Deciphering the Duality of Clock and Growth Metabolism in a Cell Autonomous System Using NMR Profiling of the Secretome
Abstract
:1. Introduction
2. Results
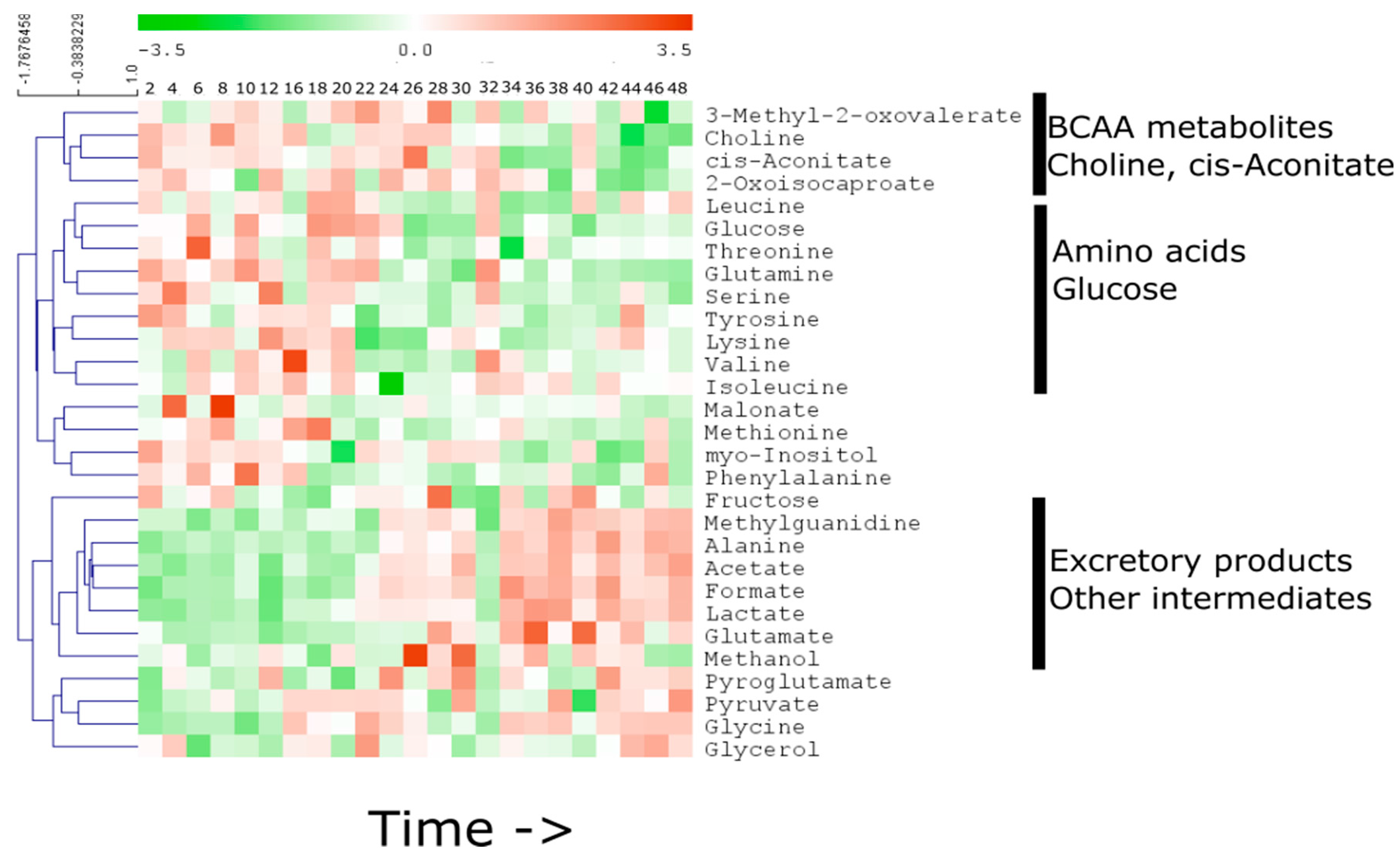
2.1. Temporal Change of the Global Secretome Composition in a Cell-Autonomous Model System
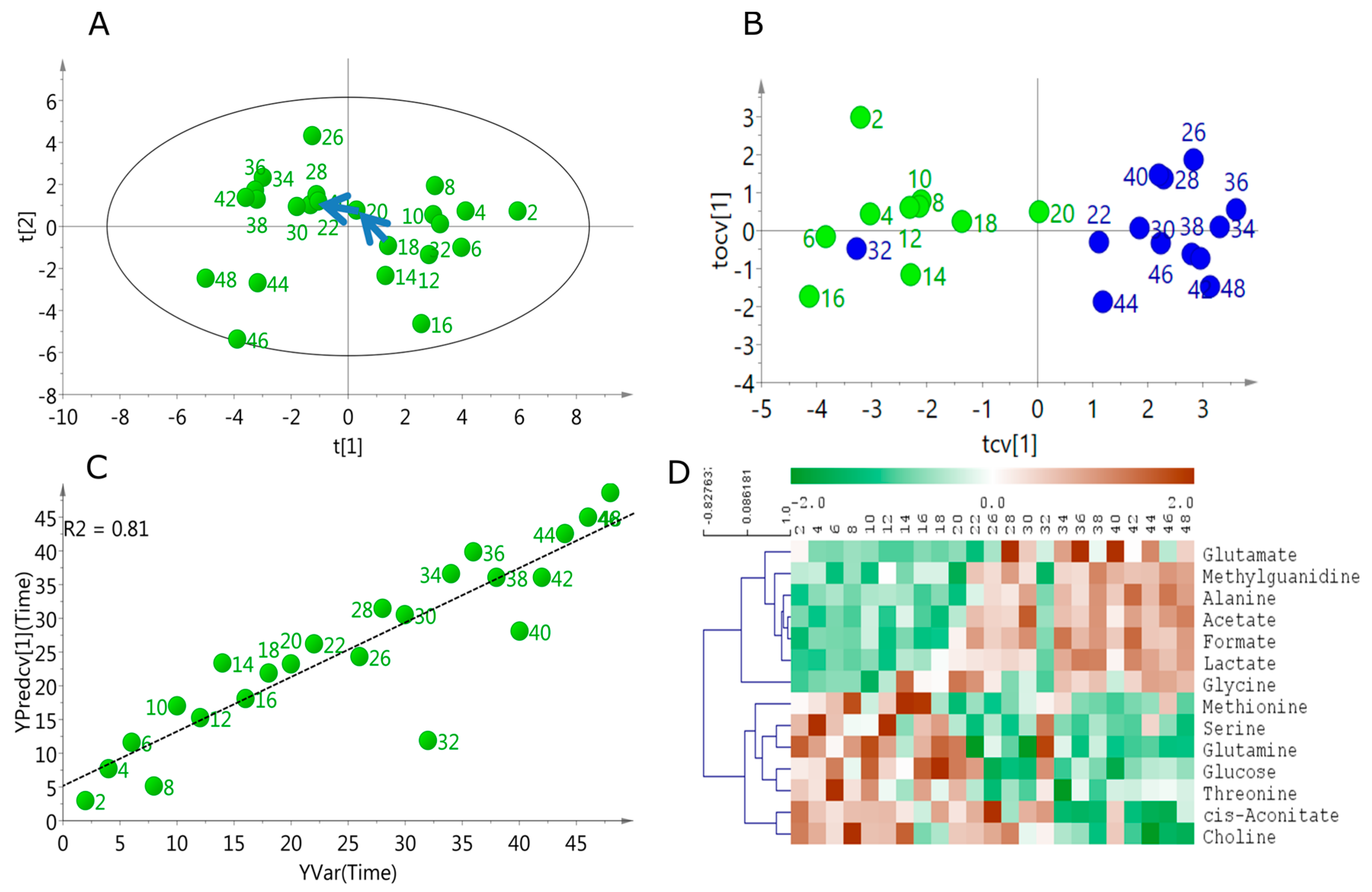
2.2. Linear Temporal Changes in the Secretome
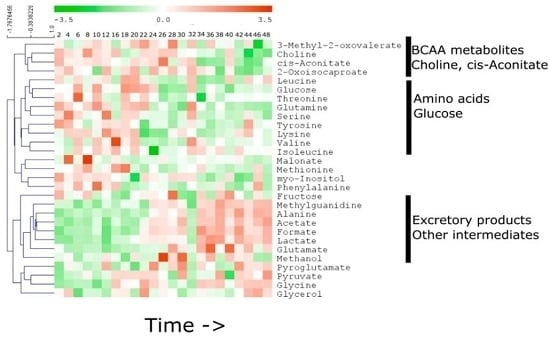
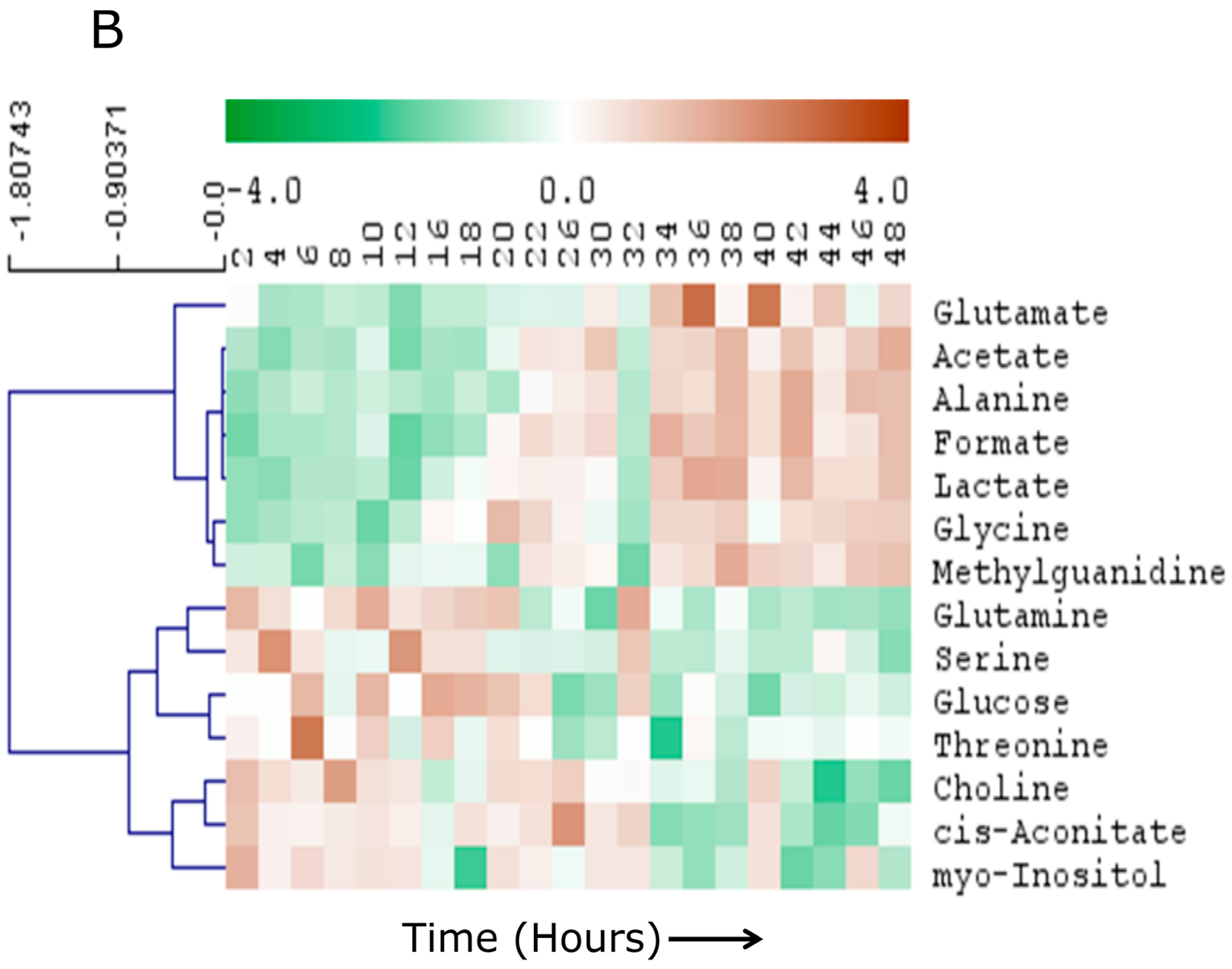
2.3. Specific Temporal Metabolic Changes in the Secretome
3. Discussion
4. Materials and Methods
4.1. U2OS Cell Collection
4.2. Metabolite Extraction from Media
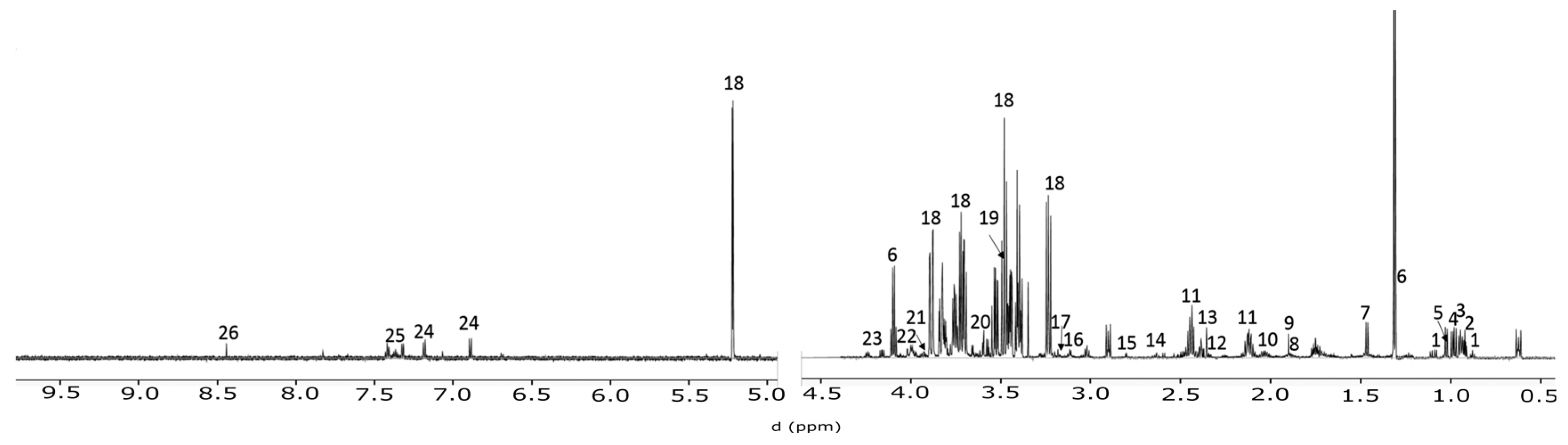
4.3. NMR Spctroscopy
4.4. Spectral Profiling and Data Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Buhr, E.D.; Takahashi, J.S. Molecular Components of the Mammalian Circadian Clock. In Handbook of Experimental Pharmacology 217 Circadian Clocks; Kramer, A., Merrow, M., Eds.; Springer: Berlin, Germany, 2013. [Google Scholar]
- Antunes, L.C.; Levandovski, R.; Dantas, G.; Caumo, W.; Hidalgo, M.P. Obesity and shift work: Chronobiological aspects. Nutr. Res. Rev. 2010, 23, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, D.A.; Gibbs, J.E.; Loudon, A.S.I. Circadian dysfunction in disease. Trends Pharmacol. Sci. 2010, 31, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Kawachi, I.; Colditz, G.A.; Stampfer, M.J.; Willett, W.C.; Manson, J.E.; Speizer, F.E.; Hennekens, C.H. Prospective Study of Shift Work and Risk of Coronary Heart Disease in Women. Circulation 1995, 92, 3178–3182. [Google Scholar] [CrossRef] [PubMed]
