Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases
Abstract
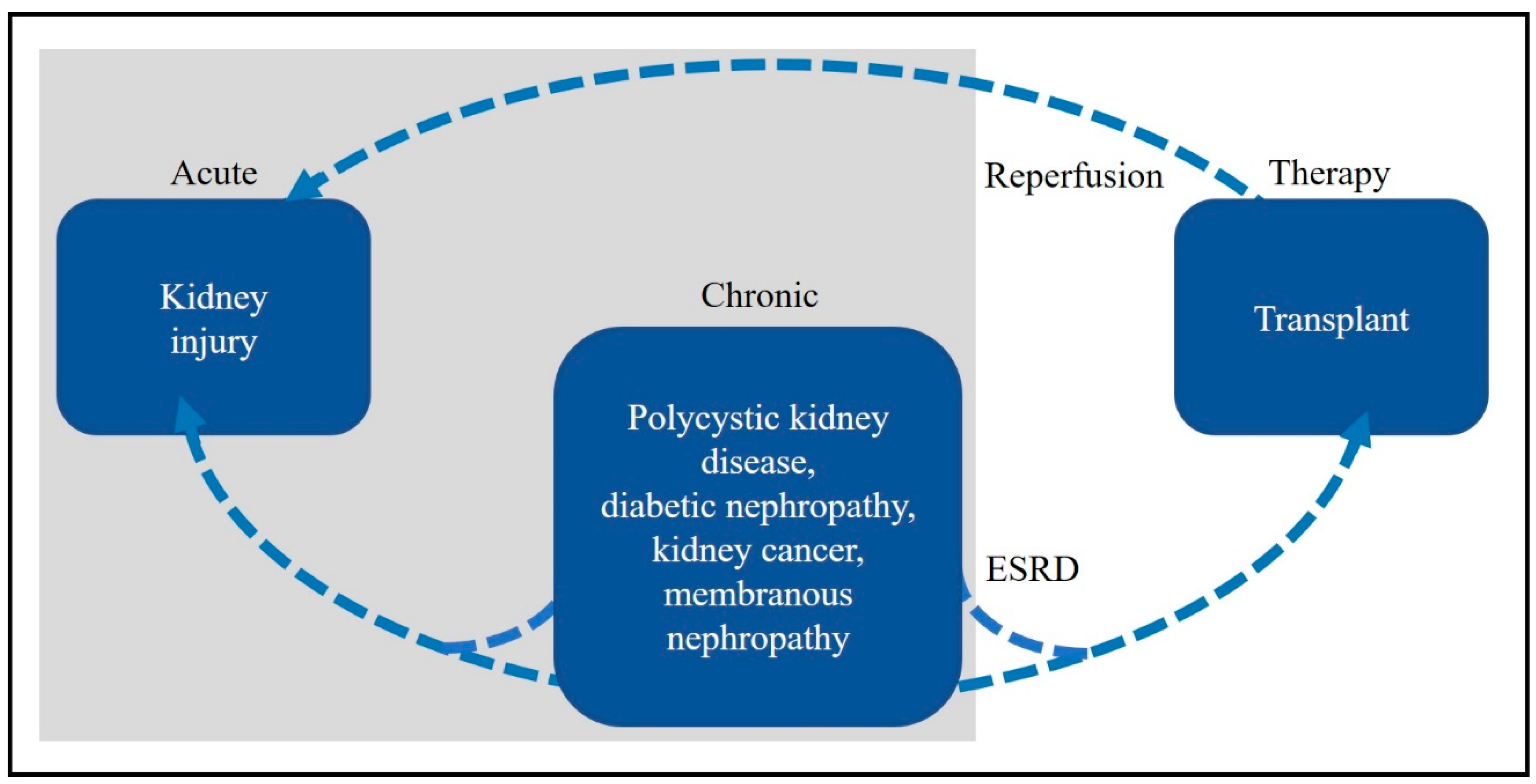
1. Introduction
2. Kidney Disease
2.1. Chronic Kidney Disease
2.2. Diabetic Nephropathy
2.3. Acute Kidney Injury (AKI)
2.4. Kidney Cancer
2.5. Kidney Transplantation
2.6. Polycystic Kidney Diseases
3. Metabolomics
3.1. Sample Collection, Preparation, Storage and Handling
3.2. Metabolite Extraction
3.3. Chromatographic Separation
3.3.1. Gas Chromatography
3.3.2. Liquid Chromatography
3.4. Mass Spectrometry
3.4.1. Ionisation
3.4.2. Mass Analysers
3.5. Data Processing and Analysis
3.6. Metabolite Identification and Interpretation of Findings
3.6.1. Identification
3.6.2. Interpretation
4. Findings from Metabolomic Studies of Kidney Disease
4.1. Purine Metabolism
4.2. Tryptophan Metabolism
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moyes, C.D.; Schulte, P.M. Principles of Animal Physiology, 2nd ed.; Benjamin Cummings: San Francisco, CA, USA, 2008. [Google Scholar]
- Giebisch, G. Kidney, Water and Electrolyte Metabolism. Annu. Rev. Physiol. 1962, 24, 357–420. [Google Scholar] [CrossRef] [PubMed]
- Blantz, R.C.; Deng, A.; Miracle, C.M.; Thomson, S.C. Regulation of kidney function and metabolism: A question of supply and demand. Trans. Am. Clin. Climatol. Assoc. 2007, 118, 23–43. [Google Scholar] [PubMed]
- Gerich, J.E. Role of the kidney in normal glucose homeostasis and in the hyperglycaemia of diabetes mellitus: Therapeutic implications. Diabet. Med. 2010, 27, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Cass, A.; Chadban, S.; Gallagher, M.; Howard, K.; Jones, A.; McDonald, S.; Snelling, P.; White, S. The Economic Impact of End-Stage Kidney Disease in Australia Projections to 2020. Available online: https://kidney.org.au/cms_uploads/docs/kha-economic-impact-of-eskd-in-australia-projections-2020.pdf (accessed on 7 February 2019).
