Integrated Analysis of Metabolomics Combined with Network Pharmacology and Molecular Docking Reveals the Effects of Processing on Metabolites of Dendrobium officinale
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
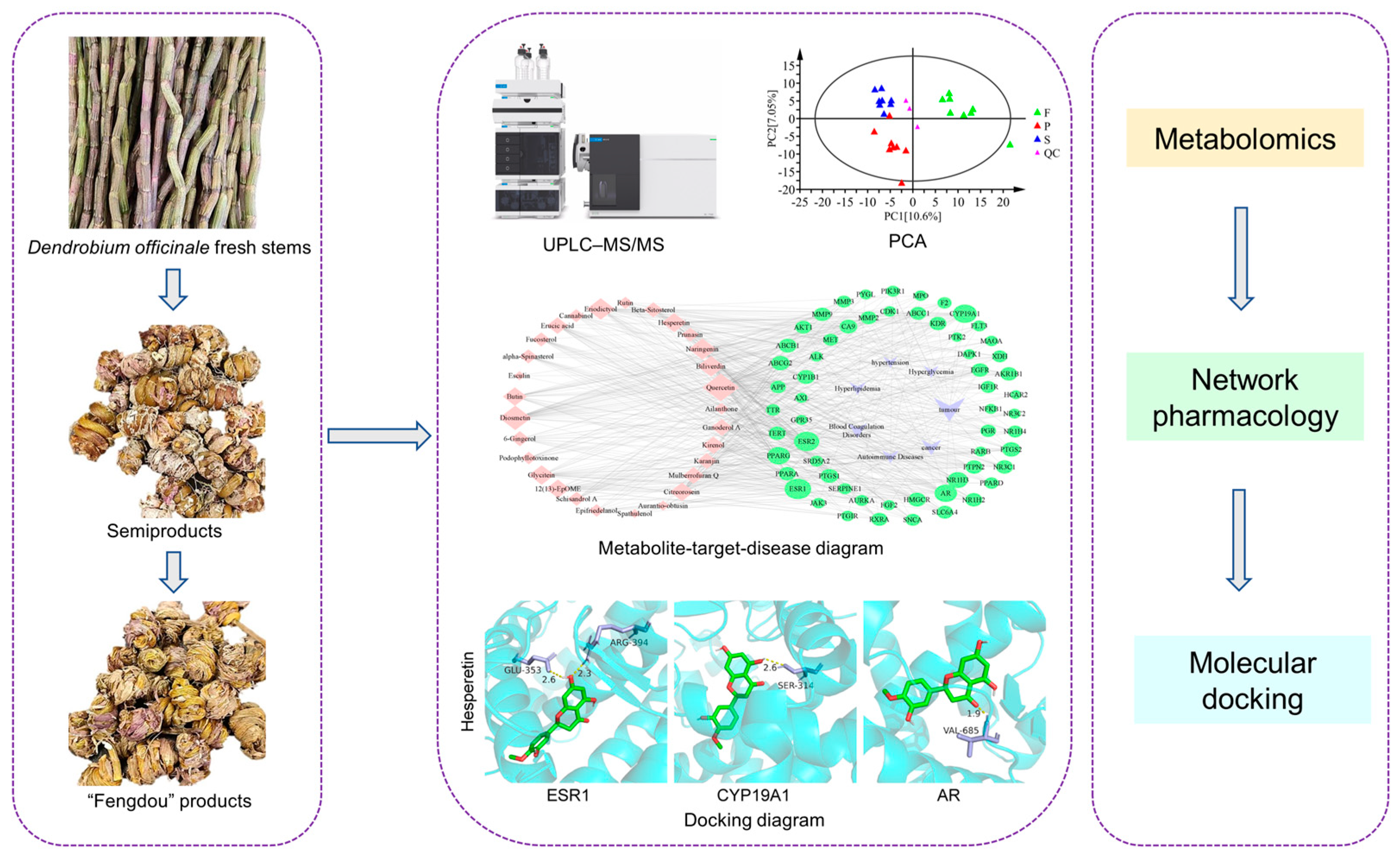
2.2. Materials
2.2.1. Raw Materials
2.2.2. Pretreatment
2.3. Determination of the Main Medicinal Components
2.3.1. Total Polysaccharide Content of D. officinale
2.3.2. Total Flavonoid Content of D. officinale
2.4. Targeted Metabolomics Based on UPLC-MS/MS
2.4.1. Sample Preparation
2.4.2. UPLC-MS/MS Analysis
2.4.3. Data Preprocessing and Annotation
2.4.4. Statistical Analysis
2.5. Network Pharmacology Analysis
2.5.1. Target Genes of Metabolites
2.5.2. Target Genes of Diseases
2.5.3. Network Construction
2.6. Molecular Docking
3. Results and Discussion
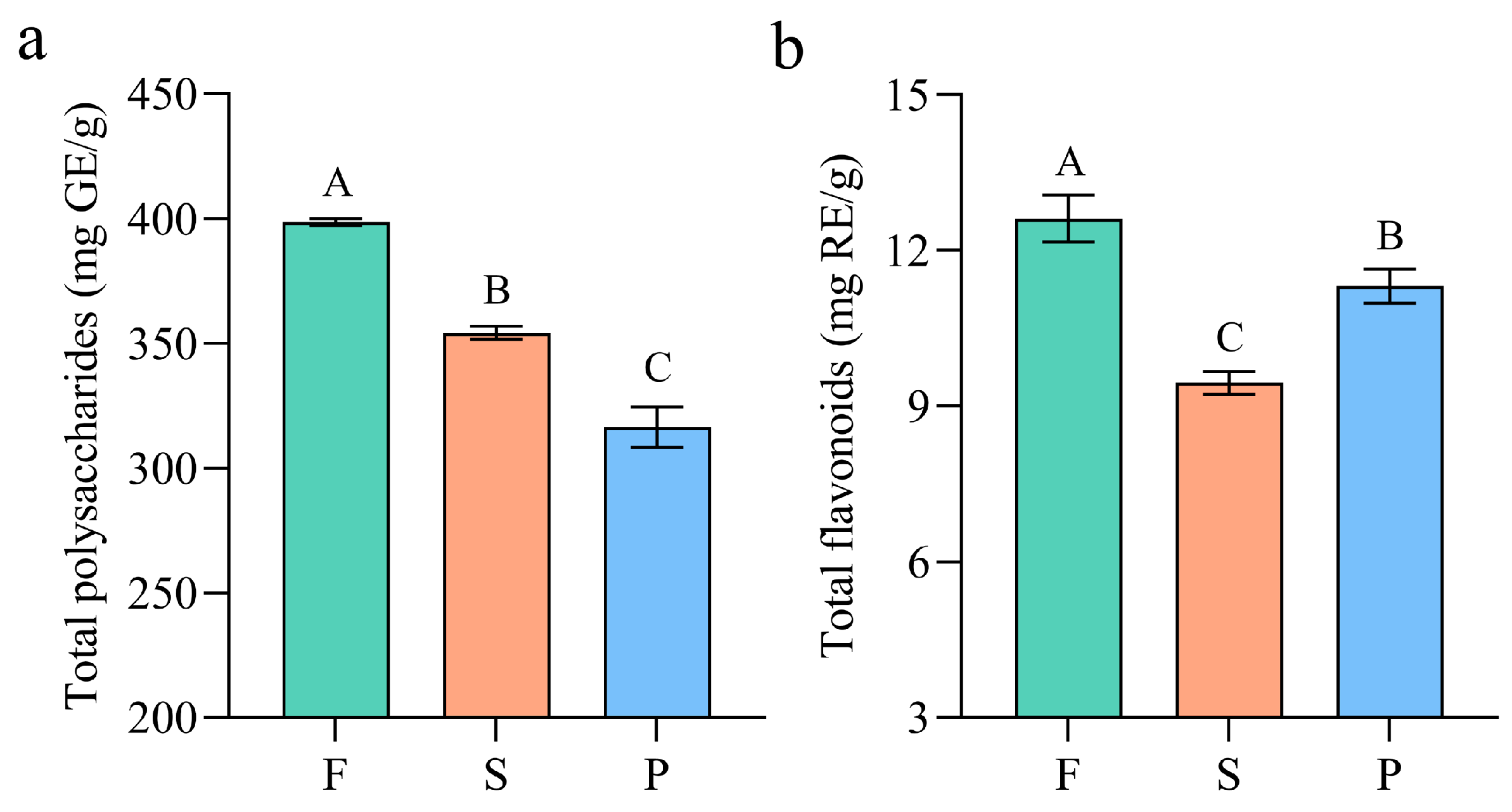
3.1. Effects of Processing on the Contents of Total Polysaccharide and Flavonoid in D. officinale
3.2. Targeted Metabolomics Analysis of D. officinale Stems at Different Processing Stages
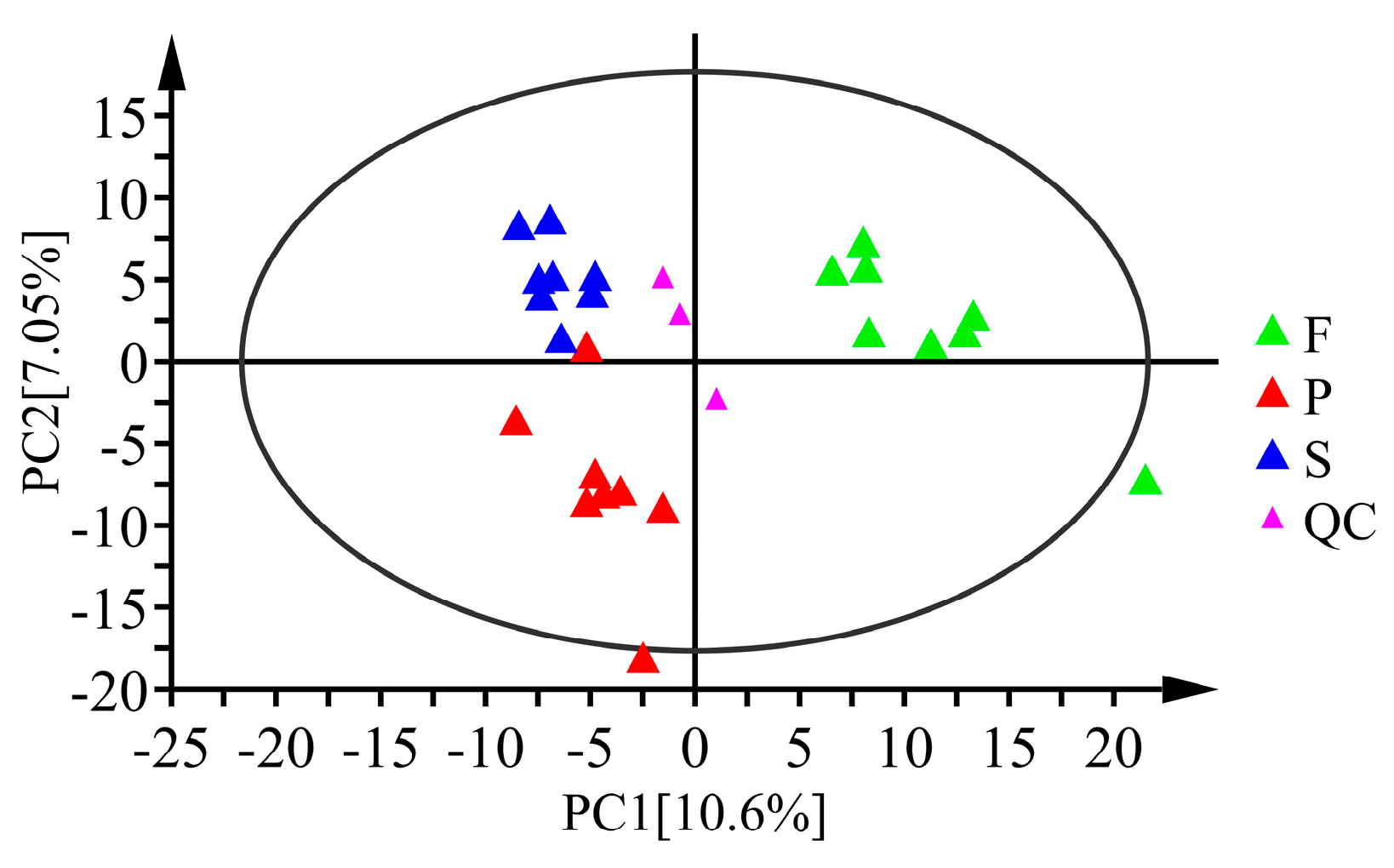
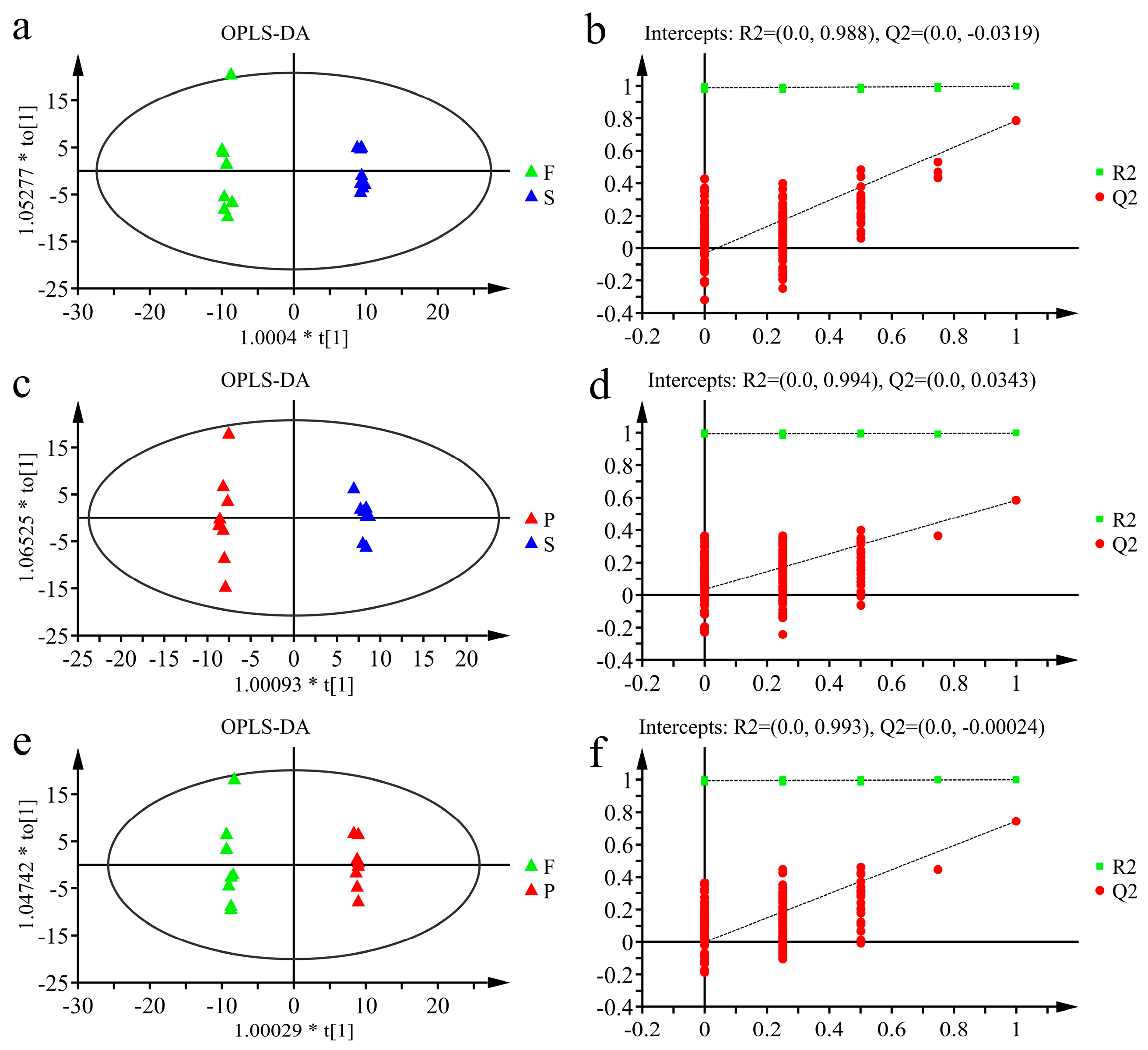
3.2.1. Multivariate Statistical Analysis
3.2.2. Identification of Differential Metabolites
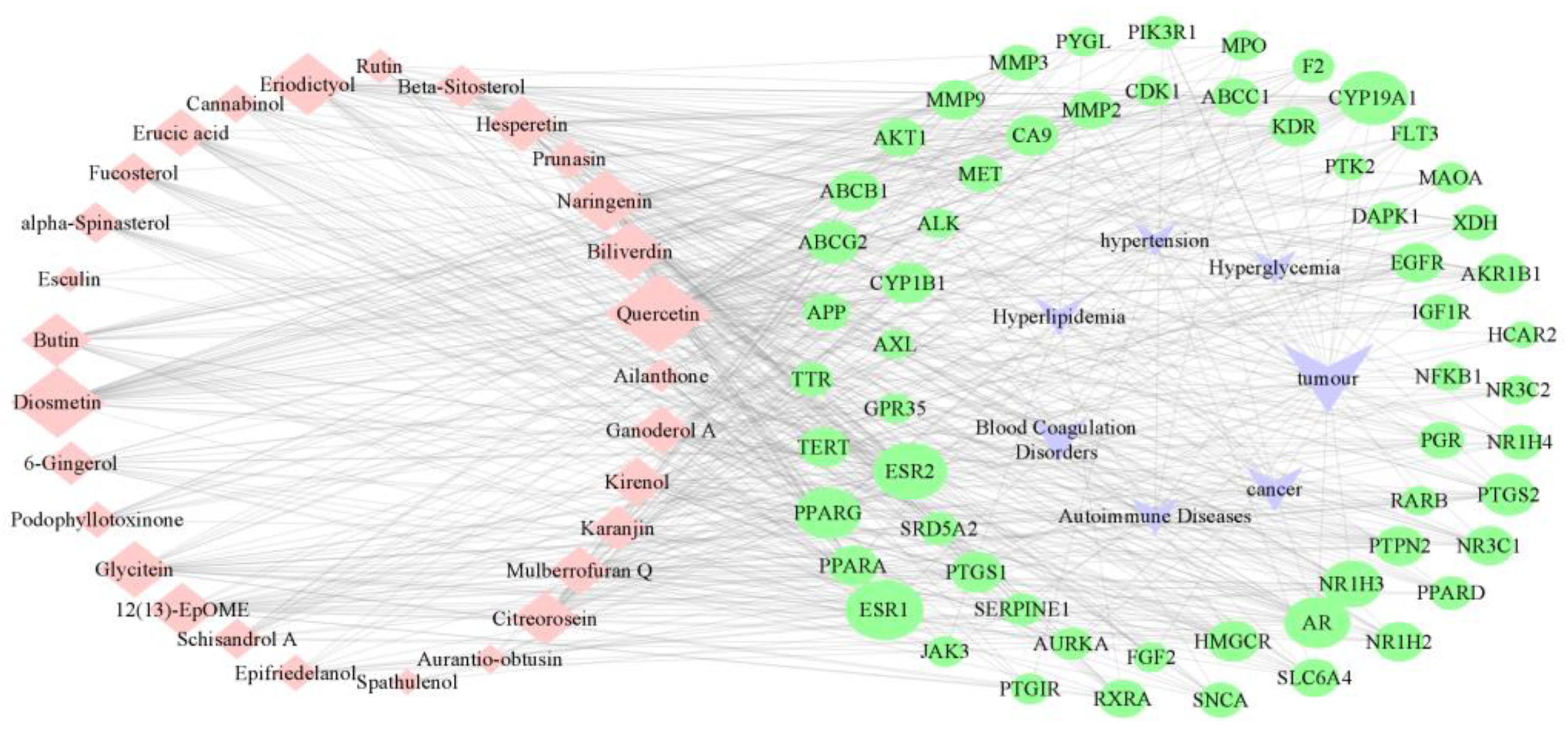
3.3. Network Pharmacology Analysis
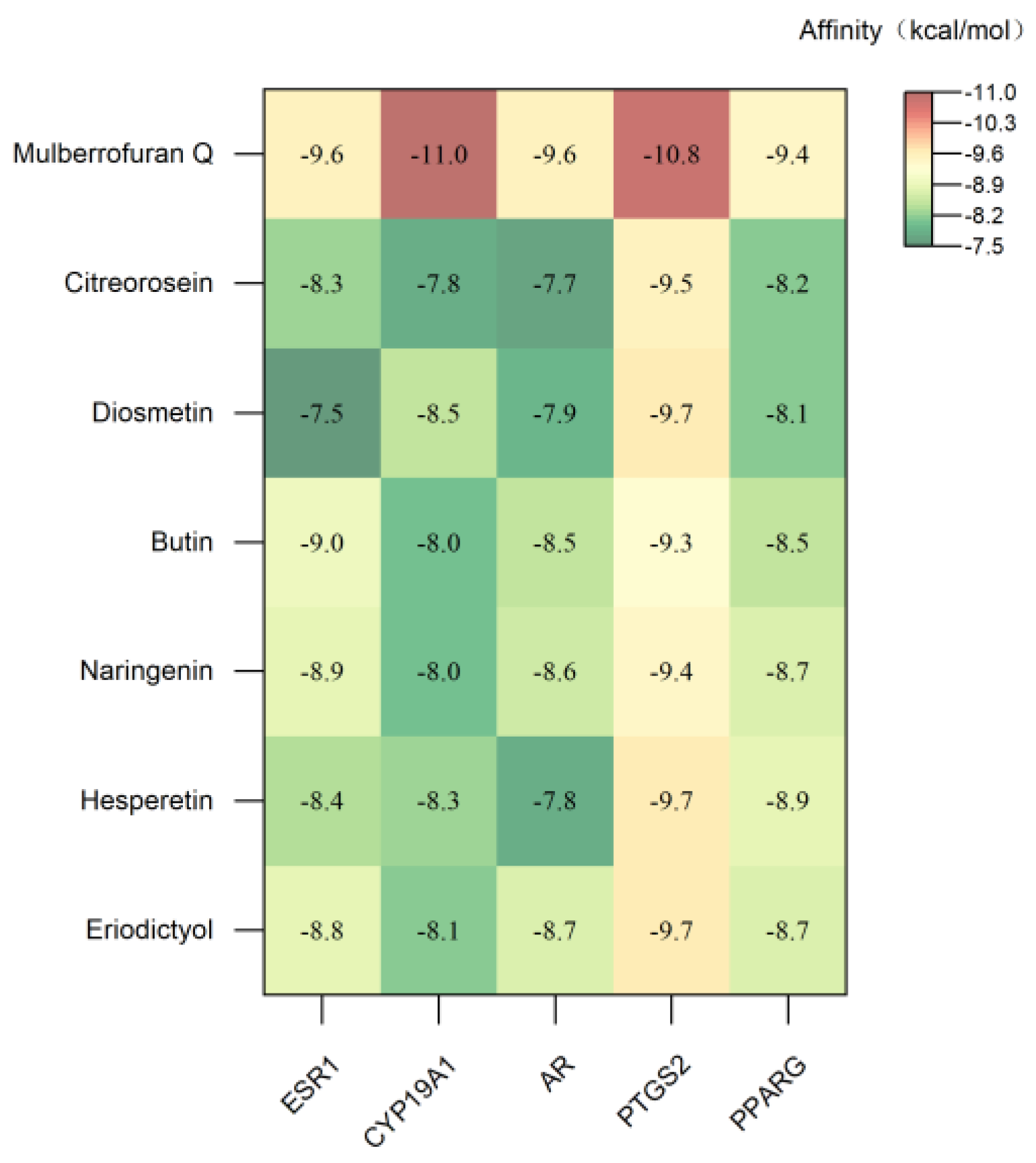
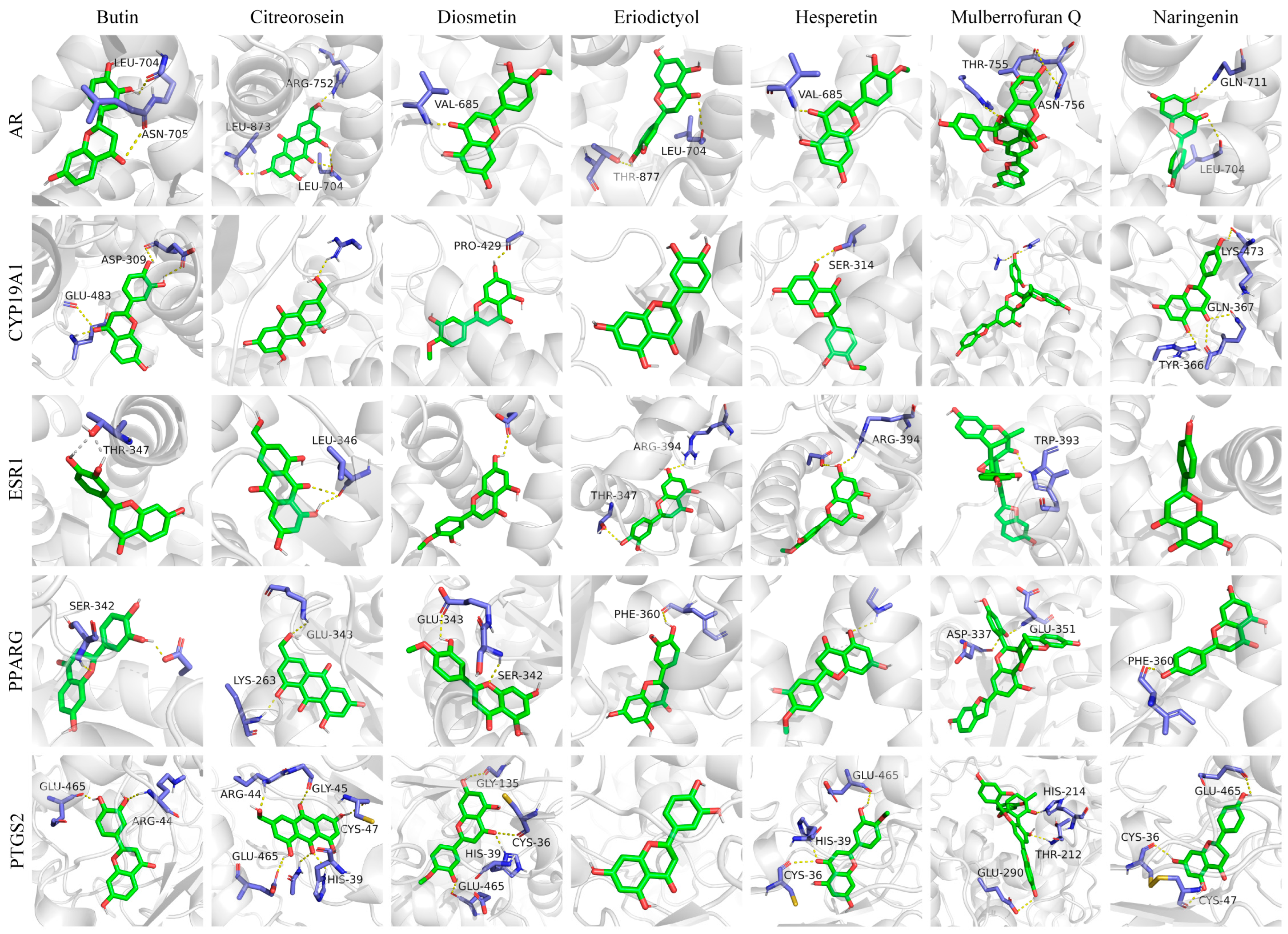
3.4. Molecular Docking Analysis
3.5. Metabolite Relative Content Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, B.; Tian, M.; Luo, J.; Ji, Y.; Xue, Q.; Ding, X. Genetic diversity assessment and ex situ conservation strategy of the endangered Dendrobium officinale (Orchidaceae) using new trinucleotide microsatellite markers. Plant Syst. Evol. 2012, 298, 1483–1491. [Google Scholar] [CrossRef]
- Hou, B.; Luo, J.; Zhang, Y.; Niu, Z.; Xue, Q.; Ding, X. Iteration expansion and regional evolution: Phylogeography of Dendrobium officinale and four related taxa in southern China. Sci. Rep. 2017, 7, 43525. [Google Scholar] [PubMed]
- Xu, J.; Han, Q.-B.; Li, S.-L.; Chen, X.-J.; Wang, X.-N.; Zhao, Z.-Z.; Chen, H.-B. Chemistry, bioactivity and quality control of Dendrobium, a commonly used tonic herb in traditional Chinese medicine. Phytochem. Rev. 2013, 12, 341–367. [Google Scholar] [CrossRef]