- Parkes, K.R. Shift work and age as interactive predictors of body mass index among offshore workers. Scand. J. Work Environ. Health 2002, 28, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Sharifian, A.; Farahani, S.; Pasalar, P.; Gharavi, M.; Aminian, O. Shift work as an oxidative stressor. J. Circadian Rhythms 2005, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.E.; DiTacchio, L.; Hayes, K.R.; Vollmers, C.; Pulivarthy, S.; Baggs, J.E.; Panda, S.; Hogenesch, J.B. Harmonics of Circadian Gene Transcription in Mammals. PLoS Genet. 2009, 5, e1000442. [Google Scholar] [CrossRef] [PubMed]
- Krishnaiah, S.Y.; Wu, G.; Growe, J.; Rhodes, S.; Coldren, F.; Venkataraman, A.; George, A.O.O.; Francey, L.J.; Er, U.; Sianati, B.; et al. Clock regulation of liver- and cell-autonomous metabolism reveals decoupling between transcription and metabolism. Cell Metabolism 2016. Under Review. [Google Scholar]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks, P.; et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Smolinska, A.; Blanchet, L.; Buydens, L.M.C.; Wijmenga, S.S. NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: A review. Anal. Chim. Acta 2012, 750, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted Profiling: Quantitative Analysis of 1H-NMR Metabolomics Data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef] [PubMed]
