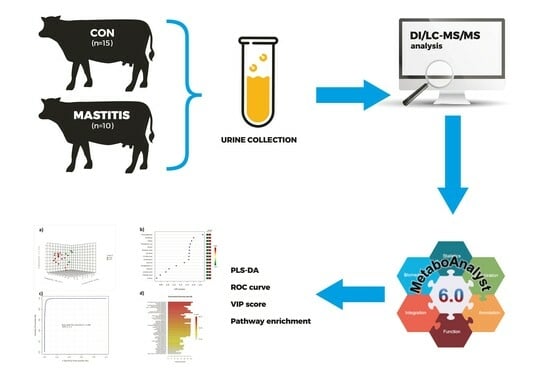
Identifying Predictive Biomarkers of Subclinical Mastitis in Dairy Cows through Urinary Metabotyping
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Diets, and Urine Samples
2.2. Mass Spectrometry (MS)-Based Compound Identification and Quantification
2.2.1. Sample Preparation
2.2.2. Tandem Mass Spectrometry (MS/MS)
2.3. Statistical Analysis
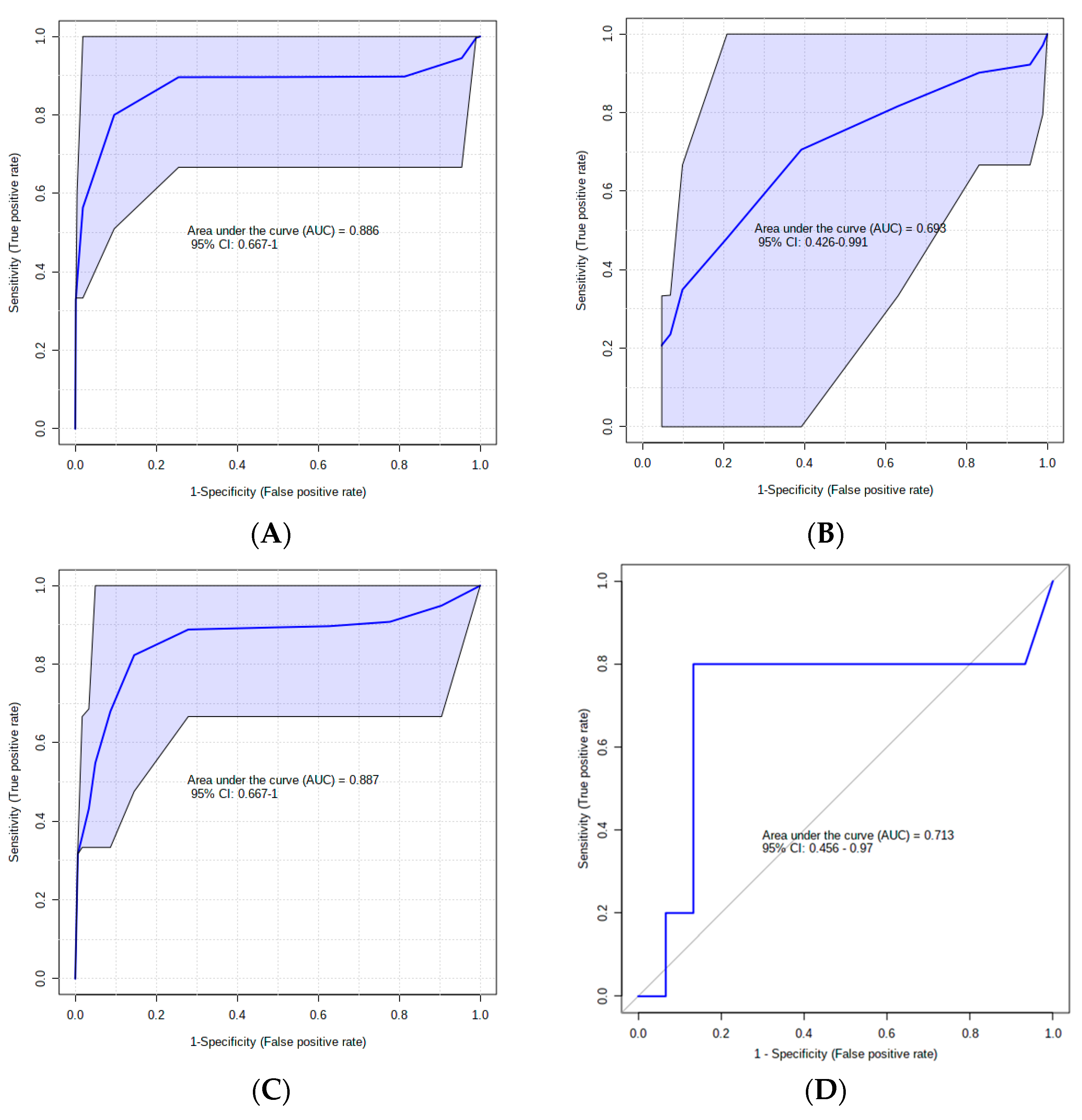
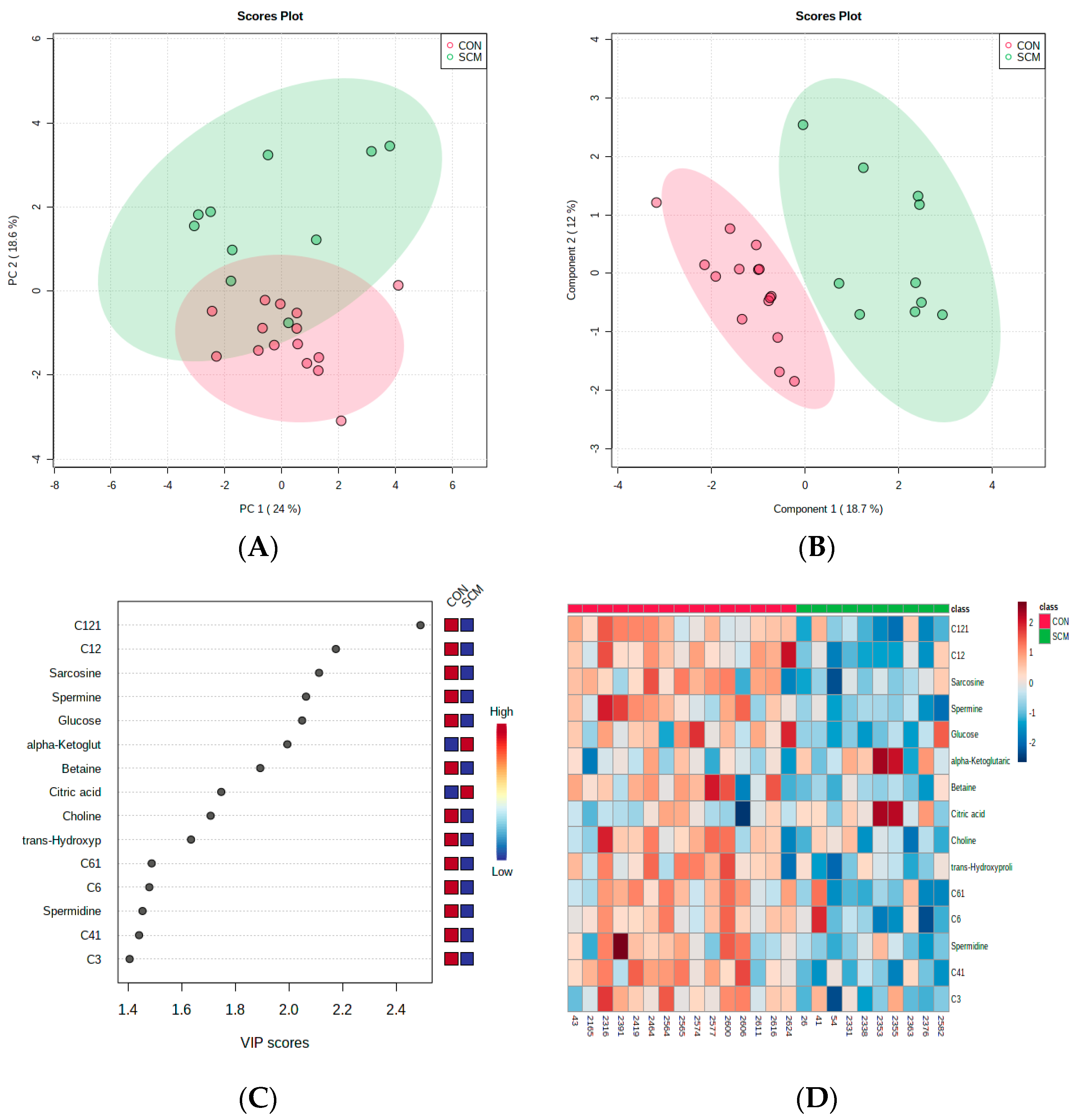
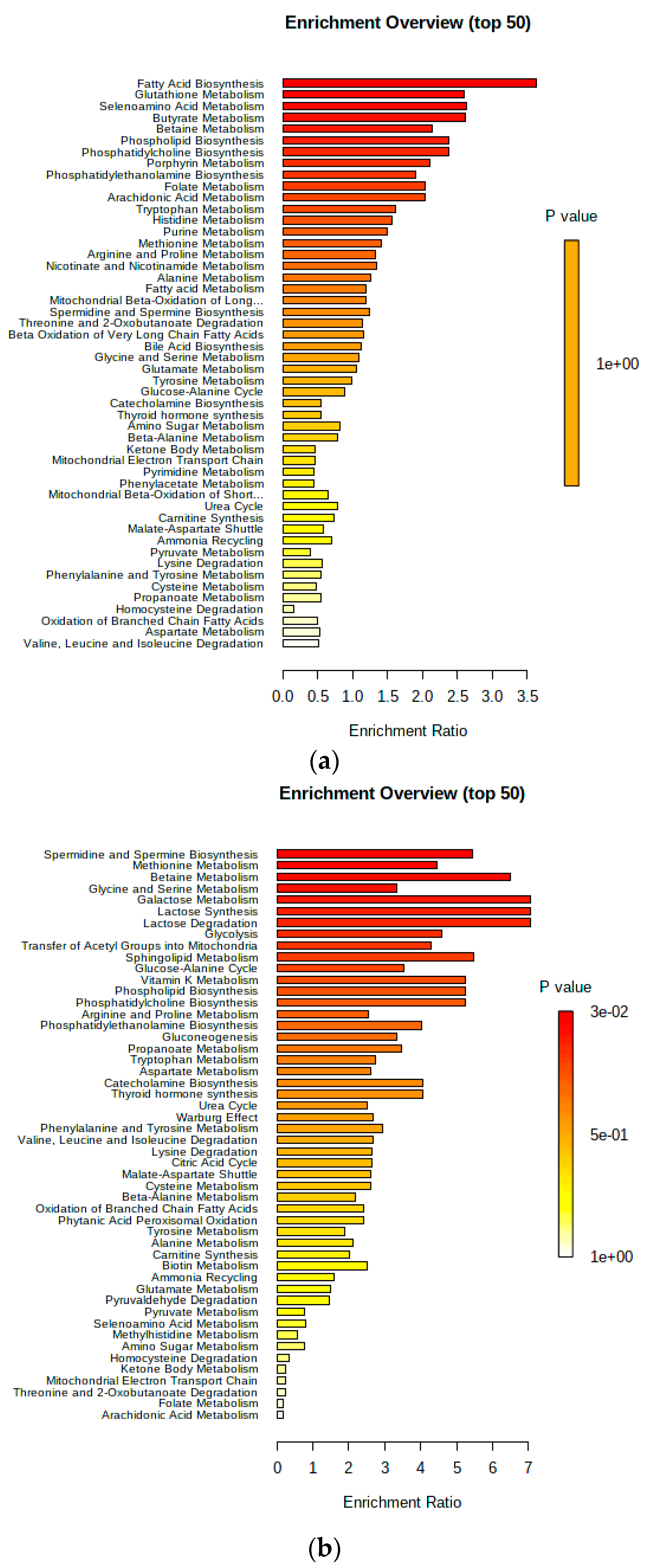
3. Results
4. Discussion
4.1. Urinary AC Alterations in Pre-SCM Cows
4.2. Changes in Urinary Amino Acids in Pre-SCM Cows
4.3. Changes in the Urinary Carbohydrate and Organic Acid Species in Pre-SCM Cows
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adkins, P.R.; Middleton, J.R. Methods for diagnosing mastitis. Vet. Clin. N. Am. 2018, 34, 479–491. [Google Scholar] [CrossRef]
- Aghamohammadi, M.; Haine, D.; Kelton, D.F.; Barkema, H.W.; Hogeveen, H.; Keefe, G.P.; Dufour, S. Herd-Level mastitis-associated costs on Canadian dairy farms. Front. Vet. Sci. 2018, 5, 100. [Google Scholar] [CrossRef]
- De Prado-Taranilla, A.I.; Holstege, M.M.; Bertocchi, L.; Appiani, A.; Becvar, O.; Davidek, J.; Bay, D.; Jimenez, L.M.; Roger, N.; Krömker, V.; et al. Incidence of milk leakage after dry-off in European dairy herds, related risk factors, and its role in new intramammary infections. J. Dairy Sci. 2020, 103, 9224–9237. [Google Scholar] [CrossRef]
- Dervishi, E.; Zhang, G.; Hailemariam, D.; Dunn, S.M.; Ametaj, B.N. Innate immunity and carbohydrate metabolism alterations precede occurrence of subclinical mastitis in transition dairy cows. J. Anim. Sci. Technol. 2015, 57, 46. [Google Scholar] [CrossRef]
- Dervishi, E.; Zhang, G.; Dunn, S.M.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. GC–MS metabolomics identifies metabolite alterations that precede subclinical mastitis in the blood of transition dairy cows. J. Proteome Res. 2016, 16, 433–446. [Google Scholar] [CrossRef]
- Zwierzchowski, G.; Zhang, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Mass-spec-based urinary metabotyping around parturition identifies screening biomarkers for subclinical mastitis in dairy cows. Res. Vet. Sci. 2020, 129, 39–52. [Google Scholar] [CrossRef]
- Martins, S.A.; Martins, V.C.; Cardoso, F.A.; Germano, J.; Rodrigues, M.; Duarte, C.; Bexiga, R.; Cardoso, S.; Freitas, P.P. Biosensors for on-farm diagnosis of mastitis. Front. Bioeng. Biotechnol. 2019, 7, 186. [Google Scholar] [CrossRef]
- Donadeu, F.X.; Howes, N.L.; Esteves, C.L.; Howes, M.P.; Byrne, T.J.; Macrae, A.I. Farmer and veterinary practices and opinions related to the diagnosis of mastitis and metabolic disease in UK dairy cows. Front. Vet. Sci. 2020, 7, 127. [Google Scholar] [CrossRef]
- Haxhiaj, K.; Li, Z.; Johnson, M.; Dunn, S.M.; Wishart, D.S.; Ametaj, B.N. Blood metabolomic phenotyping of dry cows could predict the high milk somatic cells in early lactation—Preliminary results. Dairy 2022, 3, 59–77. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef] [PubMed]
