Furanoterpene Diversity and Variability in the Marine Sponge Spongia officinalis, from Untargeted LC–MS/MS Metabolomic Profiling to Furanolactam Derivatives
Abstract
:1. Introduction
2. Results and Discussion
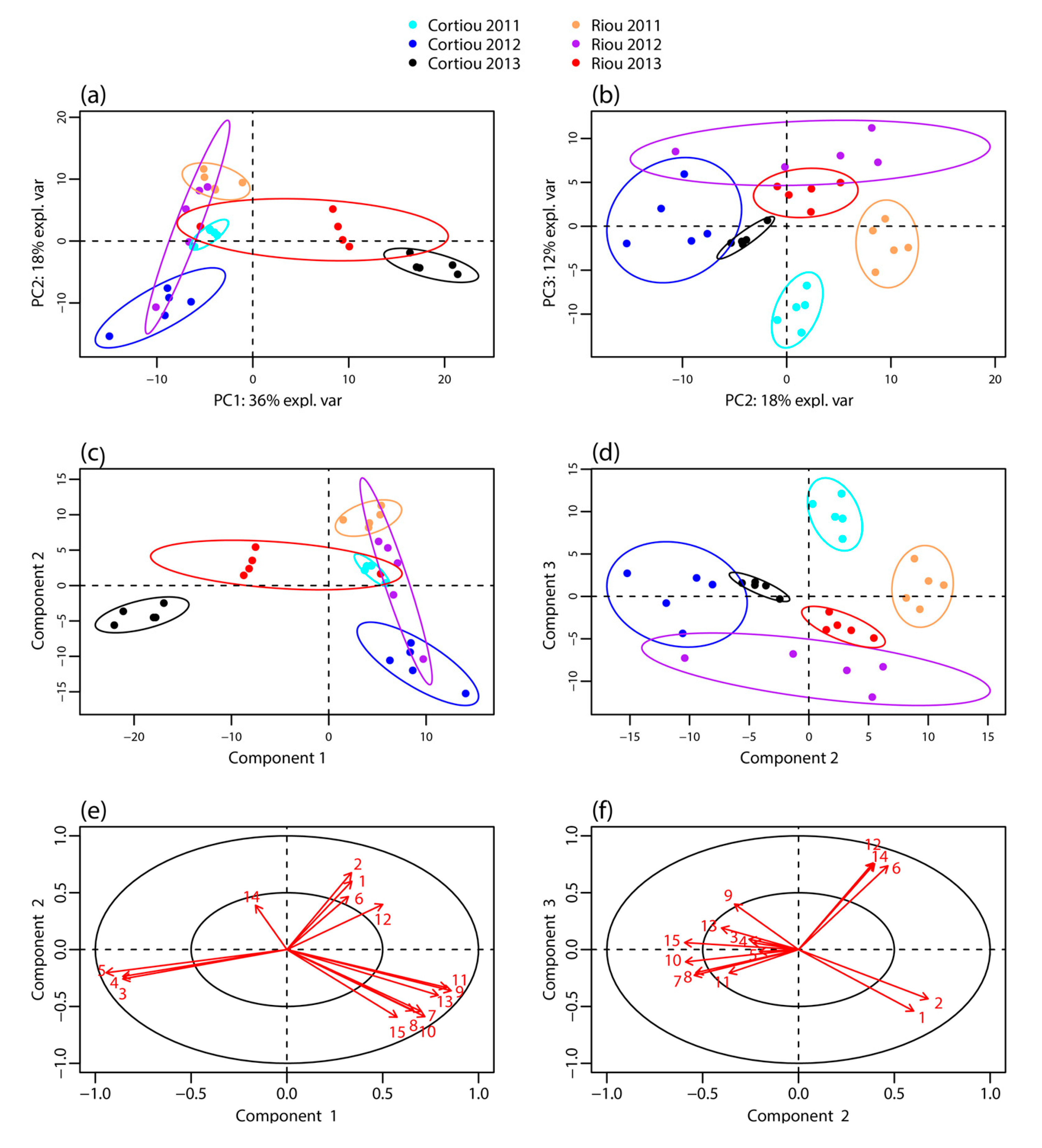
2.1. Untargeted Metabolomic Profiles of S. officinalis
2.2. Signals Involved in the Metabolomic Clustering
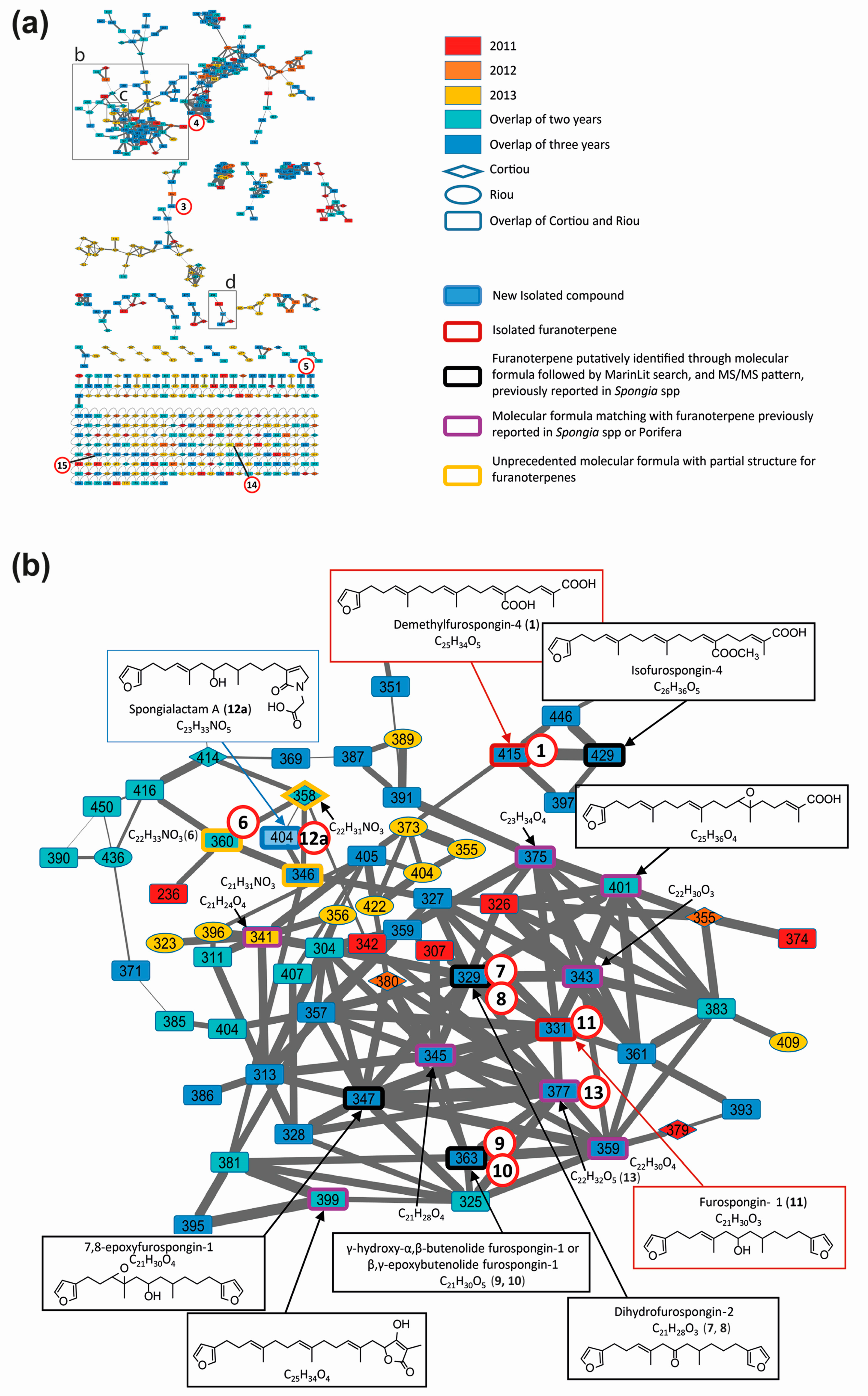
2.3. Compound Annotation through LC–MS/MS and Molecular Networking
- −
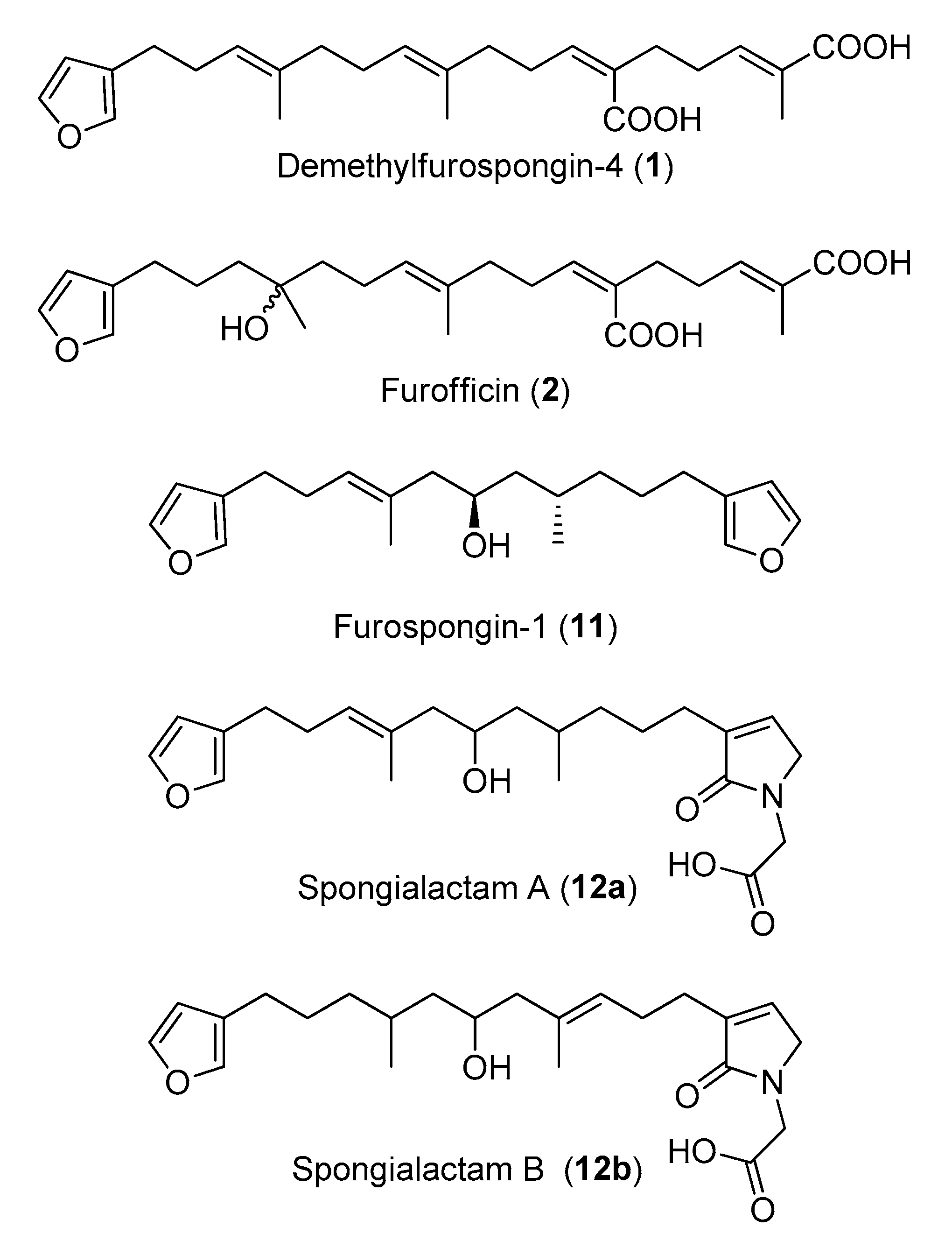
- Compound 1 was assigned to the linear furanosesterterpene demethylfurospongin-4, previously isolated from S. officinalis [12]. Its identification was confirmed by NMR on the isolated compound (Figures S5 and S6, Table S4). Therefore, the main cluster produced by molecular networking was assigned to the furanoterpene family.
- −
- Compound 2 was not observed in the network due to in-source fragmentation in positive ion mode.
- −
- Compounds 3, 4 and 5 did not appear in the furanoterpene cluster and appeared structurally unrelated. No structure could be proposed for compounds 3 and 4 based on their molecular masses and fragmentation patterns (Figure S7A,B). Compound 5 displayed a molecular formula and MS/MS spectra consistent with the coconut diethanolamide (C11 DEA), a synthetic surfactant considered as marine pollutant (Figure S7C) [22].
- −
- Compound 6 had no match in MarinLit as a furanoterpene molecule based on its assigned molecular formula C22H33NO3 and MS/MS spectrum (Figure S8A).