- Cermakian, N.; Boivin, D.B. A molecular perspective of human circadian rhythm disorders. Brain Res. Brain Res. Rev. 2003, 42, 204–220. [Google Scholar] [CrossRef]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Peirson, S.N.; Foster, R.G. Sleep and Circadian Rhythm Disruption in Psychosis. In Circadian Medicine; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2015; pp. 271–282. [Google Scholar]
- Lauretti, E.; Di Meco, A.; Merali, S.; Praticò, D. Circadian rhythm dysfunction: A novel environmental risk factor for Parkinson’s disease. Mol. Psychiatry 2016. [Google Scholar] [CrossRef] [PubMed]
- Erion, R.; King, A.N.; Wu, G.; Hogenesch, J.B.; Sehgal, A. Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. Elife 2016, 5, e13552. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.; Lahens, N.F.; Hogenesch, J.B.; Sehgal, A. Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res. 2015, 25, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.; Sonawat, H.M.; Sharma, S. Malaria parasite-infected erythrocytes inhibit glucose utilization in uninfected red cells. FEBS Lett. 2005, 579, 6151–6158. [Google Scholar] [CrossRef] [PubMed]
- Dabos, K.J.; Parkinson, J.A.; Hewage, C.; Nelson, L.J.; Sadler, I.H.; Hayes, P.C.; Plevris, J.N. 1H-NMR spectroscopy as a tool to evaluate key metabolic functions of primary porcine hepatocytes after cryopreservation. NMR Biomed. 