- Smith, H.W. Kidney. Annu. Rev. Physiol. 1939, 1, 503–528. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C. Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.H.; Kim, K. Metabolomics in the study of kidney diseases. Nat. Rev. Nephrol. 2012, 8, 22–33. [Google Scholar] [CrossRef]
- Al-Ismaili, Z.; Palijan, A.; Zappitelli, M. Biomarkers of acute kidney injury in children: Discovery, evaluation, and clinical application. Pediatr. Nephrol. 2011, 26, 29–40. [Google Scholar] [CrossRef]
- Ferenbach, D.A.; Bonventre, J.V. Acute kidney injury and chronic kidney disease: From the laboratory to the clinic. Nephrol. Ther. 2016, 12 (Suppl. S1), S41–S48. [Google Scholar] [CrossRef]
- National Kidney Foundation. K/DOQI Clinical practice guidelines for chronic kidney disease: Evaluation, classification and stratification. Am. J. Kidney Dis. 2002, 39 (Suppl. S1), S1–S266. [Google Scholar]
- Zhao, Y.-Y. Metabolomics in chronic kidney disease. Clin. Chim. Acta 2013, 422, 59–69. [Google Scholar] [CrossRef]
- Inker, L.A.; Schmid, C.H.; Tighiouart, H.; Eckfeldt, J.H.; Feldman, H.I.; Greene, T.; Kusek, J.W.; Manzi, J.; Van Lente, F.; Zhang, Y.L.; et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N. Engl. J. Med. 2012, 367, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.L.; de Azevedo, M.J.; Silveiro, S.P.; Canani, L.H.; Caramori, M.L.; Zelmanovitz, T. Diabetic Nephropathy: Diagnosis, Prevention, and Treatment. Diabetes Care 2005, 28, 164. [Google Scholar] [CrossRef]
- Van der Kloet, F.M.; Tempels, F.W.A.; Ismail, N.; van der Heijden, R.; Kasper, P.T.; Rojas-Cherto, M.; van Doorn, R.; Spijksma, G.; Koek, M.; van der Greef, J.; et al. Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study). Metabolomics 2012, 8, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Parving, H.H.; Lehnert, H.; Brochner-Mortensen, J.; Gomis, R.; Andersen, S.; Arner, P. The effect of irbesartan on the development of diabetic nephropathy in patients with type 2 diabetes. N. Engl. J. Med. 2001, 345, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Schena, F.P.; Gesualdo, L. Pathogenetic mechanisms of diabetic nephropathy. J. Am. Soc. Nephrol. 2005, 16 (Suppl. S1), S30–S33. [Google Scholar] [CrossRef]
- Forbes, J.M.; Coughlan, M.T.; Cooper, M.E. Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes 2008, 57, 1446–1454. [Google Scholar] [CrossRef]
- Bellomo, R.; Kellum, J.A.; Ronco, C. Acute kidney injury. Lancet 2012, 380, 756–766. [Google Scholar] [CrossRef]
- Bellomo, R.; Ronco, C.; Kellum, J.A.; Mehta, R.L.; Palevsky, P.; ADQI. Acute renal failure—Definition, outcome measures, animal models, fluid therapy and information technology needs: The Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) group. Crit. Care 2004, 8, R204–R212. [Google Scholar] [CrossRef]
- Wheeler, D.S.; Devarajan, P.; Ma, Q.; Harmon, K.; Monaco, M.; Cvijanovich, N.; Wong, H.R. Serum neutrophil gelatinase-associated lipocalin (NGAL) as a marker of acute kidney injury in critically ill children with septic shock. Crit. Care Med. 2008, 36, 1297–1303. [Google Scholar] [CrossRef]
- Laterza, O.F.; Price, C.P.; Scott, M.G. Cystatin C: An improved estimator of glomerular filtration rate? Clin. Chem. 2002, 48, 699–707. [Google Scholar]
- Han, W.K.; Bailly, V.; Abichandani, R.; Thadhani, R.; Bonventre, J.V. Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury. Kidney Int. 2002, 62, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Branten, A.J.; Mulder, T.P.; Peters, W.H.; Assmann, K.J.; Wetzels, J.F. Urinary excretions of glutathione S transferases alpha and pi in patients with proteinuria: Reflection of the site of tubular injury. Nephron 2000, 85, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Kamijo, A.; Sugaya, T.; Hikawa, A.; Yamanouchi, M.; Hirata, Y.; Ishimitsu, T.; Numabe, A.; Takagi, M.; Hayakawa, H.; Tabei, F.; et al. Urinary liver-type fatty acid binding protein as a useful biomarker in chronic kidney disease. Mol. Cell. Biochem. 2006, 284, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Yuan, J.; Zhao, Y.; Zha, Y. Urine interleukin-18 in prediction of acute kidney injury: A systematic review and meta-analysis. J. Nephrol. 2015, 28, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Ganti, S.; Weiss, R.H. Urine Metabolomics for kidney cancer detection and biomarker discovery. Urol. Oncol. 2011, 29, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.S.; Carvalho, M.; de Lourdes Bastos, M.; de Pinho, P.G. Biomarkers in renal cell carcinoma: A metabolomics approach. Metabolomics 2014, 10, 1210–1222. [Google Scholar] [CrossRef]
- Rini, B.I.; Campbell, S.C.; Escudier, B. Renal cell carcinoma. Lancet 2009, 373, 1119–1132. [Google Scholar] [CrossRef]
- Cohen, H.T.M.D.; McGovern, F.J.M.D. Renal-cell carcinoma. N. Engl. J. Med. 2005, 353, 2477–2490. [Google Scholar] [CrossRef]
- Hariharan, S.; McBride, M.A.; Cherikh, W.S.; Tolleris, C.B.; Bresnahan, B.A.; Johnson, C.P. Post-transplant renal function in the first year predicts long-term kidney transplant survival. Kidney Int. 2002, 62, 311–318. [Google Scholar] [CrossRef]
- Petersdorf, E.W. HLA mismatching in transplantation. Blood 2015, 125, 1058–1059. [Google Scholar] [CrossRef]