- Yuan, Y.; Tang, X.; Jia, Z.; Li, C.; Ma, J.; Zhang, J. The Effects of Ecological Factors on the Main Medicinal Components of Dendrobium officinale under Different Cultivation Modes. Forests 2020, 11, 94. [Google Scholar]
- Liu, H.; Liang, J.; Xiao, G.; Ma, L.; Wang, Q. Dendrobine Suppresses Lipopolysaccharide-induced Gut Inflammation in a Co-culture of Intestinal Epithelial Caco2 Cells and RAW264.7 Macrophages. eFood 2021, 2, 92–99. [Google Scholar]
- Zhao, Y.; Liu, Y.; Lan, X.-M.; Xu, G.-L.; Sun, Y.-Z.; Li, F.; Liu, H.-N. Effect of Dendrobium officinale Extraction on Gastric Carcinogenesis in Rats. Evid.-Based Complement. Altern. Med. 2016, 2016, 1213090. [Google Scholar]
- Liang, J.; Li, H.; Chen, J.; He, L.; Du, X.; Zhou, L.; Xiong, Q.; Lai, X.; Yang, Y.; Huang, S.; et al. Dendrobium officinale polysaccharides alleviate colon tumorigenesis via restoring intestinal barrier function and enhancing anti-tumor immune response. Pharmacol. Res. 2019, 148, 104417. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, B.; Wang, G.; Ge, S.; Lan, X.; Xu, G.; Liu, H. Dendrobium officinale Polysaccharides Inhibit 1-Methyl-2-Nitro-1-Nitrosoguanidine Induced Precancerous Lesions of Gastric Cancer in Rats through Regulating Wnt/beta-Catenin Pathway and Altering Serum Endogenous Metabolites. Molecules 2019, 24, 2660. [Google Scholar] [CrossRef]
- Pan, L.-H.; Li, X.-F.; Wang, M.-N.; Zha, X.-Q.; Yang, X.-F.; Liu, Z.-J.; Luo, Y.-B.; Luo, J.-P. Comparison of hypoglycemic and antioxidative effects of polysaccharides from four different Dendrobium species. Int. J. Biol. Macromol. 2014, 64, 420–427. [Google Scholar] [CrossRef]
- Wang, X.Y.; Yin, J.Y.; Hu, J.L.; Nie, S.P.; Xie, M.Y. Gastroprotective polysaccharide from natural sources: Review on structure, mechanism, and structure–activity relationship. Food Front. 2022, 3, 560–591. [Google Scholar] [CrossRef]
- Huang, K.; Li, Y.; Tao, S.; Wei, G.; Huang, Y.; Chen, D.; Wu, C. Purification, Characterization and Biological Activity of Polysaccharides from Dendrobium officinale. Molecules 2016, 21, 701. [Google Scholar] [CrossRef]