2002, 15, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Drago, D.; Cossetti, C.; Iraci, N.; Gaude, E.; Musco, G.; Bachi, A.; Pluchino, S. The stem cell secretome and its role in brain repair. Biochimie 2013, 95, 2271–2285. [Google Scholar] [CrossRef] [PubMed]
- Uyar, A.; Seli, E. Metabolomic assessment of embryo viability. Semin. Reprod. Med. 2014, 32, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Booth, S.C.; Weljie, A.M.; Turner, R.J. Metabolomics reveals differences of metal toxicity in cultures of Pseudomonas pseudoalcaligenes KF707 grown on different carbon sources. Front. Microbiol. 2015, 6, 827. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, E.; Saini, C.; Bauer, C.; Laroche, T.; Naef, F.; Schibler, U. Circadian Gene Expression in Individual Fibroblasts: Cell-Autonomous and Self-Sustained Oscillators Pass Time to Daughter Cells. Cell 2004, 119, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Leise, T.L. Wavelet analysis of circadian and ultradian behavioral rhythms. J. Circadian Rhythms 2013, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Baggs, J.E.; Price, T.S.; DiTacchio, L.; Panda, S.; Fitzgerald, G.A.; Hogenesch, J.B. Network features of the mammalian circadian clock. PLoS Biol. 2009, 7, e52. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Tambellini, N.P.; Zaremberg, V.; Turner, R.J.; Weljie, A.M. Evaluation of extraction protocols for simultaneous polar and non-polar yeast metabolite analysis using multivariate projection methods. Metabolites 2013, 3, 592–605. [Google Scholar] [CrossRef] [PubMed]