- Tarasenko, T.N.; Cusmano-Ozog, K.; McGuire, P.J. Tissue acylcarnitine status in a mouse model of mitochondrial β-oxidation deficiency during metabolic decompensation due to influenza virus infection. Mol. Genet. Metab. 2018, 125, 144–152. [Google Scholar] [CrossRef]
- Zhang, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. A multi-platform metabolomics approach identifies urinary metabolite signatures that differentiate ketotic from healthy dairy cows. Front. Vet. Sci. 2021, 8, 595983. [Google Scholar] [CrossRef] [PubMed]
- Rutkowsky, J.M.; Knotts, T.A.; Ono-Moore, K.D.; McCoin, C.S.; Huang, S.; Schneider, D.; Singh, S.; Adams, S.H.; Hwang, D.H. Acylcarnitines activate proinflammatory signaling pathways. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1378–E1387. [Google Scholar] [CrossRef] [PubMed]
- Minuti, A.; Zhou, Z.; Graugnard, D.E.; Rodriguez-Zas, S.L.; Palladino, A.R.; Cardoso, F.C.; Trevisi, E.; Loor, J.J. Acute mammary and liver transcriptome responses after an intramammary Escherichia coli lipopolysaccharide challenge in postpartal dairy cows. Physiol. Rep. 2015, 3, e12388. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.A.; Roe, C.R.; Stacey, T.E.; Hoppel, C.L. Urinary excretion of l-carnitine and acylcarnitines by patients with disorders of organic acid metabolism: Evidence for secondary insufficiency of l-carnitine. Pediatr. Res. 1984, 18, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Fowler, B.; Leonard, J.V.; Baumgartner, M.R. Causes of and diagnostic approach to methylmalonic acidurias. J. Inherit. Metab. Dis. 2008, 31, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Dervishi, E.; Zwierzchowski, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Urinary metabolomics around parturition identifies metabolite alterations in dairy cows affected postpartum by lameness: Preliminary study. Dairy 2020, 1, 6–19. [Google Scholar] [CrossRef]
- Zhenyukh, O.; Civantos, E.; Ruiz-Ortega, M.; Sánchez, M.S.; Vázquez, C.; Peiró, C.; Egido, J.; Mas, S. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 activation. Free. Radic. Biol. Med. 2017, 104, 165–177. [Google Scholar] [CrossRef]
- Dervishi, E.; Zhang, G.; Hailemariam, D.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Urine metabolic fingerprinting can be used to predict the risk of metritis and highlight the pathobiology of the disease in dairy cows. Metabolomics 2018, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Nystrom, G.; Pruznak, A.; Huber, D.; Frost, R.A.; Lang, C.H. Local insulin-like growth factor I prevents sepsis-induced muscle atrophy. Metabolism 2009, 58, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Koshikawa, M.; Harada, M.; Noyama, S.; Kiyono, K.; Motoike, Y.; Nomura, Y.; Nishimura, A.; Izawa, H.; Watanabe, E.; Ozaki, Y. Association between inflammation and skeletal muscle proteolysis, skeletal mass and strength in elderly heart failure patients and their prognostic implications. BMC Cardiovasc. Disord. 2020, 20, 228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Du, J.; Hu, Z.; Han, G.; Delafontaine, P.; Garcia, G.; Mitch, W.E. IL-6 and serum amyloid A synergy mediates angiotensin ii–induced muscle wasting. J. Am. Soc. Nephrol. 2009, 20, 604–612. [Google Scholar] [CrossRef]