- Khan, N.; Adhami, V.M.; Mukhtar, H. Apoptosis by dietary agents for prevention and treatment of prostate cancer. Endocr.-Relat. Cancer 2010, 17, R39–R52. [Google Scholar] [CrossRef]
- Zhao, X.; Dou, M.; Zhang, Z.; Zhang, D.; Huang, C. Protective effect of Dendrobium officinale polysaccharides on H2O2-induced injury in H9c2 cardiomyocytes. Biomed. Pharmacother. 2017, 94, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Sun, H.-M.; Jia, J.-P.; Qin, X.-M.; Li, Z.-Y. Integrative hepatoprotective efficacy comparison of raw and vinegar-baked Radix Bupleuri using nuclear magnetic resonance-based metabolomics. J. Pharm. Biomed. Anal. 2017, 138, 215–222. [Google Scholar] [CrossRef]
- Meng, Q.; Fan, H.; Li, Y.; Zhang, L. Effect of drying methods on physico-chemical properties and antioxidant activity of Dendrobium officinale. J. Food Meas. Charact. 2018, 12, 1–10. [Google Scholar] [CrossRef]
- Nie, H.; Chen, H.; Li, G.; Su, K.; Song, M.; Duan, Z.; Li, X.; Cao, X.; Huang, J.; Huang, S.; et al. Comparison of flavonoids and phenylpropanoids compounds in Chinese water chestnut processed with different methods. Food Chem. 2021, 335, 127662. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.-J.; Hua, Y.-L.; Ji, P.; Yao, W.-L.; Zhang, W.-Q.; Li, J.; Wei, Y.-M. Evaluation of the anti-inflammatory effects of volatile oils from processed products of Angelica sinensis radix by GC MS-based metabolomics. J. Ethnopharmacol. 2016, 191, 195–205. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Z.; Gmitter, F.G., Jr.; Grosser, J.W.; Wang, Y. Effects of Different Rootstocks on the Metabolites of Huanglongbing-Affected Sweet Orange Juices Using a Novel Combined Strategy of Untargeted Metabolomics and Machine Learning. J. Agric. Food Chem. 2023, 71, 1246–1257. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Xiao, M.; Qian, K.; Wang, Y.; Bao, F. GC-MS metabolomics reveals metabolic differences of the farmed Mandarin fish Siniperca chuatsi in recirculating ponds aquaculture system and pond. Sci. Rep. 2020, 10, 6090. [Google Scholar] [CrossRef]
- Wang, M.; Gong, C.; Amakye, W.K.; Ren, J. Exploring the Mechanisms of Anti-Aβ42 Aggregation Activity of Walnut-derived Peptides using Transcriptomics and Proteomics in vitro. eFood 2021, 2, 247–258. [Google Scholar] [CrossRef]