| OPLS-DA Model (Pre- and Post-Time Shift) | OPLS Model (Time-Dependent) | ||||
|---|---|---|---|---|---|
| Metabolite | VIP | P (loadings) | Metabolite | VIP | P (loadings) |
| Glutamine | 1.42 | −0.28 | Glutamine | 1.37 | −0.25 |
| Glucose | 1.28 | −0.25 | |||
| Serine | 1.20 | −0.24 | Serine | 1.08 | −0.19 |
| Threonine | 1.06 | −0.21 | Cis-Aconitate | 1.10 | −0.22 |
| Methionine | 1.06 | −0.20 | |||
| Choline | 1.05 | −0.17 | Choline | 1.36 | −0.27 |
| Glycine | 1.06 | 0.15 | Glycine | 1.25 | 0.25 |
| Glutamate | 1.06 | 0.22 | Glutamate | 1.17 | 0.22 |
| Methylguanidine | 1.17 | 0.22 | Methylguanidine | 1.35 | 0.27 |
| Alanine | 1.33 | 0.24 | Alanine | 1.55 | 0.30 |
| Acetate | 1.37 | 0.26 | Acetate | 1.55 | 0.31 |
| Lactate | 1.41 | 0.26 | Formate | 1.60 | 0.31 |
| Formate | 1.53 | 0.30 | Lactate | 1.61 | 0.32 |
| Media | |||||
|---|---|---|---|---|---|
| Increased Pre-Shift | P | FDR | Increased Post-Shift | p | FDR |
| Glutamine | 0 | 0 | Glutamate | 0.002 | 0.01 |
| Serine | 0.01 | 0.03 | Acetate | 0 | 0 |
| Glucose | 0.004 | 0.02 | Alanine | 0 | 0 |
| Threonine | 0.005 | 0.02 | Formate | 0 | 0 |
| Choline | 0.004 | 0.02 | Lactate | 0.002 | 0.01 |
| cis-Aconitate | 0.04 | 0.08 | Glycine | 0.02 | 0.04 |