- −
- Compounds 7 and 8 were assigned to the molecular formula C21H28O3, which matched with two molecules previously isolated from Spongia spp.: furospongenone [23] and dihydrofurospongin-2 [25]. The intense m/z 135 and the m/z 179 product ions observed for compound 7 were consistent with the structure of dihydrofurospongin-2 (Figure S9A). The corresponding node was thus annotated as a dihydrofurospongin-2 type. The MS/MS spectrum of compound 8 displayed a small product ion at m/z 135, together with a species at m/z 149 (Figure S9B). This compound was proposed to contain a dimethyl-allyl backbone, but could not be further identified.
- −
- Compounds 9 and 10 were assigned to the molecular formula C21H30O5 corresponding to a series of furanoterpene isomers isolated from S. officinalis, named butenolide furospongin-1 [24]. These compounds contain a furan moiety and either a γ-hydroxy-α-β-butenolide or a β-γ-epoxy butenolide moiety. The fragmentation patterns of compound 9 in positive and negative ion modes (Figure S10A,B) were compatible with butenolide furospongin-1. The chromatographic peak corresponding to this compound had a bimodal peak shape, suggesting a close elution of two isomers. The fragmentation pattern of compound 10 seemed close to that of compound 9, although much weaker, in particular in positive ion mode (Figure S10C,D). In the literature, no difference in the fragmentation pattern of butenolide furospongin-1 has been reported when a γ-hydroxy-α-β-butenolide is replaced by a β-γ-epoxy butenolide [24], hindering unambiguous identification of compounds 9 and 10.
- −
- Compound 11 was assigned to the molecular formula C21H30O3, which could correspond to different furanoterpernes isolated from species of the Spongiidae family: furospongin-1 [9], tetrahydrofurospongin-2 [25] and furospongenol [23]. The product ions detected in the MS/MS spectrum of the [M + H]+ species of this compound (Figure S11) was compatible with the three structures. NMR analysis of the purified compound permitted to assign it to furospongin-1 (Figures S12 and S13, Table S5).
- −
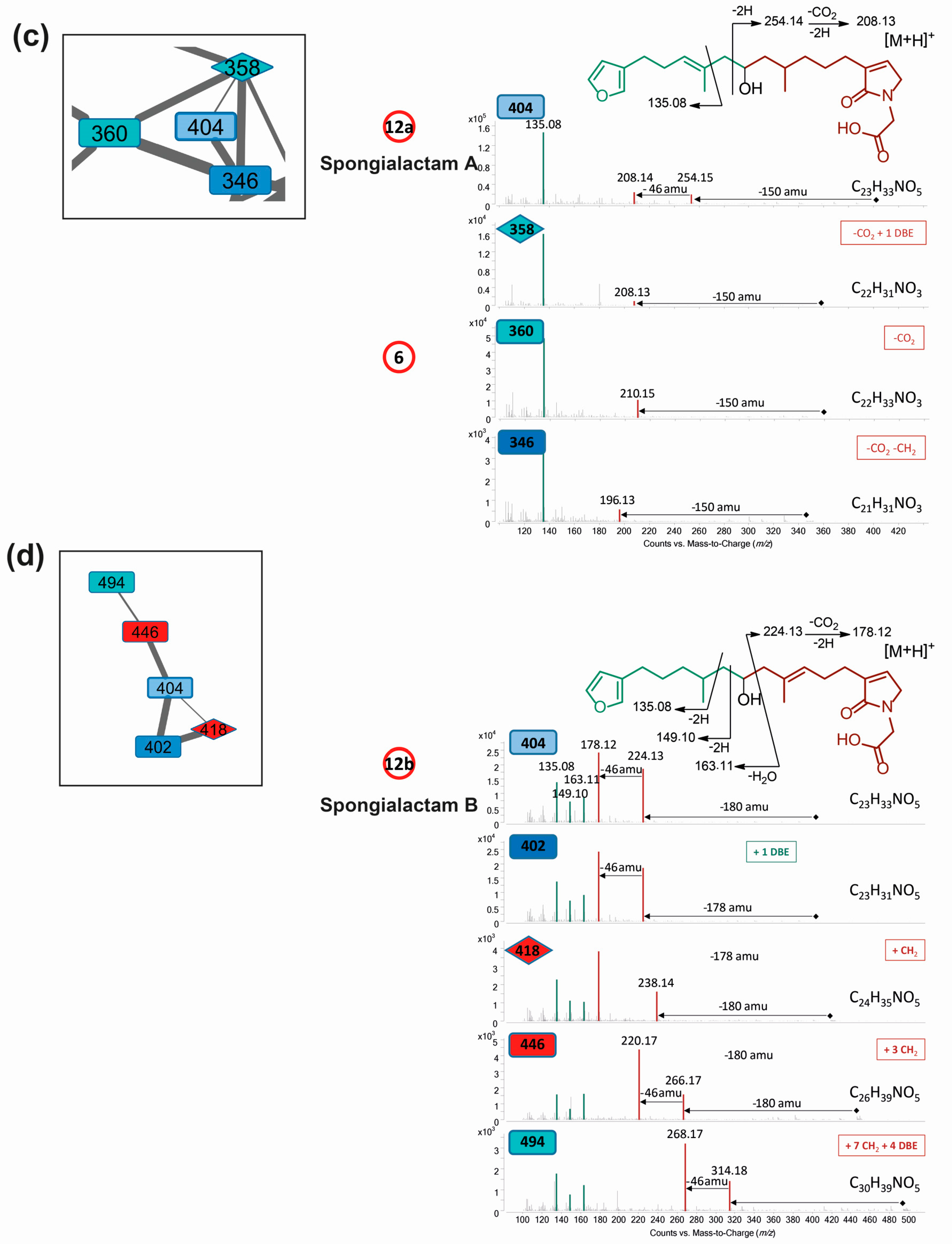
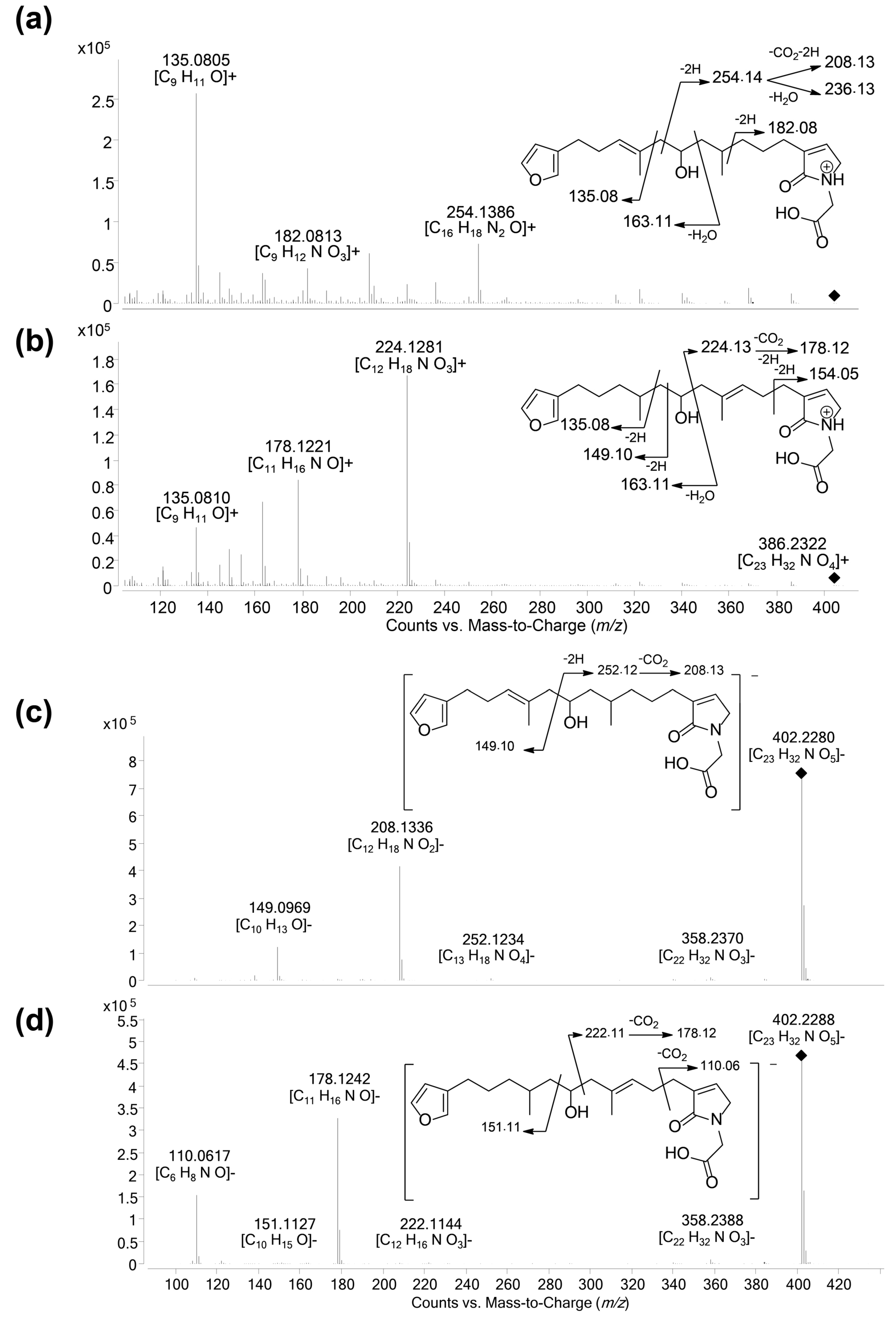
- LC–MS/MS analysis revealed that compound 12 was in fact a mixture of two isomers showing distinct fragmentation patterns, and thus featured into two independent clusters (Figure 4c,d). These compounds were therefore named 12a and 12b. Their molecular formula C23H33NO5 had no match in MarinLit as a furanoterpene molecule.
- −
- The molecular formula C22H32O5 assigned to compound 13 (Table S2, Figure S8B) matched with irciformonins B and J isolated from Ircinia formosana [28,29]. The compound fragmentation pattern was consistent with the structure of irciformonin B.
- −
- Compounds 14 and 15, assigned to the molecular formulas C27H41NO5 and C28H33NO4, respectively, appeared outside the furanoterpene cluster. However, their fragmentation patterns included the diagnostic ion m/z 135 which could imply their annotation as furanoterpenes with unusual product ions (Figure S8C,D).
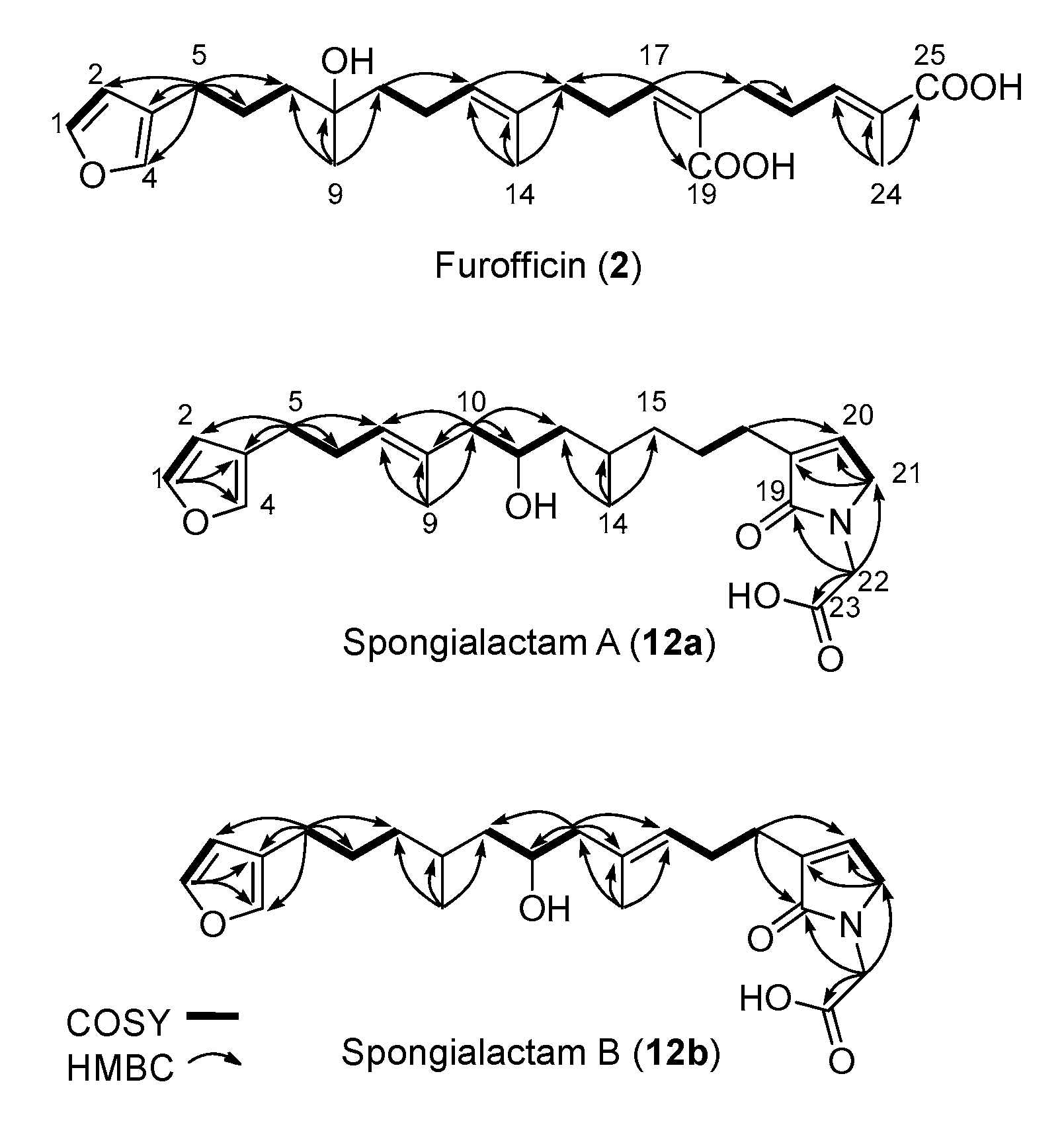
2.4. Structure Elucidation of New Furanoterpene Derivatives
2.5. Unravelling Pyrrolofuranoterpene Derivatives from S. officinalis