- Son, D.O.; Satsu, H.; Shimizu, M. Histidine inhibits oxidative stress- and TNF-α-induced interleukin-8 secretion in intestinal epithelial cells. FEBS Lett. 2005, 579, 4671–4677. [Google Scholar] [CrossRef] [PubMed]
- Satriano, J. Arginine pathways and the inflammatory response: Interregulation of nitric oxide and polyamines: Review article. Amino Acids. 2004, 26, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wang, C.; Ding, L.; Shen, Y.; Cui, H.; Wang, M. Arginine relieves the inflammatory response and enhances the casein expression in bovine mammary epithelial cells induced by lipopolysaccharide. Mediat. Inflamm. 2016, 2016, 9618795. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.Y.; Wang, Y.F.; Shen, Y.Z.; Zhou, G.; Wu, T.Y.; Zhang, X.; Wang, M.Z.; Loor, J.J.; Zhang, J. Effects of intravenous arginine infusion on inflammation and metabolic indices of dairy cows in early lactation. Animal 2020, 14, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.N.; Lopreiato, V.; Alharthi, A.; Loor, J.J. Amino acids and the regulation of oxidative stress and immune function in dairy cattle. J. Anim. Sci. 2020, 98, S175–S193. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D.; Bollenbach, A.; Hanff, E.; Kayacelebi, A.A. Asymmetric dimethylarginine (ADMA), symmetric dimethylarginine (SDMA) and homoarginine (hArg): The ADMA, SDMA and hArg paradoxes. Cardiovasc. Diabetol. 2018, 17, 1. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, Q.; Li, X.; Chen, C.; Liu, J.; Ye, Y.; Ruan, T.; Hei, Z. Asymmetric dimethylarginine and all-cause mortality: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 44692. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, C.; Liao, H.; Ishidate, K. Structure and function of choline kinase isoforms in mammalian cells. Prog. Lipid Res. 2004, 43, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Fullerton, M.D.; Wagner, L.; Yuan, Z.; Bakovic, M. Impaired trafficking of choline transporter-like protein-1 at plasma membrane and inhibition of choline transport in THP-1 monocyte-derived macrophages. Am. J. Physiol. Cell Physiol. 2006, 290, C1230–C1238. [Google Scholar] [CrossRef] [PubMed]
- Snider, S.A.; Margison, K.D.; Ghorbani, P.; LeBlond, N.D.; O’Dwyer, C.; Nunes, J.R.; Nguyen, T.; Xu, H.; Bennett, S.A.; Fullerton, M.D. Choline transport links macrophage phospholipid metabolism and inflammation. J. Biol. Chem. 2018, 293, 11600–11611. [Google Scholar] [CrossRef] [PubMed]
- Parrish, W.R.; Rosas-Ballina, M.; Gallowitsch-Puerta, M.; Ochani, M.; Ochani, K.; Yang, L.H.; Hudson, L.; Lin, X.; Patel, N.; Johnson, S.M.; et al. Modulation of TNF release by choline requires α7 subunit nicotinic acetylcholine receptor-mediated signaling. Mol. Med. 2008, 14, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Go, E.K.; Jung, K.J.; Kim, J.Y.; Yu, B.P.; Chung, H.Y. Betaine suppresses proinflammatory signaling during aging: The involvement of nuclear factor- B via nuclear factor-inducing kinase/i B kinase and mitogen-activated protein kinases. J. Gerontol. Ser. A 2005, 60, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Park, K.; Lee, S.B.; Kang, S.; Park, J.S.; Ahn, C.W.; Nam, J.S. Relationship between natural killer cell activity and glucose control in patients with type 2 diabetes and prediabetes. J. Diabetes Investig. 