- Lee, P.-C.; Terasaki, P.I.; Takemoto, S.K.; Lee, P.-H.; Hung, C.-J.; Chen, Y.-L.; Tsai, A.; Lei, H.-Y. All chronic rejection failures of kidney transplants were preceded by the development of HLA antibodies. Transplantation 2002, 74, 1192–1194. [Google Scholar] [CrossRef] [PubMed]
- Solez, K.i.m.; Axelsen, R.A.; Benediktsson, H.; Burdick, J.F.; Cohen, A.H.; Colvin, R.B.; Croker, B.P.; Droz, D.; Dunnill, M.S.; Halloran, P.F.; et al. International standardization of criteria for the histologic diagnosis of renal allograft rejection: The Banff working classification of kidney transplant pathology. Kidney Int. 1993, 44, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Schwiebert, E.M. Compelling ’metabolomic’ biomarkers may signal PKD pathogenesis. Am. J. Physiol. Renal. Physiol. 2010, 298, F1103–F1104. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.; Ibraghimov-Beskrovnaya, O.; Bukanov, N.O. Serum and urinary biomarker signatures for rapid preclinical in vivo assessment of CDK inhibition as a therapeutic approach for PKD. Cell Cycle 2008, 7, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Simms, R.J.; Haynes, A.M.; Eley, L.; Sayer, J. Nephronophthisis: A genetically diverse ciliopathy. Int. J. Nephrol. 2011, 2011, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.T. Nephronophthisis and related syndromes. Curr. Opin. Pediatr. 2015, 27, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Ariza, M.; Alvarez, V.; Marin, R.; Aguado, S.; Lopez-Larrea, C.; Alvarez, J.; Menendez, M.J.; Coto, E. A family with a milder form of adult dominant polycystic kidney disease not linked to the PKD1 (16p) or PKD2 (4q) genes. J. Med. Genet. 1997, 34, 587–589. [Google Scholar] [CrossRef]
- Arnaout, M.A. Molecular genetics and pathogenesis of autosomal dominant polycystic kidney disease. Annu. Rev. Med. 2001, 52. [Google Scholar] [CrossRef]
- Harris, P.C.; Torres, V.E. Polycystic kidney disease. Annu. Rev. Med. 2009, 60, 321–337. [Google Scholar] [CrossRef]
- Torres, V.E.; Harris, P.C. Polycystic kidney disease: Genes, proteins, animal models, disease mechanisms and therapeutic opportunities. J. Int. Med. 2007, 261, 17–31. [Google Scholar] [CrossRef]
- Daoust, M.C.; Reynolds, D.M.; Bichet, D.G.; Somlo, S. Evidence for a third genetic locus for autosomal dominant polycystic kidney disease. Genomics 1994, 25, 733–736. [Google Scholar] [CrossRef]
- Gigarel, N.; Frydman, N.; Burlet, P.; Kerbrat, V.; Tachdjian, G.; Fanchin, R.; Antignac, C.; Frydman, R.; Munnich, A.; Steffann, J. Preimplantation genetic diagnosis for autosomal recessive polycystic kidney disease. Reprod. Biomed. Online 2008, 16, 152–158. [Google Scholar] [CrossRef]
- Herman, T.E.; Siegel, M.J. Neonatal autosomal recessive polycystic kidney disease. J. Perinatol. 2008, 28, 584–585. [Google Scholar] [CrossRef] [PubMed]
- Arbeiter, A.; Buscher, R.; Bonzel, K.E.; Wingen, A.M.; Vester, U.; Wohlschlager, J.; Zerres, K.; Nurnberger, J.; Bergmann, C.; Hoyer, P.F. Nephrectomy in an autosomal recessive polycystic kidney disease (ARPKD) patient with rapid kidney enlargement and increased expression of EGFR. Nephrol. Dial. Transplant. 2008, 23, 3026–3029. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, G.F.; Rice, R.R.; Suarez, E.S. Autosomal recessive polycystic kidney disease: Radiologic-pathologic correlation. RadioGraphics 2000, 20, 837–855. [Google Scholar] [CrossRef] [PubMed]
- Guay-Woodford, L. Murine models of polycystic kidney disease: Molecular and therapeutic insights. Am. J. Physiol. Renal. Physiol. 2003, 285, 1034–1049. [Google Scholar] [CrossRef] [PubMed]
- Sumfest, J.M.; Burns, M.W.; Mitchell, M.E. Aggressive surgical and medical management of autosomal recessive polycystic kidney disease. Pediatr. Urol. 1993, 42, 309–312. [Google Scholar] [CrossRef]
- Fischer, D.C.; Jacoby, U.; Pape, L.; Ward, C.J.; Kuwertz-Broeking, E.; Renken, C.; Nizze, H.; Querfeld, U.; Rudolph, B.; Mueller-Wiefel, D.E.; et al. Activation of the AKT/mTOR pathway in autosomal recessive polycystic kidney disease (ARPKD). Nephrol. Dial. Transplant. 2009, 24, 1819–1827. [Google Scholar] [CrossRef]
- Calvet, J.P. MEK inhibition holds promise for polycystic kidney disease. J. Am. Soc. Nephrol. 2006, 17, 1498–1500. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Hempson, S.J.; Reif, G.A.; Hedge, A.-M.; Wallace, D.P. Calcium restores a normal proliferation phenotype in human polycystic kidney disease epithelial cells. J. Am. Soc. Nephrol. 2006, 17, 178–187. [Google Scholar] [CrossRef]
- Bailey, J.L.; Zheng, B.; Hu, Z.; Price, S.R.; Mitch, W.E. Chronic kidney disease causes defects in signalling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: Implications for muscle atrophy. J. Am. Soc. Nephrol. 2006, 17, 1388–1394. [Google Scholar] [CrossRef]