- Chen, L.; Huang, X.; Wang, H.; Shao, J.; Luo, Y.; Zhao, K.; Liu, Y.; Wang, S. Integrated metabolomics and network pharmacology strategy for ascertaining the quality marker of flavonoids for Sophora flavescens. J. Pharm. Biomed. Anal. 2020, 186, 113297. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.-M.; Yu, H.-D.; Yun, Y.-H.; Zhang, W.; Zhong, Q.; Chen, W.; Chen, W.; Chen, H. Comparative Metabolomic Analysis of Dendrobium officinale under Different Cultivation Substrates. Metabolites 2020, 10, 325. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Sun, X.; Miao, Y.; Qin, S.; Jiang, Y.; Zhang, X.; Huang, L. A systematic study on the chemical diversity and efficacy of the inflorescence and succulent stem of Cynomorium songaricum. Food Funct. 2021, 12, 7501–7513. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [PubMed]
- Li, H.; Guo, L.; Ding, X.; An, Q.; Wang, L.; Hao, S.; Li, W.; Wang, T.; Gao, Z.; Zheng, Y.; et al. Molecular Networking, Network Pharmacology, and Molecular Docking Approaches Employed to Investigate the Changes in Ephedrae Herba before and after Honey-Processing. Molecules 2022, 27, 4057. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, W.; Huang, C.; Li, Y.; Yu, H.; Wang, Y.; Duan, J.; Ling, Y. A Novel Chemometric Method for the Prediction of Human Oral Bioavailability. Int. J. Mol. Sci. 2012, 13, 6964–6982. [Google Scholar] [CrossRef]
- Tao, W.; Xu, X.; Wang, X.; Li, B.; Wang, Y.; Li, Y.; Yang, L. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J. Ethnopharmacol. 2013, 145, 1–10. [Google Scholar] [CrossRef]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. Swiss Target Prediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, F.; Wang, Y.; Li, C.; Zhang, X.; Li, H.; Diao, L.; Gu, J.; Wang, W.; Li, D.; et al. BATMAN-TCM: A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016, 6, 21146. [Google Scholar] [CrossRef]
- Li, Z.-T.; Zhang, F.-X.; Fan, C.-L.; Ye, M.-N.; Chen, W.-W.; Yao, Z.-H.; Yao, X.-S.; Dai, Y. Discovery of potential Q-marker of traditional Chinese medicine based on plant metabolomics and network pharmacology: Periplocae Cortex as an example. Phytomedicine 2021, 85, 153535. [Google Scholar] [CrossRef]