2.6. Metabolite Variability and Furanoterpene Signature
3. Materials and Methods
3.1. Materials
3.2. Sponge Extraction
3.3. LC–MS Analyses
3.4. NMR Analysis
3.5. Molecular Networking and Manual Dereplication
LC–MS/MS data were converted into mgf files using MassHunter® software,(Qualitative Analysis B.07.00, Agilent Technologies, Les Ulis, France). Converted data files were subjected to online GNPS workflow (http://gnps.ucsd.edu). Consensus spectra were generated through MS-Cluster with a parent ion mass tolerance of 0.5 Da and a fragment ion mass tolerance of 0.5 Da, with a minimum of 2 spectra. The networks were generated using the following settings: min pair cos: 0.7, minimum matched fragment ion: 6, network topK: 10. Resulting networks were visualized using Cytoscape 3.2.0. The preferred layout was applied. Node colors were mapped based on the source files of MS/MS spectra. The edge thickness attribute was defined to reflect cosine similarity scores, with thicker lines indicating higher similarity. Manual dereplication was performed using the MarinLit database (http://pubs.rsc.org/marinlit).
3.6. Multivariate Data Analysis
3.7. Compound Isolation and Characterization
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus, C. Systema Naturæ per Regna Tria Naturæ, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis; Laurentii Salvii: Stockholm, Sweden, 1759. [Google Scholar]
- Bergquist, P.R.; Hofheinz, W.; Hofheinz, W.; Oesterhelt, G. Sterol composition and the classification of the demospongiae. Biochem. Syst. Ecol. 1980, 8, 423–435. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, S.; Abreu, P.J. Heterocyclic terpenes: Linear furano- and pyrroloterpenoids. Nat. Prod. Rep. 2006, 23, 630–651. [Google Scholar] [CrossRef] [PubMed]
- Manzo, E.; Ciavatta, M.L.; Villani, G.; Varcamonti, M.; Sayem, S.M.; van Soest, R.; Gavagnin, M. Bioactive terpenes from Spongia officinalis. J. Nat. Prod. 2011, 74, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Máximo, P.; Ferreira, L.M.; Branco, P.; Lima, P.; Lourenco, A. The role of Spongia sp. in the discovery of marine lead compounds. Mar. Drugs 2016, 14, E139. [Google Scholar] [CrossRef] [PubMed]
- Migliuolo, A.; Notaro, G.; Piccialli, V.; Sica, D. New tetrahydroxylated sterols from the marine sponge Spongia officinalis. J. Nat. Prod. 1990, 53, 1414–1424. [Google Scholar] [CrossRef]
- Cimino, G.; De Stefano, S.; Minale, L.; Fattorusso, E. Furospongin-1, a new C-21 furanoterpene from the sponges Spongia officinalis and Hippospongia communis. Tetrahedron 1971, 27, 4673–4679. [Google Scholar] [CrossRef]
- Orhan, I.; Sener, B.; Kaiser, M.; Brun, R.; Tasdemir, D. Inhibitory activity of marine sponge-derived natural products against parasitic protozoa. Mar. Drugs 2010, 8, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.-X.; Xu, Z.-J.; Chen, H.-J.; Wu, Y.; You, J. Synthesis and configurations of (−)-furospongin-1 and (+)-dihydrofurospongin-2. Eur. J. Org. Chem. 2016, 2016, 946–957. [Google Scholar] [CrossRef]
- Garrido, L.; Zubía, E.; Ortega, M.J.; Salvá, J. New furanoterpenoids from the sponge Spongia officinalis. J. Nat. Prod. 1997, 60, 794–797. [Google Scholar] [CrossRef]