- Bukanov, N.O.; Smith, L.A.; Klinger, K.W.; Ledbetter, S.R.; Ibraghimov-Beskrovnaya, O. Long-lasting arrest of murine polycystic kidney disease with CDK inhibitor roscovitine. Nature 2006, 444, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Natoli, T.A.; Gareski, T.C.; Dackowski, W.R.; Smith, L.; Bukanov, N.O.; Piepenhagen, P.; Ibraghimov-Beskrovnaya, O. Pkd1 and Nek8 mutations affect cell-cell adhesion and cilia in cysts formed in kidney organ cultures. Am. J. Physiol. Renal. Physiol. 2007, 294, F73–F83. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics: The principles and potential applications to transplantation. Am. J. Transplant. 2005, 5, 2814–2820. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. Trends Analyt. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; Hammock, B.D.; Watkins, S.M. Metabolomics: Building on a century of biochemistry to guide human health. Metabolomics 2005, 1, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zou, L.; Ong, C.N. Experiment originated variations, and multi-peak and multi-origination phenomena in derivatization-based GC-MS metabolomics. Trends Analyt. Chem. 2010, 29, 269–280. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.C.; Nugent, A.; Brennan, L.; Gibney, M.J. Understanding the metabolome—Challenges for metabolomics. Nutr. Bull. 2008, 33, 316–323. [Google Scholar] [CrossRef]
- Chobanyan, K.; Mitschke, A.; Gutzki, F.M.; Stichtenoth, D.O.; Tsikas, D. Accurate quantification of dimethylamine (DMA) in human plasma and serum by GC-MS and GC-tandem MS as pentafluorobenzamide derivative in the positive-ion chemical ionization mode. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 851, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Lehmann, R.; Xu, G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal. Bioanal. Chem. 2015, 407, 4879–4892. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sanchez, B.; Priego-Capote, F.; Luque de Castro, M.D. Metabolomics analysis I. Selection of biological samples and practical aspects preceding sample preparation. Trends Analyt. Chem. 2010, 29, 111–119. [Google Scholar] [CrossRef]
- Alvarez-Sanchez, B.; Priego-Capote, F.; Luque de Castro, M.D. Metabolomics analysis II. Preparation of biological samples prior to detection. Trends Analyt. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Griffin, J.L.; Nicholls, A.W.; Daykin, C.A.; Heald, S.; Keun, H.C.; Schuppe-Koistinen, I.; Griffiths, J.R.; Cheng, L.L.; Rocca-Serra, P.; Rubtsov, D.V.; et al. Standard reporting requirements for biological samples in metabolomics experiments: Mammalian/in vivo experiments. Metabolomics 2007, 3, 179–188. [Google Scholar] [CrossRef]
- Sangster, T.; Major, H.; Plumb, R.; Wilson, A.J.; Wilson, I.D. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst 2006, 131, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Yanes, O.; Tautenhahn, R.; Patti, G.J.; Siuzdak, G. Expanding Coverage of the Metabolome for Global Metabolite Profiling. Anal. Chem. 2011, 83, 2152–2161. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Koek, M.M.; Jellema, R.H.; van der Greef, J.; Tas, A.C.; Hankemeier, T. Quantitative metabolomics based on gas chromatography mass spectrometry: Status and perspectives. Metabolomics 2011, 7, 307–328. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The Human Urine Metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- Fancy, S.-A.; Rumpel, K. GC-MS-based metabolomics. Methods Pharmacol. Toxicol. 2008, 317–340. [Google Scholar]
- Halket, J.M.; Waterman, D.; Przyborowska, A.M.; Patel, R.K.P.; Fraser, P.D.; Bramley, P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2004, 56, 219–243. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 2017. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012. In Press. [Google Scholar] [CrossRef]
- DeHaven, C.D.; Evans, A.M.; Dai, H.; Lawton, K.A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2010, 2, 9. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics 2006, 7, 142. [Google Scholar] [CrossRef]
- Peters, K.; Bradbury, J.; Bergmann, S.; Capuccini, M.; Cascante, M.; de Atauri, P.; Ebbels, T.; Foguet, C.; Glen, R.; Gonzalez-Beltran, A.; et al. PhenoMeNal: Processing and analysis of Metabolomics data in the Cloud. bioRxiv 2008. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. In Current Protocols in Bioinformatics; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Metabolomic Data Processing, Analysis, and Interpretation Using MetaboAnalyst. In Current Protocols in Bioinformatics; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Creek, D.J.; Dunn, W.B.; Fiehn, O.; Griffin, J.L.; Hall, R.D.; Lei, Z.; Mistrik, R.; Neumann, S.; Schymanski, E.L.; Sumner, L.W.; et al. Metabolite identification: Are you sure? And how do your peers gauge your confidence? Metabolomics 2014, 10, 350–353. [Google Scholar] [CrossRef]
- Sumner, L.W.; Lei, Z.; Nikolau, B.J.; Saito, K.; Roessner, U.; Trengove, R. Proposed quantitative and alphanumeric metabolite identification metrics. Metabolomics 2014, 10, 1047–1049. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Indentifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef]
- Arnold, M.; Raffler, J.; Pfeufer, A.; Suhre, K.; Kastenmuller, G. SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 2015, 31, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef]
- Frolkis, A.; Knox, C.; Lim, E.; Jewison, T.; Law, V.; Hau, D.D.; Liu, P.; Gautam, B.; Ly, S.; Guo, A.C.; et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010, 38, D480–D487. [Google Scholar] [CrossRef]