| myo-Inositol | 0.05 | 0.1 | Methylguanidine | 0.005 | 0.02 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sengupta, A.; Krishnaiah, S.Y.; Rhoades, S.; Growe, J.; Slaff, B.; Venkataraman, A.; Olarerin-George, A.O.; Van Dang, C.; Hogenesch, J.B.; Weljie, A.M. Deciphering the Duality of Clock and Growth Metabolism in a Cell Autonomous System Using NMR Profiling of the Secretome. Metabolites 2016, 6, 23. https://doi.org/10.3390/metabo6030023
Sengupta A, Krishnaiah SY, Rhoades S, Growe J, Slaff B, Venkataraman A, Olarerin-George AO, Van Dang C, Hogenesch JB, Weljie AM. Deciphering the Duality of Clock and Growth Metabolism in a Cell Autonomous System Using NMR Profiling of the Secretome. Metabolites. 2016; 6(3):23. https://doi.org/10.3390/metabo6030023
Chicago/Turabian StyleSengupta, Arjun, Saikumari Y. Krishnaiah, Seth Rhoades, Jacqueline Growe, Barry Slaff, Anand Venkataraman, Anthony O. Olarerin-George, Chi Van Dang, John B. Hogenesch, and Aalim M. Weljie. 2016. "Deciphering the Duality of Clock and Growth Metabolism in a Cell Autonomous System Using NMR Profiling of the Secretome" Metabolites 6, no. 3: 23. https://doi.org/10.3390/metabo6030023
APA StyleSengupta, A., Krishnaiah, S. Y., Rhoades, S., Growe, J., Slaff, B., Venkataraman, A., Olarerin-George, A. O., Van Dang, C., Hogenesch, J. B., & Weljie, A. M. (2016). Deciphering the Duality of Clock and Growth Metabolism in a Cell Autonomous System Using NMR Profiling of the Secretome. Metabolites, 6(3), 23. https://doi.org/10.3390/metabo6030023