- Bauvais, C.; Zirah, S.; Piette, L.; Chaspoul, F.; Domart-Coulon, I.; Chapon, V.; Gallice, P.; Rebuffat, S.; Pérez, T.; Bourguet-Kondracki, M.L. Sponging up metals: Bacteria associated with the marine sponge Spongia officinalis. Mar. Environ. Res. 2015, 104, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Stabili, L.; Licciano, M.; Longo, C.; Corriero, G.; Mercurio, M. Evaluation of microbiological accumulation capability of the commercial sponge Spongia officinalis var. adriatica (Schmidt) (Porifera, Demospongiae). Water Res. 2008, 42, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Pérez, T.; Longet, D.; Schembri, T.; Rebouillon, P.; Vacelet, J. Effects of 12 years’ operation of a sewage treatment plant on trace metal occurrence within a Mediterranean commercial sponge (Spongia officinalis, Demospongiae). Mar. Pollut. Bull. 2005, 50, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Berthet, B.; Mouneyrac, C.; Pérez, T.; Amiard-Triquet, C. Metallothionein concentration in sponges (Spongia officinalis) as a biomarker of metal contamination. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2005, 141, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of LC/MS data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.; Riu, J.; Ventura, F.; Boleda, R.; Scheding, R.; Schroder, H.F.; Nistor, C.; Emneus, J.; Eichhorn, P.; Knepper, T.P.; et al. Inter-laboratory comparison of liquid chromatographic techniques and enzyme-linked immunosorbent assay for the determination of surfactants in wastewaters. J. Chromatogr. A 2000, 889, 195–209. [Google Scholar] [CrossRef]
- Kazlauskas, R.; Murphy, P.T.; Quinn, R.J.; Wells, R.J. Two new unsymmetrically oxygenated C21 furanoterpenes from a sponge. Tetrahedron Lett. 1976, 17, 1333–1334. [Google Scholar] [CrossRef]
- Cimino, G.; De Stefano, S.; Minale, L. Oxidized furanoterpenes from the sponge Spongia officinalis. Experientia 1974, 30, 18–20. [Google Scholar] [CrossRef]
- Cimino, G.; De Stefano, S.; Minale, L.; Fattorusso, E. Minor C-21 furanoterpenes from the sponges Spongia officinalis and Hippospongia communis. Tetrahedron 1972, 28, 267–273. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-C.; Lo, K.-L.; Lin, Y.-C.; Khalil, A.T.; Kuo, Y.-H.; Shih, P.-S. Novel linear C22-sesterterpenoids from sponge Ircinia formosana. Tetrahedron Lett. 2006, 47, 4007–4010. [Google Scholar] [CrossRef]
- Shen, Y.-C.; Shih, P.-S.; Lin, Y.-S.; Lin, Y.-C.; Kuo, Y.-H.; Kuo, Y.-C.; Khalil, A.T. Irciformonins E–K, C22-Trinorsesterterpenoids from the Sponge Ircinia formosana. Helv. Chim. Acta 2009, 92, 2101–2110. [Google Scholar] [CrossRef]
- Balansa, W.; Islam, R.; Fontaine, F.; Piggott, A.M.; Zhang, H.; Xiao, X.; Webb, T.I.; Gilbert, D.F.; Lynch, J.W.; Capon, R.J. Sesterterpene glycinyl-lactams: A new class of glycine receptor modulator from Australian marine sponges of the genus Psammocinia. Org. Biomol. Chem. 2013, 11, 4695–4701. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hong, J.; Lee, C.O.; Im, K.S.; Kim, N.D.; Choi, J.S.; Jung, J.H. Cytotoxic pyrrolo- and furanoterpenoids from the sponge Sarcotragus species. J. Nat. Prod. 2002, 65, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Balansa, W.; Islam, R.; Fontaine, F.; Piggott, A.M.; Zhang, H.; Webb, T.I.; Gilbert, D.F.; Lynch, J.W.; Capon, R.J. Ircinialactams: Subunit-selective glycine receptor modulators from Australian sponges of the family Irciniidae. Bioorg. Med. Chem. 2010, 18, 2912–2919. [Google Scholar] [CrossRef] [PubMed]