2019, 10, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, W.M.; Yokota, C.N.; Curi, R.; Alba-Loureiro, T.C. Obesity and Type 2 Diabetes mellitus induce lipopolysaccharide tolerance in rat neutrophils. Sci. Rep. 2018, 8, 17534. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Takahashi, I.; Iwane, K.; Okubo, N.; Nishimura, M.; Matsuzaka, M.; Wada, N.; Miwa, T.; Umeda, T.; Nakaji, S. The influence of blood glucose on neutrophil function in individuals without diabetes. Luminescence 2013, 28, 569–573. [Google Scholar] [CrossRef]
- Hrdlička, V.; Barek, J.; Navrátil, T. Differential pulse voltammetric determination of homovanillic acid as a tumor biomarker in human urine after hollow fiber-based liquid-phase microextraction. Talanta 2021, 221, 121594. [Google Scholar] [CrossRef]
- O’Neill, H.A. A review on the involvement of catecholamines in animal behaviour. S. Afr. J. Anim. Sci. 2019, 49, 1. [Google Scholar] [CrossRef]
- Bonifačić, D.; Aralica, M.; Sotošek Tokmadžić, V.; Rački, V.; Tuškan-Mohar, L.; Kučić, N. Values of vanillylmandelic acid and homovanillic acid in the urine as potential prognostic biomarkers in ischaemic stroke patients. Biomarkers 2017, 22, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Iacobazzi, V.; Menga, A.; Avantaggiati, M.L.; Palmieri, F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFα- and IFNγ-triggered inflammation. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2014, 1839, 1217–1225. [Google Scholar] [CrossRef]
- Infantino, V.; Iacobazzi, V.; Palmieri, F.; Menga, A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013, 440, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Convertini, P.; Cucci, L.; Panaro, M.A.; Di Noia, M.A.; Calvello, R.; Palmieri, F.; Iacobazzi, V. The mitochondrial citrate carrier: A new player in inflammation. Biochem. J. 2011, 438, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Lovaas, E.; Carlin, G. Spermine: An anti-oxidant and anti-inflammatory agent. Free Radic. Biol. Med. 1991, 11, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Theoharides, T.C. Polyamines spermidine and spermine as modulators of calcium-dependent immune processes. Life Sci. 1980, 27, 703–713. [Google Scholar] [CrossRef]
- Naik, S.; Lagishetty, C. Polyamines: Potential anti-inflammatory agents and their possible mechanism of action. Indian J. Pharmacol. 2008, 40, 121. [Google Scholar] [CrossRef]


| Ingredient | Weight/Cow (kg) | DM (%) | Final DMI (kg) 1 |
|---|---|---|---|
| Hay | 5.50 | 85.14% | 4.68 |
| Oats | 5.75 | 36.20% | 2.08 |
| Corn | 8.84 | 30.30% | 2.68 |
| Protein | 2.00 | 93.00% | 1.86 |
| Ground Barley | 0.75 | 97.26% | 0.66 |
| Minerals | 0.42 | 97.26% | 0.41 |
| Total | 23.36 | 53.17% | 12.37 |