- Jewison, T.; Su, Y.; Disfany, F.M.; Liang, Y.; Knox, C.; Maciejewski, A.; Poelzer, J.; Huynh, J.; Zhou, Y.; Arndt, D.; et al. SMPDB 2.0: Big Improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014, 42, D478–D484. [Google Scholar] [CrossRef]
- Brunk, E.; Sahoo, S.; Zielinski, D.C.; Altunkaya, A.; Dräger, A.; Mih, N.; Gatto, F.; Nilsson, A.; Preciat Gonzalez, G.A.; Aurich, M.K.; et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018, 36, 272. [Google Scholar] [CrossRef]
- Menezes, L.F.; Germino, G.G. Systems biology of polycystic kidney disease: A critical review. WIREs Syst. Biol. Med. 2015, 7, 39–52. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Qiu, S.; Wang, X. Metabolomics insights into pathophysiological mechanisms of nephrology. Int. Urol. Nephrol. 2014, 46, 1025–1030. [Google Scholar] [CrossRef]
- Rhee, E.P. Metabolomics and Renal Disease. Curr. Opin. Nephrol. Hypertens. 2015, 24, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Kalim, S.; Rhee, E.P. An overview of renal metabolomics. Kidney Int. 2017, 91, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Hocher, B.; Adamski, J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Breit, M.; Weinberger, K.M. Metabolic biomarkers for chronic kidney disease. Arch. Biochem. Biophys. 2016, 589, 62–80. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Aronov, P.; Zakharkin, S.O.; Anderson, D.; Perroud, B.; Thompson, I.M.; Weiss, R.H. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol. Cell. Proteomics 2008, 8, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Abbiss, H.; Maker, G.; Gummer, J.; Sharman, M.J.; Phillips, J.K.; Boyce, M.; Trengove, R.D. The development of a non-targeted metabolomics method to investigate urine in a rat model of polycystic kidney disease. Nephrology 2012, 17, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Boelaert, J.; t’Kindt, R.; Schepers, E.; Jorge, L.; Glorieux, G.; Neirynck, N.; Lynen, F.; Sandra, P.; Vanholder, R.; Sandra, K. State-of-the-art non-targeted metabolomics in the study of chronic kidney disease. Metabolomics 2014, 10, 425–442. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Lei, P.; Chen, D.-Q.; Feng, Y.-L.; Bai, X. Renal metabolic profiling of early renal injury and renoprotective effects of Poria cocos epidermis using UPLC Q-TOF/HSMS/MSE. J. Pharm. Biomed. Anal. 2013, 81, 202–209. [Google Scholar] [CrossRef]
- Sun, J.; Shannon, M.; Ando, Y.; Schnackenberg, L.K.; Khan, N.A.; Portilla, D.; Beger, R.D. Serum metabolomic profiles from patients with acute kidney injury: A pilot study. J. Chromatogr. B 2012, 893, 107–113. [Google Scholar] [CrossRef]
- Shah, V.O.; Townsend, R.R.; Feldman, H.I.; Pappan, K.L.; Kensicki, E.; Vander Jagt, D.L. Plasma Metabolomic Profiles in Different Stages of CKD. Clin. J. Am. Soc. Nephrol. 2013, 8, 363–370. [Google Scholar] [CrossRef]
- Luck, M.; Bertho, G.; Bateson, M.; Karras, A.; Yartseva, A.; Thervet, E.; Damon, C.; Pallet, N. Rule-Mining for the Early Prediction of Chronic Kidney Disease Based on Metabolomics and Multi-Source Data. PLoS ONE 2016, 11, e0166905. [Google Scholar] [CrossRef] [PubMed]
- Rhee, E.P.; Clish, C.B.; Wenger, J.; Roy, J.; Elmariah, S.; Pierce, K.A.; Bullock, K.; Anderson, A.H.; Gerszten, R.E.; Feldman, H.I. Metabolomics of Chronic Kidney Disease Progression: A Case-Control Analysis in the Chronic Renal Insufficiency Cohort Study. Am. J. Nephrol. 2016, 43, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Sekula, P.; Goek, O.-N.; Quaye, L.; Barrios, C.; Levey, A.S.; Römisch-Margl, W.; Menni, C.; Yet, I.; Gieger, C.; Inker, L.A.; et al. A Metabolome-Wide Association Study of Kidney Function and Disease in the General Population. J. Am. Soc. Nephrol. 2016, 27, 1175–1188. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Metabolomics and Stages of Chronic Kidney Disease. In Biomarkers in Kidney Disease; Patel, V.B., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–14. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Cheng, X.-L.; Wei, F.; Bai, X.; Tan, X.-J.; Lin, R.-C.; Mei, Q. Intrarenal Metabolomic Investigation of Chronic Kidney Disease and its TGF-β1 Mechanism in Induced-adenine Rats using UPLC Q-TOF/HSMS/MSE. J. Proteome Res. 2013, 12, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Atzori, L.; Mussap, M.; Noto, A.; Barberini, L.; Puddu, M.; Coni, E.; Murgia, F.; Lussu, M.; Fanos, V. Clinical metabolomics and urinary NGAL for the early prediction of chronic kidney disease in healthy adults born ELBW. J. Matern.-Fetal Neonatal Med. 2011, 24, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Nkuipou-Kenfack, E.; Duranton, F.; Gayrard, N.; Argilés, À.; Lundin, U.; Weinberger, K.M.; Dakna, M.; Delles, C.; Mullen, W.; Husi, H.; et al. Assessment of Metabolomic and Proteomic Biomarkers in Detection and Prognosis of Progression of Renal Function in Chronic Kidney Disease. PLoS ONE 2014, 9, e96955. [Google Scholar] [CrossRef]