- Balansa, W.; Islam, R.; Gilbert, D.F.; Fontaine, F.; Xiao, X.; Zhang, H.; Piggott, A.M.; Lynch, J.W.; Capon, R.J. Australian marine sponge alkaloids as a new class of glycine-gated chloride channel receptor modulator. Bioorg. Med. Chem. 2013, 21, 4420–4425. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mansoor, T.A.; Hong, J.; Lee, C.O.; Sim, C.J.; Im, K.S.; Kim, N.D.; Jung, J.H. New cytotoxic sesterterpenoids and norsesterterpenoids from two sponges of the genus Sarcotragus. J. Nat. Prod. 2003, 66, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Rho, J.-R.; Seo, Y.; Lee, H.-S.; Cho, K.W.; Sim, C.J. Sarcotragins A and B, new sesterterpenoid alkaloids from the sponge Sarcotragus sp. Tetrahedron Lett. 2001, 42, 3005–3007. [Google Scholar] [CrossRef]
- Rochfort, S.J.; Atkin, D.; Hobbs, L.; Capon, R.J. Hippospongins A–F: New furanoterpenes from a Southern Australian marine sponge Hippospongia sp. J. Nat. Prod. 1996, 59, 1024–1028. [Google Scholar] [CrossRef]
- Issa, H.H.; Tanaka, J.; Higa, T. New cytotoxic furanosesterterpenes from an Okinawan marine sponge, Ircinia sp. J. Nat. Prod. 2003, 66, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, M.P.; Paul, V.J.; Slattery, M. Biogeographic comparisons of chemical and structural defenses of the Pacific gorgonians Annella mollis and A. reticulata. Mar. Ecol. Prog. Ser. 2000, 207, 263–272. [Google Scholar] [CrossRef]
- Fahey, S.J.; Garson, M.J. Geographic variation of natural products of tropical nudibranch Asteronotus cespitosus. J. Chem. Ecol. 2002, 28, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Jumaryatno, P.; Stapleton, B.L.; Hooper, J.N.; Brecknell, D.J.; Blanchfield, J.T.; Garson, M.J. A comparison of sesquiterpene scaffolds across different populations of the tropical marine sponge Acanthella cavernosa. J. Nat. Prod. 2007, 70, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Noyer, C.; Thomas, O.P.; Becerro, M.A. Patterns of chemical diversity in the Mediterranean sponge Spongia lamella. PLoS ONE 2011, 6, e20844. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Pérez, T.; Ereskovsky, A.V.; Banaigs, B. Secondary Metabolome Variability and Inducible Chemical Defenses in the Mediterranean Sponge Aplysina cavernicola. J. Chem. Ecol. 2016, 42, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.E.; Murphy, P.T.; Berquist, P.R.; Evans, E.A. Environmentally induced variation in diterpene composition of the marine sponge Rhopaloeides odorabile. Biochem. Syst. Ecol. 1987, 15, 595–606. [Google Scholar] [CrossRef]
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, J.A. Diversity and biotechnological potential of microorganisms associated with marine sponges. Appl. Microbiol. Biotechnol. 2014, 98, 7331–7347. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef] [PubMed]