| Ingredient | Weight/Cow (kg) | DM (%) | Final DMI (kg) |
|---|---|---|---|
| Hay dairy | 2.50 | 88.50 | 2.21 |
| Grass silage | 10.75 | 31.80 | 3.42 |
| Oats | 5.99 | 36.20 | 2.17 |
| Barley-Dakota | 11.50 | 40.00 | 4.80 |
| Corn | 13.52 | 31.50 | 4.26 |
| Whey | 2.75 | 17.00 | 0.47 |
| Protein | 4.75 | 93.30 | 4.43 |
| Energy dairy | 4.25 | 88.00 | 3.74 |
| Ground Barley | 1.75 | 88.00 | 1.54 |
| Mineral & Fat | 1.26 | 97.26 | 1.23 |
| Total | 59.02 | 47.56 | 28.07 |
| Metabolites (μM) | MEAN ± SEM | Fold Change | SCM/CON | p-Value | |
|---|---|---|---|---|---|
| Pre-SCM 1 (n = 10) | CON 2 (n = 15) | ||||
| Creatinine | 13,903 ± 1679 | 8732 ± 1477 | 1.59 | up | 0.01 |
| Glycine | 302 ± 169 | 196 ± 149 | 1.54 | up | 0.78 |
| Alanine | 265 ± 60 | 171 ± 52.8 | 1.55 | up | 0.48 |
| Serine | 103 ± 17.3 | 56 ± 15.2 | 1.84 | up | 0.11 |
| Histamine | 0.1315 ± 0.0244 | 0.0915 ± 0.0214 | 1.44 | up | 0.28 |
| Proline | 6.14 ± 1.56 | 5.54 ± 1.37 | 1.11 | up | 0.42 |
| Valine | 18.48 ± 3.41 | 8.04 ± 3 | 2.30 | up | 0.02 |
| Threonine | 84.4 ± 18.3 | 46.4 ± 16.1 | 1.82 | up | 0.2 |
| Taurine | 655 ± 211 | 420 ± 186 | 1.56 | up | 0.55 |
| Putrescine | 0.724 ± 0.144 | 0.299 ± 0.127 | 2.42 | up | 0.01 |
| trans-Hydroxyproline | 1.69 ± 0.598 | 1 ± 0.526 | 1.69 | up | 0.86 |
| Leucine | 12.4 ± 1.94 | 6.3 ± 1.7 | 1.97 | up | 0.01 |
| Isoleucine | 19.34 ± 6.6 | 7.83 ± 5.8 | 2.47 | up | 0.28 |
| Asparagine | 13.1 ± 1.79 | 8.47 ± 1.57 | 1.55 | up | 0.05 |
| Aspartic acid | 174 ± 36.8 | 127 ± 32.4 | 1.37 | up | 0.43 |
| Glutamine | 347 ± 75.1 | 154 ± 66 | 2.25 | up | 0.15 |
| Glutamic acid | 107 ± 31.2 | 80 ± 27.5 | 1.34 | up | 0.64 |
| Methionine | 3.33 ± 0.29 | 2.7 ± 0.255 | 1.23 | up | 0.06 |
| Histidine | 108.2 ± 24.6 | 53.1 ± 21.6 | 2.04 | up | 0.17 |
| alpha-Aminoadipic acid | 135 ± 26.6 | 72 ± 23.4 | 1.88 | up | 0.25 |
| Phenylalanine | 16.38 ± 2.72 | 9.87 ± 2.4 | 1.66 | up | 0.09 |
| Methionine-sulfoxide | 3.81 ± 0.948 | 1.86 ± 0.834 | 2.05 | up | 0.17 |
| Arginine | 15.46 ± 2.05 | 8.15 ± 1.8 | 1.90 | up | 0.01 |
| Acetyl-ornithine | 76.1 ± 12.5 | 44 ± 11 | 1.73 | up | 0.08 |
| Citrulline | 8.54 ± 2.36 | 3.5 ± 2.07 | 2.44 | up | 0.07 |
| Serotonin | 1.79 ± 0.297 | 1.37 ± 0.262 | 1.31 | up | 0.44 |
| Tyrosine | 27.4 ± 5 | 16.4 ± 4.4 | 1.67 | up | 0.11 |
| Asymmetric dimethylarginine | 9.48 ± 1.21 | 2.61 ± 1.07 | 3.63 | up | <0.001 |
| Total dimethylarginine | 33.8 ± 4.03 | 18.3 ± 3.55 | 1.85 | up | 0.007 |
| Tryptophan | 36 ± 8.65 | 19.3 ± 7.61 | 1.87 | up | 0.28 |
| Kynurenine | 1.585 ± 0.449 | 0.967 ± 0.395 | 1.64 | up | 0.71 |
| Carnosine | 21.7 ± 4.39 | 15.5 ± 3.86 | 1.40 | up | 0.4 |
| Ornithine | 25.8 ± 4.44 | 14 ± 3.91 | 1.84 | up | 0.07 |
| Lysine | 82.1 ± 14.1 | 43.4 ± 12.4 | 1.89 | up | 0.08 |
| Spermidine | 0.2248 ± 0.0871 | 0.0746 ± 0.0766 | 3.01 | up | 0.05 |
| Spermine | 0.0868 ± 0.0171 | 0.0791 ± 0.015 | 1.10 | up | 0.43 |
| Sarcosine | 4.69 ± 1.17 | 1.27 ± 1.03 | 3.69 | up | 0.17 |
| Tyramine | 0.183 ± 0.042 | 0.111 ± 0.0501 | 1.65 | up | 0.26 |
| Creatine | 5098 ± 663 | 2354 ± 583 | 2.17 | up | 0.001 |
| Betaine | 265.2 ± 67.6 | 74.9 ± 59.4 | 3.54 | up | 0.02 |
