. [Google Scholar] [CrossRef]
- Gong, Z.G.; Zhao, W.; Zhang, J.; Wu, X.; Hu, J.; Yin, G.C.; Xu, Y.J. Metabolomics and eicosanoid analysis identified serum biomarkers for distinguishing hepatocellular carcinoma from hepatitis B virus-related cirrhosis. Oncotarget 2017, 8, 63890–63900. [Google Scholar] [CrossRef]
- Chen, T.; Xie, G.; Wang, X.; Fan, J.; Qiu, Y.; Zheng, X.; Qi, X.; Cao, Y.; Su, M.; Wang, X.; et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell. Proteom. 2011, 10, M110 004945. [Google Scholar] [CrossRef]
- Fitian, A.I.; Nelson, D.R.; Liu, C.; Xu, Y.L.; Ararat, M.; Cabrera, R. Integrated metabolomic profiling of hepatocellular carcinoma in hepatitis C cirrhosis through GC/MS and UPLC/MS-MS. Liver Int. 2014, 34, 1428–1444. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Lv, S.; Yin, P.; Zhao, X.; Lu, X.; Zhang, F.; Xu, G. Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal. Chim. Acta 2009, 650, 3–9. [Google Scholar] [CrossRef]
- Yin, P.; Wan, D.; Zhao, C.; Chen, J.; Zhao, X.; Wang, W.; Lu, X.; Yang, S.; Gu, J.; Xu, G. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol. Biosyst. 2009, 5, 868–876. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, N.; Cao, Y.F.; Zhu, Z.T.; Gao, P. Differential diagnosis between hepatocellular carcinoma and cirrhosis by serum amino acids and acylcarnitines. Int. J. Clin. Exp. Pathol. 2018, 11, 1763–1769. [Google Scholar]
- Lu, X.; Nie, H.; Li, Y.Q.; Zhan, C.; Liu, X.; Shi, X.Y.; Shi, M.; Zhang, Y.B.; Li, Y. Comprehensive characterization and evaluation of hepatocellular carcinoma by LC-MS based serum metabolomics. Metabolomics 2015, 11, 1381–1393. [Google Scholar] [CrossRef]
- Elsemman, I.E.; Mardinoglu, A.; Shoaie, S.; Soliman, T.H.; Nielsen, J. Systems biology analysis of hepatitis C virus infection reveals the role of copy number increases in regions of chromosome 1q in hepatocellular carcinoma metabolism. Mol. Biosyst. 2016, 12, 1496–1506. [Google Scholar] [CrossRef]
- Zhou, L.; Ding, L.; Yin, P.; Lu, X.; Wang, X.; Niu, J.; Gao, P.; Xu, G. Serum metabolic profiling study of hepatocellular carcinoma infected with hepatitis B or hepatitis C virus by using liquid chromatography-mass spectrometry. J. Proteome Res. 2012, 11, 5433–5442. [Google Scholar] [CrossRef]
- Lin, X.; Gao, J.; Zhou, L.; Yin, P.; Xu, G. A modified k-TSP algorithm and its application in LC-MS-based metabolomics study of hepatocellular carcinoma and chronic liver diseases. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 966, 100–108. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, X.; Zhang, Y.; Zhang, K.; Zhan, C.; Shi, X.; Li, Y.; Zhao, J.; Bai, Y.; Wang, Y.; et al. Metabolic profiling analysis upon acylcarnitines in tissues of hepatocellular carcinoma revealed the inhibited carnitine shuttle system caused by the downregulated carnitine palmitoyltransferase 2. Mol. Carcinog. 2019. [Google Scholar] [CrossRef]
- Lin, M.; Lv, D.; Zheng, Y.; Wu, M.; Xu, C.; Zhang, Q.; Wu, L. Downregulation of CPT2 promotes tumourigenesis and chemoresistance to cisplatin in hepatocellular carcinoma. OncoTargets Ther. 2018, 11, 3101–3110. [Google Scholar] [CrossRef]
- Chen, S.; Wang, C.; Cui, A.; Yu, K.; Huang, C.; Zhu, M.; Chen, M. Development of a genetic and clinical data-based (gc) risk score for predicting survival of hepatocellular carcinoma patients after tumour resection. Cell Physiol. Biochem. 2018, 48, 491–502. [Google Scholar] [CrossRef]

| Reference | Sample | Platform | Main Findings |
|---|---|---|---|
| [23] | Urine 27 CIR 33 HCC 26 HC | LC-MS Non-targeted | HCC vs. CIR: MCACs and SCACs increased. CIR & HCC vs. HC: LCAC and MCAC decreased. HCC vs. CIR: carnitine C4:0 elevated |
| [24] | Tissues 50 HCT 50 DNT Serum 81 CH 78 CIR 139 HCC | LC-MS CE-MS Non-targeted Targeted | HCT vs. DNT: carnitine (C2/C0) upregulated, propionylcarnitine to carnitine (C3/C0) downregulated, SCAC decreased, LCAC increased HCC vs. CIR and CH: propionylcarnitine decreased, acetylcarnitine elevated |
| [35] | Sera 136 CHB 104 CIR 95 HCC | LC-MS Targeted | CIR and HCC vs. CHB: carnitine, 2 SCAC, and 4 MCAC decreased. CIR vs. CHB and HCC: AC C18:2 higher, AC C3-OH lower |