- Pinero, J.; Manuel Ramirez-Anguita, J.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Stein, T.I.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Bing, Z.; Han, J.; An, X.; Liu, X.; Li, R.; Wang, C.; Sun, X.; Yang, L.; Yang, K. Study on the anti-tumor mechanism related to immune microenvironment of Bombyx Batryticatus on viral and non-viral infections of hepatocellular carcinoma. Biomed. Pharmacother. 2020, 124, 109838. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.H.; Sun, F.; Yan, B.; Li, J.Y.; Xin, D.L. Data mining and systematic pharmacology to reveal the mechanisms of traditional Chinese medicine in Mycoplasma pneumoniae pneumonia treatment. Biomed. Pharmacother. 2020, 125, 109900. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- El-Banna, A.A.; Darwish, R.S.; Ghareeb, D.A.; Yassin, A.M.; Abdulmalek, S.A.; Dawood, H.M. Metabolic profiling of Lantana camara L. using UPLC-MS/MS and revealing its inflammation-related targets using network pharmacology-based and molecular docking analyses. Sci. Rep. 2022, 12, 14828. [Google Scholar] [CrossRef]
- Yue, H.; Zeng, H.; Ding, K. A review of isolation methods, structure features and bioactivities of polysaccharides from Dendrobium species. Chin. J. Nat. Med. 2020, 18, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Li, C.Y.; Hu, C.; Gong, P.S.; Zhao, S.H. Purification and Structural Characterization of Dendrobium officinale Polysaccharides and Its Activities. Chem. Biodivers. 2021, 18, e2001023. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, L.; Chang, H.; Cai, B.; Gao, H.; Chen, G.; Hou, W.; Jappar, Z.; Yan, Y. Research progress on extraction, purification, structure and biological activity of Dendrobium officinale polysaccharides. Front. Nutr. 2022, 9, 965073. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zuo, J.; Zhang, H.; Zu, M.; Yu, M.; Liu, S. Transcriptome and metabolome profiling unveil the accumulation of flavonoids in Dendrobium officinale. Genomics 2022, 114, 110324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Liu, J.; Liang, J.; Si, J.; Wu, S. Dendrobium officinale leaves as a new antioxidant source. J. Funct. Foods 2017, 37, 400–415. [Google Scholar] [CrossRef]