| Choline | 92.8 ± 21.5 | 32.1 ± 18.9 | 2.89 | up | 0.007 |
| Trimethylamine N-oxide | 6150 ± 1895 | 3677 ± 1667 | 1.67 | up | 0.38 |
| Methylhistidine | 198 ± 52.3 | 370 ± 59.4 | 0.54 | down | 0.05 |
| Lactic acid | 125 ± 53.9 | 112 ± 47.4 | 1.12 | up | 0.93 |
| beta-Hydroxybutyric acid | 400 ± 471 | 416 ± 415 | 0.96 | down | 0.47 |
| alpha-Ketoglutaric acid | 25.7 ± 45.1 | 34.8 ± 39.7 | 0.74 | down | 0.47 |
| Citric acid | 856 ± 801 | 778 ± 684 | 1.10 | up | 0.84 |
| Butyric acid | 28.4 ± 14.7 | 29.5 ± 12.9 | 0.96 | down | 0.35 |
| p-hydroxyhippuric acid | 36.7 ± 14.1 | 37.9 ± 12.4 | 0.97 | down | 0.93 |
| Succinic acid | 30.3 ± 9.86 | 20.6 ± 8.67 | 1.47 | up | 0.69 |
| Pyruvic acid | 8.71 ± 1.79 | 6.33 ± 1.58 | 1.38 | up | 0.16 |
| Isobutyric acid | 7.23 ± 1.66 | 5.56 ± 1.46 | 1.30 | up | 0.59 |
| Hippuric acid | 14,438 ± 2073 | 13,225 ± 1823 | 1.09 | up | 0.65 |
| Methylmalonic acid | 29.4 ± 6.68 | 17.9 ± 5.87 | 1.64 | up | 0.31 |
| Homovanillic acid | 14.67 ± 1.46 | 8.83 ± 1.28 | 1.66 | up | <0.001 |
| Indole acetic acid | 67.5 ± 20.7 | 51.3 ± 18.2 | 1.32 | up | 0.96 |
| Uric acid | 5014 ± 883 | 4279 ± 776 | 1.17 | up | 0.44 |
| Glucose | 3369 ± 462 | 1955 ± 406 | 1.72 | up | 0.002 |
| C0 | 2.516 ± 0.386 | 0.893 ± 0.339 | 2.82 | up | 0.01 |
| C2 | 0.714 ± 0.0915 | 0.305 ± 0.0805 | 2.34 | up | 0.001 |
| C3:1 | 0.0319 ± 0.00428 | 0.0258 ± 0.00376 | 1.24 | up | 0.02 |
| C3 | 0.0402 ± 0.00722 | 0.037 ± 0.00635 | 1.09 | up | 0.33 |
| C4:1 | 0.0871 ± 0.00819 | 0.0631 ± 0.00721 | 1.38 | up | 0.04 |
| C4 | 0.58 ± 0.1096 | 0.129 ± 0.0964 | 4.50 | up | 0.002 |
| C3OH | 0.0855 ± 0.0092 | 0.0642 ± 0.00809 | 1.33 | up | 0.11 |
| C5:1 | 0.251 ± 0.0226 | 0.147 ± 0.0199 | 1.71 | up | 0.001 |
| C5 | 0.1598 ± 0.0273 | 0.0929 ± 0.024 | 1.72 | up | 0.18 |
| C4OH | 0.0898 ± 0.00984 | 0.0653 ± 0.00865 | 1.38 | up | 0.05 |
| C6:1 | 0.057 ± 0.0122 | 0.0817 ± 0.0108 | 0.70 | down | 0.27 |
| C6 | 0.072 ± 0.0134 | 0.0872 ± 0.0118 | 0.83 | down | 0.77 |
| C5OH | 0.1372 ± 0.0131 | 0.0849 ± 0.0115 | 1.62 | up | 0.002 |
| C5:1DC | 0.0438 ± 0.00413 | 0.0377 ± 0.00363 | 1.16 | up | 0.23 |
| C5DC | 0.0469 ± 0.00595 | 0.0323 ± 0.00523 | 1.45 | up | 0.03 |
| C8 | 0.0556 ± 0.00556 | 0.0356 ± 0.00489 | 1.56 | up | 0.003 |
| C5MDC | 0.0514 ± 0.00366 | 0.0466 ± 0.00322 | 1.10 | up | 0.02 |
| C9 | 0.147 ± 0.023 | 0.106 ± 0.0203 | 1.39 | up | 0.21 |
| C7DC | 0.0437 ± 0.00899 | 0.0385 ± 0.00791 | 1.14 | up | 0.5 |
| C10:2 | 0.0578 ± 0.00732 | 0.0437 ± 0.00644 | 1.32 | up | 0.19 |
| C10:1 | 0.171 ± 0.0177 | 0.15 ± 0.0156 | 1.14 | up | 0.15 |
| C10 | 0.135 ± 0.0125 | 0.104 ± 0.011 | 1.30 | up | 0.01 |
| C12:1 | 0.14 ± 0.0387 | 0.127 ± 0.034 | 1.10 | up | 0.43 |
| C12 | 0.1037 ± 0.00933 | 0.0943 ± 0.0082 | 1.10 | up | 0.01 |
| Metabolites (μM) | MEAN ± SEM | Fold Change | SCM/CON | p-Value | |
|---|---|---|---|---|---|
| Pre-SCM 1 (n = 10) | CON 2 (n = 15) | ||||
| Creatinine | 14,300 ± 1521 | 10,569 ± 1140 | 1.35 | up | 0.01 |
| Glycine | 105 ± 28.3 | 67.5 ± 21.2 | 1.56 | up | 0.1 |
| Alanine | 97.5 ± 11.7 | 82.5 ± 8.8 | 1.18 | up | 0.23 |