- Mutsaers, H.A.M.; Engelke, U.F.H.; Wilmer, M.J.G.; Wetzels, J.F.M.; Wevers, R.A.; van den Heuvel, L.P.; Hoenderop, J.G.; Masereeuw, R. Optimized Metabolomic Approach to Identify Uremic Solutes in Plasma of Stage 3–4 Chronic Kidney Disease Patients. PLoS ONE 2013, 8, e71199. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Wei, F.; Vaziri, N.D.; Cheng, X.-L.; Bai, X.; Lin, R.-C.; Zhao, Y.-Y. Metabolomics insights into chronic kidney disease and modulatory effect of rhubarb against tubulointerstitial fibrosis. Sci. Rep. 2015, 5, 14472. [Google Scholar] [CrossRef]
- Qi, S.; Ouyang, X.; Wang, L.; Peng, W.; Wen, J.; Dai, Y. A Pilot Metabolic Profiling Study in Serum of Patients with Chronic Kidney Disease Based on 1H-NMR-Spectroscopy. Clin. Transl. Sci. 2012, 5, 379–385. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Feng, Y.-L.; Bai, X.; Tan, X.-J.; Lin, R.-C.; Mei, Q. Ultra Performance Liquid Chromatography-Based Metabonomic Study of Therapeutic Effect of the Surface Layer of Poria cocos on Adenine-Induced Chronic Kidney Disease Provides New Insight into Anti-Fibrosis Mechanism. PLoS ONE 2013, 8, e59617. [Google Scholar] [CrossRef]
- Goek, O.-N.; Prehn, C.; Sekula, P.; Römisch-Margl, W.; Döring, A.; Gieger, C.; Heier, M.; Koenig, W.; Wang-Sattler, R.; Illig, T.; et al. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol. Dial. Transplant. 2013, 28, 2131–2138. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.F.; Wang, S.; Stothers, C.; Avance, J.; Denson, D.; Harris, R.; Voziyan, P. Alterations of urinary metabolite profile in model diabetic nephropathy. Biochem. Biophys. Res. Commun. 2015, 456, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Karl, B.; Mathew, A.V.; Gangoiti, J.A.; Wassel, C.L.; Saito, R.; Pu, M.; Sharma, S.; You, Y.-H.; Wang, L.; et al. Metabolomics Reveals Signature of Mitochondrial Dysfunction in Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2013, 24, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhang, H.; Zhao, T.; Zhang, X.; Lu, J.; Yin, T.; Liang, Q.; Wang, Y.; Luo, G.; Lan, H.; et al. Intrarenal metabolomics reveals the association of local organic toxins with the progression of diabetic kidney disease. J. Pharm. Biomed. Anal. 2012, 60, 32–43. [Google Scholar] [CrossRef]
- You, Y.-H.; Quach, T.; Saito, R.; Pham, J.; Sharma, K. Metabolomics Reveals a Key Role for Fumarate in Mediating the Effects of NADPH Oxidase 4 in Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, V.-P.; Tynkkynen, T.; Soininen, P.; Forsblom, C.; Peltola, T.; Kangas, A.J.; Groop, P.-H.; Ala-Korpela, M. Sphingomyelin is associated with kidney disease in type 1 diabetes (The FinnDiane Study). Metabolomics 2012, 8, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, V.-P.; Soininen, P.; Forsblom, C.; Parkkonen, M.; Ingman, P.; Kaski, K.; Groop, P.-H.; FinnDiane Study, G.; Ala-Korpela, M. 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Mol. Syst. Biol. 2008, 4, 167. [Google Scholar] [CrossRef]
- Makinen, V.P.; Soininen, P.; Kangas, A.J.; Forsblom, C.; Tolonen, N.; Thorn, L.M.; Viikari, J.; Raitakari, O.T.; Savolainen, M.; Groop, P.H.; et al. Triglyceride-cholesterol imbalance across lipoprotein subclasses predicts diabetic kidney disease and mortality in type 1 diabetes: The FinnDiane Study. J. Intern. Med. 2013, 273, 383–395. [Google Scholar] [CrossRef]
- Barrios, C.; Zierer, J.; Würtz, P.; Haller, T.; Metspalu, A.; Gieger, C.; Thorand, B.; Meisinger, C.; Waldenberger, M.; Raitakari, O.; et al. Circulating metabolic biomarkers of renal function in diabetic and non-diabetic populations. Sci. Rep. 2018, 8, 15249. [Google Scholar] [CrossRef]
- Kind, T.; Tolstikov, V.; Fiehn, O.; Weiss, R.H. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal. Biochem. 2007, 363, 185–195. [Google Scholar] [CrossRef]
- Kim, K.; Taylor, S.L.; Ganti, S.; Guo, L.; Osier, M.V.; Weiss, R.H. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. Omics 2011, 15, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Serkova, N.; Florian Fuller, T.; Klawitter, J.; Freise, C.E.; Niemann, C.U. 1H-NMR–based metabolic signatures of mild and severe ischemia/reperfusion injury in rat kidney transplants. Kidney Int. 2005, 67, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Stenlund, H.; Madsen, R.; Vivi, A.; Calderisi, M.; Lundstedt, T.; Tassini, M.; Carmellini, M.; Trygg, J. Monitoring kidney-transplant patients using metabolomics and dynamic modeling. Chemom. Intell. Lab. Syst. 2009, 98, 45–50. [Google Scholar] [CrossRef]
- Suhre, K.; Schwartz, J.E.; Sharma, V.K.; Chen, Q.; Lee, J.R.; Muthukumar, T.; Dadhania, D.M.; Ding, R.; Ikle, D.N.; Bridges, N.D.; et al. Urine Metabolite Profiles Predictive of Human Kidney Allograft Status. J. Am. Soc. Nephrol. 2016, 27, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, W.; Li, R.; Wang, M.; Chen, C.; Zeng, R.; Deng, Y. Systematic variations associated with renal disease uncovered by parallel metabolomics of urine and serum. BMC Syst. Biol. 2012, 6, S14. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; Ganti, S.; Bukanov, N.O.; Chapman, A.; Fiehn, O.; Osier, M.; Kim, K.; Weiss, R.H. A metabolomics approach using juvenile cystic mice to identify urinary biomarkers and altered pathways in polycystic kidney disease. Am. J. Physiol. Renal. Physiol. 2010, 298, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Toyohara, T.; Suzuki, T.; Akiyama, Y.; Yoshihara, D.; Takeuchi, Y.; Mishima, E.; Kikuchi, K.; Suzuki, C.; Tanemoto, M.; Ito, S.; et al. Metabolomic profiling of the autosomal dominant polycystic kidney disease rat model. Clin. Exp. Nephrol. 2011, 15, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Gronwald, W.; Klein, M.S.; Zeltner, R.; Schulze, B.D.; Reinhold, S.W.; Deutschmann, M.; Immervoll, A.K.; Boger, C.A.; Banas, B.; Eckardt, K.U.; et al. Detection of autosomal dominant polycystic kidney disease by NMR spectroscopic fingerprinting of urine. Kidney Int. 2011, 79, 1244–1253. [Google Scholar] [CrossRef]