- Ravisankar, S.; Queiroz, V.A.V.; Awika, J.M. Rye flavonoids—Structural profile of the flavones in diverse varieties and effect of fermentation and heat on their structure and antioxidant properties. Food Chem. 2020, 324, 126871. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, P.; Tang, K.; Shen, G.; Chen, H.; Zhang, Z.; Zhao, W.; Shang, Q.; Zhu, G.; Tan, R.; et al. Network Pharmacology Integrated with Molecular Docking Explores the Mechanisms of Naringin against Osteoporotic Fracture by Regulating Oxidative Stress. Evid.-Based Complement. Altern. Med. 2021, 2021, 6421122. [Google Scholar] [CrossRef]
- Wong, F.; Krishnan, A.; Zheng, E.J.; Stark, H.; Manson, A.L.; Earl, A.M.; Jaakkola, T.; Collins, J.J. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 2022, 18, e11081. [Google Scholar] [CrossRef]
- Li, L.Z.; Wang, H.Y.; Huang, J.H.; Liu, K.; Feng, X.J.; Wang, X.M.; Zhu, L.J.; He, X.L.; Zheng, X.; Li, H.L.; et al. The Mechanism of Dendrobium officinale as a Treatment for Hyperlipidemia Based on Network Pharmacology and Experimental Validation. Evid.-Based Complement. Altern. Med. 2022, 2022, 5821829. [Google Scholar] [CrossRef]
- Tao, S.; Li, J.; Wang, H.; Ding, S.; Han, W.; He, R.; Ren, Z.; Wei, G. Anti-colon Cancer Effects of Dendrobium officinale Kimura & Migo Revealed by Network Pharmacology Integrated with Molecular Docking and Metabolomics Studies. Front. Med. 2022, 9, 879986. [Google Scholar]
- Mueller, M.; Lukas, B.; Novak, J.; Simoncini, T.; Genazzani, A.R.; Jungbauer, A. Oregano: A source for peroxisome proliferator-activated receptor gamma antagonists. J. Agric. Food Chem. 2008, 56, 11621–11630. [Google Scholar] [CrossRef]
- D’Arrigo, G.; Gianquinto, E.; Rossetti, G.; Cruciani, G.; Lorenzetti, S.; Spyrakis, F. Binding of Androgen- and Estrogen-Like Flavonoids to Their Cognate (Non)Nuclear Receptors: A Comparison by Computational Prediction. Molecules 2021, 26, 1613. [Google Scholar]
- Doostdar, H.; Burke, M.D.; Mayer, R.T. Bioflavonoids: Selective substrates and inhibitors for cytochrome P450 CYP1A and CYP1B1. Toxicology 2000, 144, 31–38. [Google Scholar] [CrossRef]