| Serine | 77 ± 10.39 | 69 ± 7.79 | 1.12 | up | 0.1 |
| Histamine | 0.0994 ± 0.0154 | 0.0642 ± 0.0116 | 1.55 | up | 0.12 |
| Proline | 3.63 ± 0.531 | 3.94 ± 0.398 | 0.92 | down | 0.44 |
| Valine | 11.1 ± 1.185 | 10.3 ± 0.888 | 1.08 | up | 0.28 |
| Threonine | 70.7 ± 10.08 | 50.9 ± 7.56 | 1.39 | up | 0.01 |
| Taurine | 439 ± 136 | 395 ± 102 | 1.11 | up | 0.71 |
| Putrescine | 0.915 ± 0.503 | 1.167 ± 0.377 | 0.78 | down | 0.84 |
| trans-Hydroxyproline | 1.22 ± 0.425 | 1.92 ± 0.318 | 0.64 | down | 0.3 |
| Leucine | 9.1 ± 0.99 | 9.13 ± 0.742 | 1.00 | up | 0.38 |
| Isoleucine | 7.72 ± 0.745 | 5.98 ± 0.558 | 1.29 | up | 0.007 |
| Asparagine | 13.46 ± 1.48 | 9.67 ± 1.11 | 1.39 | up | 0.01 |
| Aspartic acid | 190 ± 31.7 | 131 ± 23.7 | 1.45 | up | 0.17 |
| Glutamine | 285 ± 46.8 | 206 ± 35.1 | 1.38 | up | 0.04 |
| Glutamic acid | 78.6 ± 11.7 | 53.6 ± 8.8 | 1.47 | up | 0.09 |
| Methionine | 3.34 ± 0.252 | 3.25 ± 0.189 | 1.03 | up | 0.4 |
| Histidine | 76.6 ± 9.85 | 60.4 ± 7.38 | 1.27 | up | 0.05 |
| alpha-Aminoadipic acid | 79.9 ± 14.5 | 72.9 ± 10.8 | 1.10 | up | 0.31 |
| Phenylalanine | 13.1 ± 1.08 | 10.4 ± 0.81 | 1.26 | up | 0.03 |
| Methionine-sulfoxide | 3.12 ± 0.478 | 3.13 ± 0.358 | 1.00 | up | 0.83 |
| Arginine | 11.3 ± 1.316 | 10.1 ± 0.986 | 1.12 | up | 0.48 |
| Acetyl-ornithine | 57 ± 6.82 | 47.4 ± 5.11 | 1.20 | up | 0.08 |
| Citrulline | 3.47 ± 1.265 | 6.94 ± 0.948 | 0.50 | down | 0.17 |
| Serotonin | 1.66 ± 0.202 | 1.33 ± 0.151 | 1.25 | up | 0.13 |
| Tyrosine | 19.4 ± 2.38 | 20 ± 1.78 | 0.97 | down | 0.8 |
| Asymmetric dimethylarginine | 8.39 ± 1.024 | 6.26 ± 0.768 | 1.34 | up | 0.02 |
| Total dimethylarginine | 36.2 ± 3.36 | 26.4 ± 2.52 | 1.37 | up | 0.008 |
| Tryptophan | 19.8 ± 3.17 | 17.8 ± 2.38 | 1.11 | up | 0.58 |
| Kynurenine | 0.735 ± 0.0835 | 0.696 ± 0.0625 | 1.06 | up | 0.94 |
| Carnosine | 14.7 ± 1.61 | 11.1 ± 1.21 | 1.32 | up | 0.03 |
| Ornithine | 16.7 ± 2.1 | 15.5 ± 1.58 | 1.08 | up | 0.6 |
| Lysine | 59.7 ± 5.68 | 48.5 ± 4.26 | 1.23 | up | 0.07 |
| Spermidine | 0.0772 ± 0.0256 | 0.1113 ± 0.0192 | 0.69 | down | 0.36 |
| Spermine | 0.1219 ± 0.0113 | 0.0596 ± 0.015 | 2.05 | up | 0.008 |
| Sarcosine | 3.04 ± 2.02 | 6.92 ± 1.51 | 0.44 | down | 0.04 |
| Tyramine | 0.133 ± 0.0237 | 0.113 ± 0.0188 | 1.18 | up | 0.82 |
| Creatine | 5737 ± 1558 | 6460 ± 1168 | 0.89 | down | 0.69 |
| Betaine | 134 ± 95.7 | 364 ± 71.7 | 0.37 | down | 0.06 |
| Choline | 18 ± 12.98 | 56.9 ± 9.73 | 0.32 | down | 0.05 |
| Trimethylamine N-oxide | 5083 ± 1353 | 1338 ± 1014 | 3.80 | up | 0.03 |
| Methylhistidine | 373 ± 33.7 | 246 ± 25.2 | 1.52 | up | 0.001 |
| Lactic acid | 256 ± 83.6 | 107 ± 62.6 | 2.39 | up | 0.13 |
| beta-Hydroxybutyric acid | 116 ± 60.2 | 135 ± 45.1 | 0.86 | down | 0.6 |
| alpha-Ketoglutaric acid | 129 ± 66.6 | 17.8 ± 49.9 | 7.27 | up | 0.07 |
| Citric acid | 1911 ± 723 | 91 ± 542 | 21.00 | up | 0.04 |
| Butyric acid | 11.59 ± 2.18 | 7.43 ± 1.63 | 1.56 | up | 0.09 |
| p-hydroxyhippuric acid | 44.9 ± 17.2 | 53.9 ± 12.9 | 0.83 | down | 0.96 |
| Succinic acid | 42.3 ± 14.7 | 17.9 ± 11 | 2.36 | up | 0.22 |
| Pyruvic acid | 21.87 ± 6.85 | 7.63 ± 5.13 | 2.87 | up | 0.08 |