- Indraningrat, A.A.; Smidt, H.; Sipkema, D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 2016, 14, E87. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- González González, A.; López Rodríguez, M.; San Martín Barrientos, A. On the stereochemistry and biogenesis of C21 linear furanoterpenes in Ircinia sp. J. Nat. Prod. 1983, 46, 256–261. [Google Scholar] [CrossRef]
- El Sayed, K.A.; Mayer, A.M.S.; Kelly, M.; Hamann, M.T. The biocatalytic transformation of furan to amide in the bioactive marine natural product palinurin. J. Org. Chem. 1999, 64, 9258–9260. [Google Scholar] [CrossRef]
- Lê Cao, K.A.; Martin, P.G.; Robert-Granie, C.; Besse, P. Sparse canonical methods for biological data integration: Application to a cross-platform study. BMC Bioinf. 2009, 10, 34. [Google Scholar] [CrossRef] [PubMed]
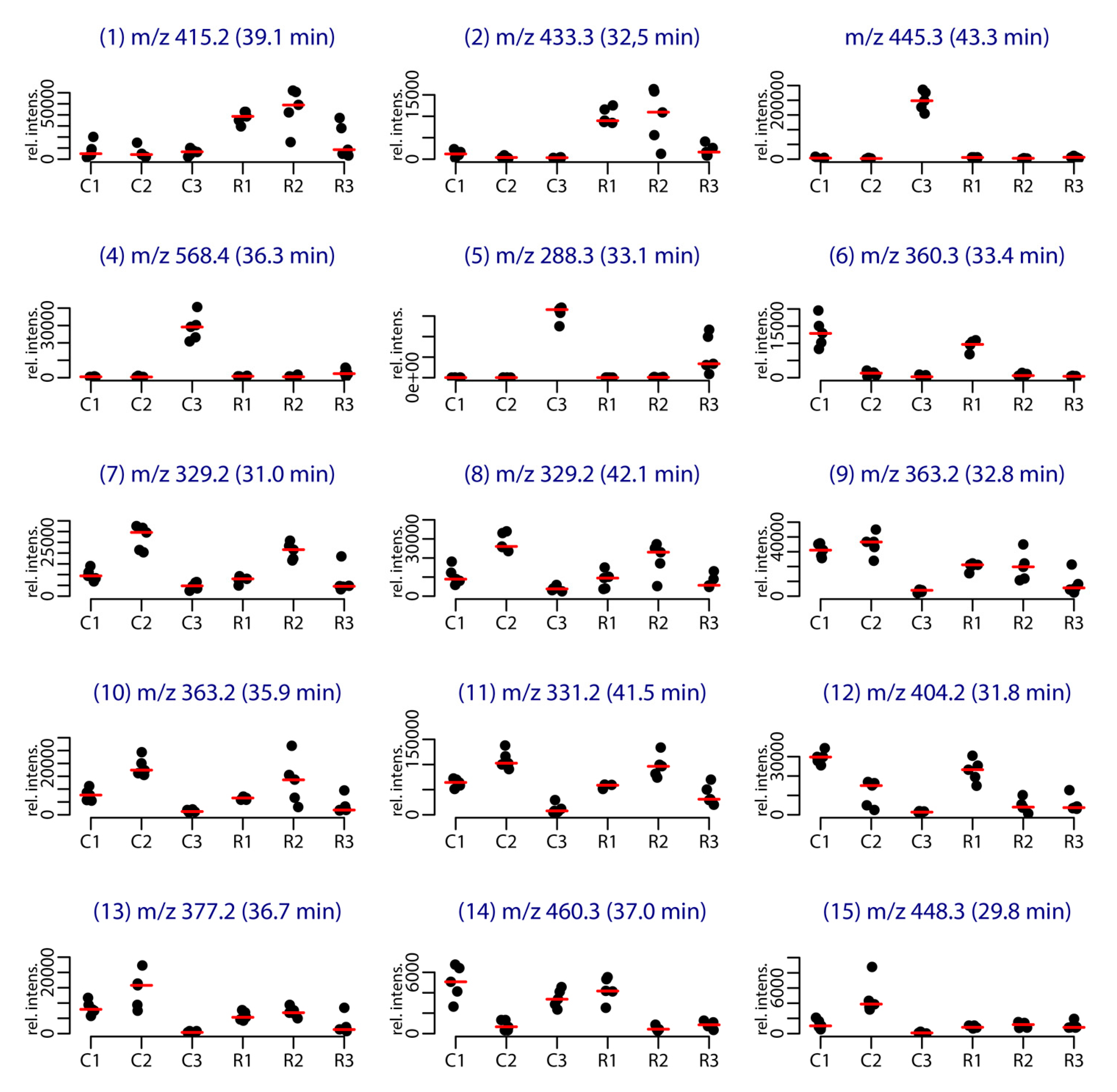

| Compound | Retention Time (min) | m/z | Identification/Annotation | Reference |
|---|---|---|---|---|
| 1 | 39.1 | 415.2 | Demethylfurospongin-4 | [12] |
| 2 | 32.5 | 433.3 | Furofficin, new compound | this study |
| 3 | 43.3 | 445.3 | Unknown | - |
| 4 | 36.3 | 568.4 | Unknown | - |
| 5 | 33.1 | 288.3 | Coconut C11 diethanolamide (a) | [22] |
| 6 | 33.4 | 360.3 | Furanoterpene, C22H33NO3 | - |
| 7 | 31.0 | 329.2 | Dihydrofurospongin-2 (a) | [23] |
| 8 | 42.1 | 329.2 | Furanoterpene, C21H28O3 | - |
| 9 | 32.8 | 363.2 | Isomers of γ-hydroxy-α,β-butenolide or β-γ-epoxy butenolide furospongin-1 (a) | [24] |
| 10 | 35.9 | 363.2 | Furanoterpene, C21H30O5 | - |
| 11 | 41.5 | 331.2 | Furospongin-1 | [9,23,25] |
| 12 | 31.8 | 404.2 | Two new isomers: Spongialactam A (12a) and Spongialactam B (12b) | this study |
| 13 | 36.7 | 377.2 | Irciformonin B (a) | - |
| 14 | 37.0 | 460.3 | Furanoterpene, C27H41NO5 | - |
| 15 | 29.8 | 448.3 | Furanoterpene, C28H33NO4 | - |
| Furofficin (2) | Spongialactam A (12a) | Spongialactam B (12b) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δCa | δH (mult, J in Hz) | COSY | HMBC (1H͢͢-13C) | δCa | δH (mult, J in Hz) | COSY | HMBC (1H͢͢-13C) | δCa | δH (mult, J in Hz) | COSY | HMBC (1H͢͢-13C) | |
| 1 | 143.6 | 7.38 (dd, 1.7 ; 1.7) | 2, 4 | 2, 3, 4 | 143.7 | 7.37 (t, 1.7) | 2,4 | 3, 4 | 143.7 | 7.37 (t, 1.7) | 2, 4 | 2, 3, 4 |
| 2 | 111.6 | 6.30 (dd, 1.7 ; 0.7) | 1, 4 | 1, 3, 4 | 112,0 | 6.31 (bd 0.9) | 1, 4 | 1, 4 | 111.8 | 6.29 (bd, 0.9) | 1, 4 | 1, 4 |
| 3 | 126.0 | - | - | - | 126.05* | - | - | - | 126.4* | - | - | - |
| 4 | 139.8 | 7.27 (dd, 1.5 ; 0.9) | 1, 2, 5 | 2, 3, 1 | 140.0 | 7.25 (quint, 0.8) | 1, 2, 5 | 1, 2 | 139.9 | 7.25 (m) | 1, 2, 5 | 1, 2, 3 |
| 5 | 25.9 | 2.42 (brt, 7.4) | 4, 6 | 2, 3, 4, 6, 7 | 25.8 | 2.46 (t, 7.4) | 4, 6 | 2, 3, 6, 7 | 25.7 | 2.40 (t, 7.4) | 4, 6 | 2, 3, 4, 6, 7 |
| 6 | 25.3 | 1.60 (m) | 5, 7 | 5, 7 | 29.5 | 2.27 (m) | 5, 7 | 7 | 28.5 | 1.59 (m) | 5, 7 | 7 |
| 7a | 41.8 | 1.48 (m) | 6 | 8, 9 | 127.8 | 5.21 (bt, 7.1) | 6 | 9, 10 | 38.5 | 1.21 (m) | 6, 7b | - |
| 7b | 1.32 (m) | 6, 7a, 9 | - | |||||||||
| 8 | 72.8 | - | - | - | 133.9* | - | - | - | 29.9 | 1.71 (m) | 9, 10a | - |
| 9 | 26.5 | 1.13 (s) | - | 7, 8, 10 | 16.5 | 1.61 (brs) | 7 | 7, 8, 10 | 19.5 | 0.89 (d, 6.6) | 7b, 8 | 7, 8, 10 |
| 10a | 42.3 | 1.45 (m) | 11 | 7, 8, 9, 11, 12 | 50.0 | 2.05 (dd, 13.5 ; 6.3) | 10b, 11 | 7, 8, 9, 11 | 45.2 | 1.14 (ddd, 13.8, 9.8, 3.5) | 10b, 11 | - |