| [105] | SK-Hep1 cells underexpressing G6PD (Sk-Gi) and control cells (Sk-Sc) after dehydroepiandrosterone (DHEA) treatment | LC-MS Non-targeted | Carnitine and acyl derivatives declined in DHEA-treated Sk-Gi cells |
| [108] | Rat sera 52 HCC 28 HCHuman sera 262 HCC 76 CIR 74 HBV | LC-MS Non-targeted | HCC vs. HC (rat): palmityl-l-carnitine decreased (with the age of the animals), increased in week 8 |
| [115] | HBV-related HCC tissue 10 CTT 10 ANT 10 DNT | LC-MSNon-targeted | DAT vs. CTT: arachidyl carnitine lower |
| [116] | Sera 30 HC 30 CHB 30 CIR 30 HCC | LC-MS Non-targeted | CHB vs. CIR vs. HCC: C16:1-CN increased (with severity of chronic liver diseases), MCAC and SCAC decreased (including C10-CN, C10:1-CN, C8-CN, and C6-CN) CIR and HCC vs. HC: C14-CN increased |
| [117] | Sera 30 CHB 30 CIR 30 HCC 30 HC | LC-MS Non-targeted | HCC vs. CHB: pimelylcarnitine and acetylcarnitine rise HCC vs. CIR: pimelylcarnitine and carnitine rise CIR and HCC vs. CHB and HC: C16:1-CN and C18:1-CN accumulated |
| [119] | Tissue 50 HCC (38 males and 12 females). Serum 18 HCC 20 CIR 20 HC | LC-MS Non-targeted | CTT vs. ANT: LCAC accumulated, MCAC and SCAC decreased. Acetylcarnitine reduced (gradually) HCC vs. CIR and HC: serum acetylcarnitine reduced |
| [120] | Urine 18 HCC 10 CIR 15 HC | NMR Non-targeted | HCC vs. CIR and HC: carnitine raised |
| [121] | Urine and serum 16 HCC 14 CIR 17 HC | NMR Non-targeted | HCC vs. CIR and HC: urinary carnitine elevated |
| [122] | Urine 42 HCC 47 CIR 7 HC | NMR Non-targeted | HCC vs. HC: carnitine increased |
| [123] | Sera 40 HCC 49 CIR | LC-MS Targeted Non-targeted | HCC vs. CIR: LCAC, oleoyl carnitine, palmitoyl carnitine, and linoelaidyl carnitine down-regulation |
| [124] | Serum 71 HCC 80 CIR 26 HC | NMR and LC-MS Non-targeted | HCC vs. CIR and HC: acylcarnitine increased, MCAC and LCAC decreased (including decanoyl-l-carnitine and palmitoylcarnitine) CIR and HCC vs. HC: carnitine decreased (progressive) |
| [125] | Serum 39 HC 49 HBV-CIR 51 HCC | LC-MS Non-targeted Targeted | HBV-CIR and HCC vs. HC: carnitine altered (stepwise) |
| [126] | Serum and urine 82 HCC 71 HC | LC-MS Non-targeted | HCC vs. HC: serum carnitine 1.36 times |
| [127] | Serum 30 HCC, 27 CIR 30 HC | LC-MS Non-targeted | HCC vs. CIR: acetylcarnitine, glutarylcarnitine and succinylcarnitine upregulated CIR vs. HC: MCAC overexpression HCC vs. CIR: LCAC downward |
| [128] | Urine 24 HC 21 HCC | LC-MS Non-targeted | HCC vs. HC: carnitine C8:1, carnitine C9:0, carnitine C9:1, carnitine C10:1 and carnitine C10:3 reduced, acetylcarnitine increased |
| [129] | Serum 24 HCC 25 CIR 25 HC | LC-MS Non-targeted | HCC and CIR vs. HC: carnitine, carnitine fragment and acetylcarnitine increased |
| [130] | Serum 92 male and 44 female CIR 41 male and 9 female HCC | LC-MS Targeted | HCC vs. CIR: decanoylcarnitine, decenoylcarnitine, propionylcarnitine/methionine, Methylmalonylcarnitine, 3-Hydroxy-isovalerylcarnitine/carnitine increased; octadecenoylcarnitine, malonylcarnitine/decanoylcarnitine, butyrylcarnitine/octanoylcarnitine decreased |
| [131] | Serum 267 HCC 48 HBV 272 HC | LC-MS Non-targeted | HCC vs. HC: SCAC and MCAC decreased, LCAC increased |
| [133] | 22 HBV 6 HCV 38 HBV-associated HCC 31 HCV-associated HCC 31 HC | LC-MS Targeted Non-targeted | HCC vs. HC: C18:1-CN and C18:2-CN increased. |
| [134] | Serum 30 HC, 30 CHB, 30 CIR 30 HCC | LC-MS Non-targeted | CIR and HCC vs. HC and CHB: C16:1-CN and C16:0-CN elevated CHB, CIR and HCC vs. HC: C10-CN decreased, LCAC elevated, MCAC and SCAC exhibited the opposite trend |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Gao, D.; Jiang, Y. Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma. Metabolites 2019, 9, 36. https://doi.org/10.3390/metabo9020036
Li S, Gao D, Jiang Y. Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma. Metabolites. 2019; 9(2):36. https://doi.org/10.3390/metabo9020036
Chicago/Turabian StyleLi, Shangfu, Dan Gao, and Yuyang Jiang. 2019. "Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma" Metabolites 9, no. 2: 36. https://doi.org/10.3390/metabo9020036
APA StyleLi, S., Gao, D., & Jiang, Y. (2019). Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma. Metabolites, 9(2), 36. https://doi.org/10.3390/metabo9020036