| Isobutyric acid | 5.14 ± 0.753 | 2.71 ± 0.564 | 1.90 | up | 0.009 |
| Hippuric acid | 20,896 ± 3944 | 17,503 ± 2955 | 1.19 | up | 0.32 |
| Methylmalonic acid | 25.2 ± 4.18 | 10.5 ± 3.13 | 2.40 | up | 0.001 |
| Homovanillic acid | 13.55 ± 1.88 | 8.01 ± 1.41 | 1.69 | up | 0.03 |
| Indole acetic acid | 58.5 ± 9.75 | 36.4 ± 7.3 | 1.61 | up | 0.06 |
| Uric acid | 4707 ± 748 | 3036 ± 561 | 1.55 | up | 0.05 |
| Glucose | 387 ± 555 | 973 ± 416 | 0.40 | down | 0.35 |
| C0 | 1.77 ± 0.167 | 1.15 ± 0.125 | 1.54 | up | 0.01 |
| C2 | 0.685 ± 0.1239 | 0.685 ± 0.0928 | 1.00 | up | 0.75 |
| C3:1 | 0.0501 ± 0.00367 | 0.047 ± 0.00275 | 1.07 | up | 0.9 |
| C3 | 0.0492 ± 0.00432 | 0.0573 ± 0.00324 | 0.86 | down | 0.17 |
| C4:1 | 0.0729 ± 0.0089 | 0.0767 ± 0.00667 | 0.95 | down | 0.86 |
| C4 | 0.484 ± 0.0904 | 0.423 ± 0.0677 | 1.14 | up | 0.24 |
| C3OH | 0.0695 ± 0.00729 | 0.0714 ± 0.00546 | 0.97 | down | 0.83 |
| C5:1 | 0.259 ± 0.0338 | 0.152 ± 0.0253 | 1.70 | up | 0.005 |
| C5 | 0.155 ± 0.025 | 0.154 ± 0.0187 | 1.01 | up | 0.88 |
| C4OH | 0.0785 ± 0.00747 | 0.0739 ± 0.0056 | 1.06 | down | 0.52 |
| C6:1 | 0.0691 ± 0.0081 | 0.0841 ± 0.00607 | 0.82 | down | 0.28 |
| C6 | 0.0857 ± 0.0145 | 0.1026 ± 0.0108 | 0.84 | down | 0.61 |
| C5OH | 0.14 ± 0.0152 | 0.109 ± 0.0114 | 1.28 | up | 0.02 |
| C5:1DC | 0.0453 ± 0.00418 | 0.0349 ± 0.00313 | 1.30 | up | 0.07 |
| C5DC | 0.048 ± 0.00412 | 0.0282 ± 0.00309 | 1.70 | up | <0.001 |
| C8 | 0.0483 ± 0.00476 | 0.0428 ± 0.00357 | 1.13 | up | 0.33 |
| C5MDC | 0.0483 ± 0.0046 | 0.0482 ± 0.00345 | 1.00 | up | 0.79 |
| C9 | 0.152 ± 0.0188 | 0.11 ± 0.0141 | 1.38 | up | 0.04 |
| C7DC | 0.0488 ± 0.00611 | 0.0281 ± 0.00458 | 1.74 | up | 0.005 |
| C10:2 | 0.0466 ± 0.00574 | 0.0514 ± 0.0043 | 0.91 | down | 0.96 |
| C10:1 | 0.204 ± 0.0213 | 0.176 ± 0.016 | 1.16 | up | 0.23 |
| C10 | 0.12 ± 0.0151 | 0.136 ± 0.0113 | 0.88 | down | 0.84 |
| C12:1 | 0.083 ± 0.0315 | 0.229 ± 0.0236 | 0.36 | down | 0.002 |
| C12 | 0.0444 ± 0.0184 | 0.091 ± 0.0138 | 0.49 | up | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zwierzchowski, G.; Haxhiaj, K.; Wójcik, R.; Wishart, D.S.; Ametaj, B.N. Identifying Predictive Biomarkers of Subclinical Mastitis in Dairy Cows through Urinary Metabotyping. Metabolites 2024, 14, 205. https://doi.org/10.3390/metabo14040205
Zwierzchowski G, Haxhiaj K, Wójcik R, Wishart DS, Ametaj BN. Identifying Predictive Biomarkers of Subclinical Mastitis in Dairy Cows through Urinary Metabotyping. Metabolites. 2024; 14(4):205. https://doi.org/10.3390/metabo14040205
Chicago/Turabian StyleZwierzchowski, Grzegorz, Klevis Haxhiaj, Roman Wójcik, David S. Wishart, and Burim N. Ametaj. 2024. "Identifying Predictive Biomarkers of Subclinical Mastitis in Dairy Cows through Urinary Metabotyping" Metabolites 14, no. 4: 205. https://doi.org/10.3390/metabo14040205
APA StyleZwierzchowski, G., Haxhiaj, K., Wójcik, R., Wishart, D. S., & Ametaj, B. N. (2024). Identifying Predictive Biomarkers of Subclinical Mastitis in Dairy Cows through Urinary Metabotyping. Metabolites, 14(4), 205. https://doi.org/10.3390/metabo14040205