| 10b | 2.15 (dd, 13.1 ; 6.8) | 10a, 11 | 7, 8, 9, 11, 12 | 1.37 (ddd, 14.3, 9.5, 3.9) | 10a, 11 | - | ||||||
| 11 | 23.2 | 2.00 (m) | 10, 12 | 10, 12, 13 | 68.1 | 3.76 (m) | 10a, 10b, 12a | - | 68.0 | 3.77 (m) | 10, 12 | 11 |
| 12a | 125.4 | 5.15 (td, 7.1 ; 1.0) | 11, 14 | 11, 14, 15 | 45.4 | 1.13 (ddd, 14.1 ; 10.0 ; 3.4) | 11, 12b | - | 49.7 | 2.06 (dd, 13.3, 6.3) | 11, 12b | 11, 13, 14, 15 |
| 12b | 1.36 (ddd, 13.9 ; 9.9 ; 3.9) | 12a, 13 | - | 2.14 (dd, 13.4, 7.6) | 11, 12a | 10, 11, 13, 14, 15 | ||||||
| 13 | 135.5b | - | - | - | 30.1 | 1.72 (m) | 12b, 14 | - | 134.2b | - | - | - |
| 14 | 15.6 | 1.61 (brs) | 12 | 12, 13, 15 | 19.5 | 0.88 (d, 6.6) | 13 | 12, 13, 15 | 16.1 | 1.63 (brs) | 15, 16 | 12, 13, 15 |
| 15a | 40.4 | 2.05 (m) | 16 | 12, 13, 14, 16, 17 | 38.8 | 1.24 (m) | 15b, 13 | - | 127.2 | 5.21 (m) | 14, 16 | - |
| 15b | 1.34 (m) | 15a | - | |||||||||
| 16 | 28.8 | 2.35 (m) | 15, 17 | 13, 15, 17, 18 | 26.2 | 1.58 (m) | 17 | - | 27.0 | 2.29 (m) | 15 | 13, 15, 17 |
| 17 | 129.5 | 5.31 (t) | 16 | 15, 16, 19, 20 | 27.0 | 2.24 (m) | 16, 20, 21 | 18, 20 | 26.8 | 2.30 (m) | 20, 21 | 16, 18, 19, 20 |
| 18 | 140.2b | - | - | - | 140.1b | - | - | - | 138.2b | - | - | - |
| 19 | 177.5b | - | - | - | 173.8b | - | - | - | 173.8b | - | - | - |
| 20 | 35.3 | 2.31 (m) | 21, 22 | 21 | 137.7 | 6.83 (m) | 17, 21 | 19, 21 | 137.9 | 6.83 (m) | 17, 21 | 19, 21 |
| 21 | 28.9 | 2.29 (m) | 20, 22, 24 | 20 | 53.0 | 4.06 (d, 1.7) | - | 18, 20 | 53.0 | 4.05 (d, 1.6) | 17, 20 | 18, 20 |
| 22 | 139.2 | 6.61 (td, 8.7 ; 1.6) | 20, 21, 24 | 24 | 46.7 | 4.05 (brs) | - | 19, 21, 23 | 46.9 | 4.03 (brs) | - | 19, 21, 23 |
| 23 | 129.9b | - | - | - | 175.5b | - | - | - | 175.9b | - | - | - |
| 24 | 13.0 | 1.81 (brs) | 21, 22 | 22, 23, 25 | ||||||||
| 25 | 174.9b | - | - | - | ||||||||
- a
- 13C assignments supported by HSQC experiment.
- b
- 13C assignments supported by HMBC experiment.
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauvais, C.; Bonneau, N.; Blond, A.; Pérez, T.; Bourguet-Kondracki, M.-L.; Zirah, S. Furanoterpene Diversity and Variability in the Marine Sponge Spongia officinalis, from Untargeted LC–MS/MS Metabolomic Profiling to Furanolactam Derivatives. Metabolites 2017, 7, 27. https://doi.org/10.3390/metabo7020027
Bauvais C, Bonneau N, Blond A, Pérez T, Bourguet-Kondracki M-L, Zirah S. Furanoterpene Diversity and Variability in the Marine Sponge Spongia officinalis, from Untargeted LC–MS/MS Metabolomic Profiling to Furanolactam Derivatives. Metabolites. 2017; 7(2):27. https://doi.org/10.3390/metabo7020027
Chicago/Turabian StyleBauvais, Cléa, Natacha Bonneau, Alain Blond, Thierry Pérez, Marie-Lise Bourguet-Kondracki, and Séverine Zirah. 2017. "Furanoterpene Diversity and Variability in the Marine Sponge Spongia officinalis, from Untargeted LC–MS/MS Metabolomic Profiling to Furanolactam Derivatives" Metabolites 7, no. 2: 27. https://doi.org/10.3390/metabo7020027
APA StyleBauvais, C., Bonneau, N., Blond, A., Pérez, T., Bourguet-Kondracki, M.-L., & Zirah, S. (2017). Furanoterpene Diversity and Variability in the Marine Sponge Spongia officinalis, from Untargeted LC–MS/MS Metabolomic Profiling to Furanolactam Derivatives. Metabolites, 7(2), 27. https://doi.org/10.3390/metabo7020027