- Hwang, V.J.; Kim, J.; Rand, A.; Yang, C.; Sturdivant, S.; Hammock, B.; Bell, P.D.; Guay-Woodford, L.M.; Weiss, R.H. The cpk model of recessive PKD shows glutamine dependence associated with the production of the oncometabolite 2-hydroxyglutarate. Am. J. Physiol. Renal. Physiol. 2015, 309, F492. [Google Scholar] [CrossRef]
- Tolun, A.A.; Zhang, H.; Il’yasova, D.; Sztáray, J.; Young, S.P.; Millington, D.S. Allantoin in human urine quantified by ultra-performance liquid chromatography–tandem mass spectrometry. Anal. Biochem. 2010, 402, 191–193. [Google Scholar] [CrossRef]
- Small, D.M.; Coombes, J.S.; Bennett, N.; Johnson, D.W.; Gobe, G.C. Oxidative stress, anti-oxidant therapies and chronic kidney disease. Nephrology 2012, 17, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Heyes, M.P.; Saito, K.; Crowley, J.S.; Davis, L.E.; Demitrack, M.A.; Der, M.; Dilling, L.A.; Elia, J.; Kruesi, M.J.; Lackner, A.; et al. Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain 1992, 115 Pt 5, 1249–1273. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, K.; Domaniewski, T.; Mysliwiec, M.; Pawlak, D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis 2009, 204, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Plasma kynurenic acid/tryptophan ratio: A sensitive and reliable biomarker for the assessment of renal function. Renal Failure 2013, 35, 648–653. [Google Scholar] [CrossRef] [PubMed]
| Stage | GFR or eGFR (mL/min/1.73 m2) |
|---|---|
| 1 | ≥ 90 |
| 2 | 60–89 |
| 3 | 30–59 |
| 3a | 45–59 |
| 3b | 30–44 |
| 4 | 15–29 |
| 5 | <15 |
| Kidney Disease | Reference | Model | n | Sample Type | Analytical Platform |
|---|---|---|---|---|---|
| Acute kidney injury | Sun, 2012 [123] | Human | 17 | Serum | LC-MS |
| Chronic kidney disease | Shah, 2013 [124] | Human | 10 | Plasma | GC-MS, LC-MS |
| Zhao, 2013 [122] | Rat | 8 | Kidney tissue | LC-MS | |
| Boelaert, 2014 [121] | Human | 20 | Serum | GC-MS, LC-MS | |
| Luck, 2016 [125] | Human | 110 | Urine | NMR | |
| Rhee, 2016 [126] | Human | 200 | Plasma | LC-MS | |
| Sekula, 2016 [127] | Human | 991 | Serum | GC-MS, LC-MS | |
| Kobayashi, 2015 [128] | Human | 69 | Plasma | LC-MS | |
| Zhao, 2013 [129] | Rat | 12 | Kidney tissue | LC-MS | |
| Atzori, 2011 [130] | Human | 13 | Urine | NMR | |
| Nkuipou-Kenfack, 2014 [131] | Human | 10 | Plasma, urine | LC-MS | |
| Mutsaers, 2013 [132] | Human | ≥4 | Plasma | NMR | |
| Zhang, 2015 [133] | Rat | 8 | Urine | LC-MS | |
| Qi, 2012 [134] | Human | 20 | Serum | NMR | |
| Zhao, 2013 [135] | Rat | 8 | Serum | LC-MS | |
| Goek, 2013 [136] | Human | 87 | Serum | LC-MS, FIA-MS | |
| Diabetic nephropathy | Van der Kloet, 2012 [15] | Human | 26 | Urine | GC-MS, LC-MS |
| Stec, 2015 [137] | Mouse | 11 | Urine | NMR | |
| Sharma, 2013 [138] | Human | 12 | Plasma, urine | GC-MS | |
| Zhao, 2012 [139] | Rat | 12 | Kidney tissue | GC-MS, LC-MS | |
| You, 2015 [140] | Mouse | 11 | Urine | GC-MS | |
| Makinen, 2012 [141] | Human | 86 | Serum | NMR | |
| Makinen, 2008 [142] | Human | 137 | Serum | NMR | |
| Makinen, 2013 [143] | Human | 63 | Serum | NMR | |
| Barrios, 2018 [144] | Human | 926 | Serum | NMR | |
| Kidney cancer | Kind, 2007 [145] | Human | 6 | Urine | GC-MS, LC-MS |
| Kim, 2011 [146] | Human | 11 | Urine | LC-MS | |
| Kidney transplantation | Serkova, 2005 [147] | Rat | 6 | Kidney tissue, whole blood | NMR |
| Stenlund, 2009 [148] | Human | 19 | Urine | NMR | |
| Suhre, 2016 [149] | Human | 241 | Urine, kidney tissue | GC-MS, LC-MS | |
| Membranous nephropathy | Gao, 2012 [150] | Human | 14 | Serum, urine | GC-MS |
| Polycystic kidney disease | Taylor, 2010 [151] | Mouse | 9 | Urine | GC-MS |
| Toyohara, 2011 [152] | Mouse | 5 | Plasma | CE-MS | |
| Abbiss, 2012 [120] | Rat | 6 | Urine | GC-MS | |
| Gronwald, 2011 [153] | Human | 10 | Urine | NMR | |
| Hwang, 2015 [154] | Mouse, human | 2 | Cells, plasma, tissue, urine | GC-MS, LC-MS |
| Metabolites | Sub Class | Kidney Disease | ||||||
|---|---|---|---|---|---|---|---|---|
| CKD | DN | PKD | AKI | T | MN | KC | ||
| allantoin | imidazoles | [122,129] | [139] | [120,152] | [147] | |||
| quinolinic acid | pyridinecarboxylic acids & derivatives | [121] | [149] | [150] | [146] | |||
| 2-hydroxyglutarate | short-chain hydroxy acids & derivatives | [15,139] | [154] | [150] | ||||
| 2-oxoglutaric acid | gamma-keto acids & derivatives | [137] | [120,152] | [146] | ||||
| aconitic acid | tricarboxylic acids & derivatives | [138] | [152] | [150] | ||||
| ADMA | amino acids, peptides & analogues | [131,136] | [152] | [123] | ||||