- Xie, B.; Pan, D.; Liu, H.; Liu, M.; Shi, X.; Chu, X.; Lu, J.; Zhu, M.; Xia, B.; Wu, J. Diosmetin Protects against Obesity and Metabolic Dysfunctions through Activation of Adipose Estrogen Receptors in Mice. Mol. Nutr. Food Res. 2021, 65, e2100070. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.S.; Lee, S.H.; Lee, K.A. A Comparative Study of Hesperetin, Hesperidin and Hesperidin Glucoside: Antioxidant, Anti-Inflammatory, and Antibacterial Activities In Vitro. Antioxidants 2022, 11, 1618. [Google Scholar] [PubMed]
- Hermawan, A.; Ikawati, M.; Khumaira, A.; Putri, H.; Jenie, R.I.; Angraini, S.M.; Muflikhasari, H.A. Bioinformatics and In Vitro Studies Reveal the Importance of p53, PPARG and Notch Signaling Pathway in Inhibition of Breast Cancer Stem Cells by Hesperetin. Adv. Pharm. Bull. 2021, 11, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Suh, S.J.; Li, X.; Hwang, S.L.; Li, Y.; Hwangbo, K.; Park, S.J.; Murakami, M.; Lee, S.H.; Jahng, Y.; et al. Citreorosein, a naturally occurring anthraquinone derivative isolated from Polygoni cuspidati radix, attenuates cyclooxygenase-2-dependent prostaglandin D2 generation by blocking Akt and JNK pathways in mouse bone marrow-derived mast cells. Food Chem. Toxicol. 2012, 50, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chae, S.; Kang, K.A.; Piao, M.J.; Ko, D.O.; Wang, Z.H.; Park, D.B.; Park, J.W.; You, H.J.; Hyun, J.W. Protective effect of butin against hydrogen peroxide-induced apoptosis by scavenging reactive oxygen species and activating antioxidant enzymes. Mol. Cell. Biochem. 2008, 318, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Kim, I.S.; Kim, S.A.; Na, C.S.; Hong, C.Y.; Dong, M.S.; Yoo, H.H. Inhibitory effect of Rhus verniciflua Stokes extract on human aromatase activity; butin is its major bioactive component. Bioorg. Med. Chem. Lett. 2014, 24, 1730–1733. [Google Scholar]
- Saltos, M.B.V.; Puente, B.F.N.; Faraone, I.; Milella, L.; Tommasi, N.D.; Braca, A. Inhibitors of α-amylase and α-glucosidase from Andromachia igniaria Humb. & Bonpl. Phytochem. Lett. 2015, 14, 45–50. [Google Scholar]
- Park, S.J.; Lee, Y.H.; Lee, K.H.; Kim, T.J. Effect of eriodictyol on the development of atopic dermatitis-like lesions in ICR mice. Biol. Pharm. Bull. 2013, 36, 1375–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Lee, J.J.; Kim, Y.; Kim, I.S.; Han, J.H.; Lee, S.G.; Ahn, M.J.; Jung, S.H.; Myung, C.S. Effect of eriodictyol on glucose uptake and insulin resistance in vitro. J. Agric. Food Chem. 2012, 60, 7652–7658. [Google Scholar] [CrossRef] [PubMed]