| carnitine | quaternary ammonium salts | [124,125] | [15] | [152] | ||||
| citrate | tricarboxylic acids & derivatives | [124,125] | [138,139] | [152,153,154] | ||||
| creatinine | amino acids, peptides & analogues | [121,125,127,132,133,135] | [152] | [123] | ||||
| hippuric acid | benzoic acids & derivatives | [122,128,132] | [15,137,139] | [120,152] | ||||
| kynurenic acid | quinoline carboxylic acids | [121] | [15] | [149] | ||||
| LysoPC (16:1) | Glycerophosphocholines | [135] | [139] | [123] | ||||
| malic acid | beta hydroxy acids & derivatives | [124] | [139] | [154] | ||||
| methionine | amino acids, peptides & analogues | [121,126] | [139] | [123] | ||||
| myo-inositol | alcohols & polyols | [127,132,134] | [139] | [152] | [149] | |||
| threonic acid | carbohydrates & conjugates | [124] | [139] | [150] | ||||
| trimethylamine oxide | aminoxides | [132] | [152] | [149] | ||||
| tryptophan | indolyl carboxylic acids & derivatives | [121,133,135] | [15] | [123] | ||||
| uric acid | purines & purine derivatives | [121,122,126] | [139] | [120] | ||||
| valine | amino acids, peptides & analogues | [130,135] | [144] | [149] | ||||
| 2-furoylglycine | amino acids, peptides & analogues | [121] | [146] | |||||
| 3-indoxyl sulfate | arylsulfates | [137] | [152] | |||||
| 3-methylhistidine | amino acids, peptides & analogues | [132,133] | [152] | |||||
| 4-pyridoxic acid | pyridinecarboxylic acids & derivatives | [121] | [139] | |||||
| 4-hydroxymandelate | 1-hydroxy-2-unsubstituted benzenoids | [126] | [149] | |||||
| acetylcarnitine | fatty acid esters | [152] | [123] | |||||
| alanine | amino acids, peptides & analogues | [121,134] | [139,144] | |||||
| arachidonic acid | fatty acids & conjugates | [122,124,129] | [139] | |||||
| arginine | amino acids, peptides & analogues | [122,126] | [123] | |||||
| citrulline | amino acids, peptides & analogues | [124,131] | [152] | |||||
| cytosine | pyrimidines & pyrimidine derivatives | [126] | [150] | |||||
| fructose | carbohydrates & conjugates | [139] | [146] | |||||
| fumaric acid | dicarboxylic acids & derivatives | [140] | [154] | |||||
| gentisate | benzoic acids & derivatives | [149] | [146] | |||||
| glucose | carbohydrates & conjugates | [134] | [139] | |||||
| glutamic acid | amino acids, peptides & analogues | [15] | [154] | |||||
| glutamine | amino acids, peptides & analogues | [121,130] | [154] | |||||
| glycine | amino acids, peptides & analogues | [125,130,134] | [152] | |||||
| homocysteine | amino acids, peptides & analogues | [133] | [123] | |||||
| hypoxanthine | purines & purine derivatives | [122] | [153] | |||||
| indole acetic acid | indolyl carboxylic acids & derivatives | [121] | [15] | |||||
| indoxyl sulfate | arylsulfates | [121,122,128,129] | [139] | |||||
| lactic acid | alpha hydroxy acids & derivatives | [134] | [139] | |||||
| leucine | amino acids, peptides & analogues | [131] | [149] | |||||
| lysine | amino acids, peptides & analogues | [121] | [139] | |||||
| Lyso PC (16:0) | Glycerophosphocholines | [135] | [123] | |||||
| Lyso PC (18:0) | Glycerophosphocholines | [135] | [123] | |||||
| Lyso PC (18:2) | Glycerophosphocholines | [135] | [123] | |||||
| Lyso PC (20:4) | *§ | [135] | [139] | |||||
| N,N-dimethylglycine | amino acids, peptides & analogues | [132] | [152] | |||||
| ornithine | amino acids, peptides & analogues | [124] | [139] | |||||
| pantothenic acid | polyols | [121] | [152] | |||||
| phenylacetylglycine | amino acids, peptides & analogues | [129] | [137] | |||||
| phenylalanine | amino acids, peptides & analogues | [126,133] | [123] | |||||
| phosphate | non-metal phosphates | [124] | [139] | |||||
| pipecolate | amino acids, peptides & analogues | [152] | [149] | |||||
| proline | amino acids, peptides & analogues | [121,131] | [149] | |||||
| pseudouridine | nucleoside & nucleotide analogues*# | [121,127,132] | [15] | |||||
| pyroglutamic acid | amino acids, peptides & analogues | [139] | [123] | |||||
| taurine | organosulfonic acids & derivatives | [134] | [139] | |||||
| tetracosahexaenoate | fatty acids & conjugates | [122] | [139] | |||||
| threonine | amino acids, peptides & analogues | [126] | [139] | |||||
| trigonelline | alkaloids & derivatives*¥ | [125,132] | [153] | |||||
| urea | ureas | [121] | [139] | |||||
| xanthosine | purine nucleosides*# | [121] | [149] | |||||
| xylitol | carbohydrates & conjugates | [149] | [150] | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbiss, H.; Maker, G.L.; Trengove, R.D. Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases. Metabolites 2019, 9, 34. https://doi.org/10.3390/metabo9020034
Abbiss H, Maker GL, Trengove RD. Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases. Metabolites. 2019; 9(2):34. https://doi.org/10.3390/metabo9020034
Chicago/Turabian StyleAbbiss, Hayley, Garth L. Maker, and Robert D. Trengove. 2019. "Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases" Metabolites 9, no. 2: 34. https://doi.org/10.3390/metabo9020034
APA StyleAbbiss, H., Maker, G. L., & Trengove, R. D. (2019). Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases. Metabolites, 9(2), 34. https://doi.org/10.3390/metabo9020034