| Time/min | Mobile Phase A/% | Mobile Phase B/% |
|---|---|---|
| 0 | 98 | 2 |
| 0.5 | 98 | 2 |
| 10 | 50 | 50 |
| 11 | 5 | 95 |
| 13 | 5 | 95 |
| 13.1 | 98 | 98 |
| 15 | 98 | 98 |
| Class | F vs. S | S vs. P | F vs. P | |||
|---|---|---|---|---|---|---|
| Down | Up | Down | Up | Down | Up | |
| Flavonoids | 12 | 8 | 0 | 6 | 0 | 9 |
| Terpenoids | 13 | 2 | 0 | 5 | 3 | 3 |
| Alkaloids | 6 | 4 | 0 | 7 | 5 | 9 |
| Coumarins | 6 | 0 | 1 | 2 | 0 | 2 |
| Phenols | 7 | 1 | 0 | 3 | 4 | 3 |
| Lipids | 5 | 1 | 0 | 2 | 4 | 1 |
| Steroids | 8 | 0 | 0 | 4 | 2 | 0 |
| Anthraquinones | 0 | 0 | 1 | 0 | 1 | 1 |
| Organic acids | 3 | 0 | 0 | 0 | 2 | 0 |
| Others | 2 | 1 | 0 | 3 | 0 | 2 |
| Significantly differential metabolites | 62 | 17 | 2 | 32 | 21 | 30 |
| Total | 109 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Zuo, S.-M.; Liu, M.; Wang, T.; Li, Z.; Yun, Y.-H.; Zhang, W. Integrated Analysis of Metabolomics Combined with Network Pharmacology and Molecular Docking Reveals the Effects of Processing on Metabolites of Dendrobium officinale. Metabolites 2023, 13, 886. https://doi.org/10.3390/metabo13080886
Xu L, Zuo S-M, Liu M, Wang T, Li Z, Yun Y-H, Zhang W. Integrated Analysis of Metabolomics Combined with Network Pharmacology and Molecular Docking Reveals the Effects of Processing on Metabolites of Dendrobium officinale. Metabolites. 2023; 13(8):886. https://doi.org/10.3390/metabo13080886
Chicago/Turabian StyleXu, Lilan, Si-Min Zuo, Mei Liu, Tao Wang, Zizheng Li, Yong-Huan Yun, and Weimin Zhang. 2023. "Integrated Analysis of Metabolomics Combined with Network Pharmacology and Molecular Docking Reveals the Effects of Processing on Metabolites of Dendrobium officinale" Metabolites 13, no. 8: 886. https://doi.org/10.3390/metabo13080886
APA StyleXu, L., Zuo, S.-M., Liu, M., Wang, T., Li, Z., Yun, Y.-H., & Zhang, W. (2023). Integrated Analysis of Metabolomics Combined with Network Pharmacology and Molecular Docking Reveals the Effects of Processing on Metabolites of Dendrobium officinale. Metabolites, 13(8), 886. https://doi.org/10.3390/metabo13080886







