Production of Saffron Apocarotenoids in Nicotiana benthamiana Plants Genome-Edited to Accumulate Zeaxanthin Precursor
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Agroinfiltration
2.3. Virus Inoculation
2.4. Extraction and Analysis of Carotenoids
2.5. Extraction and Analysis of Crocins
2.6. Statistical Analyses
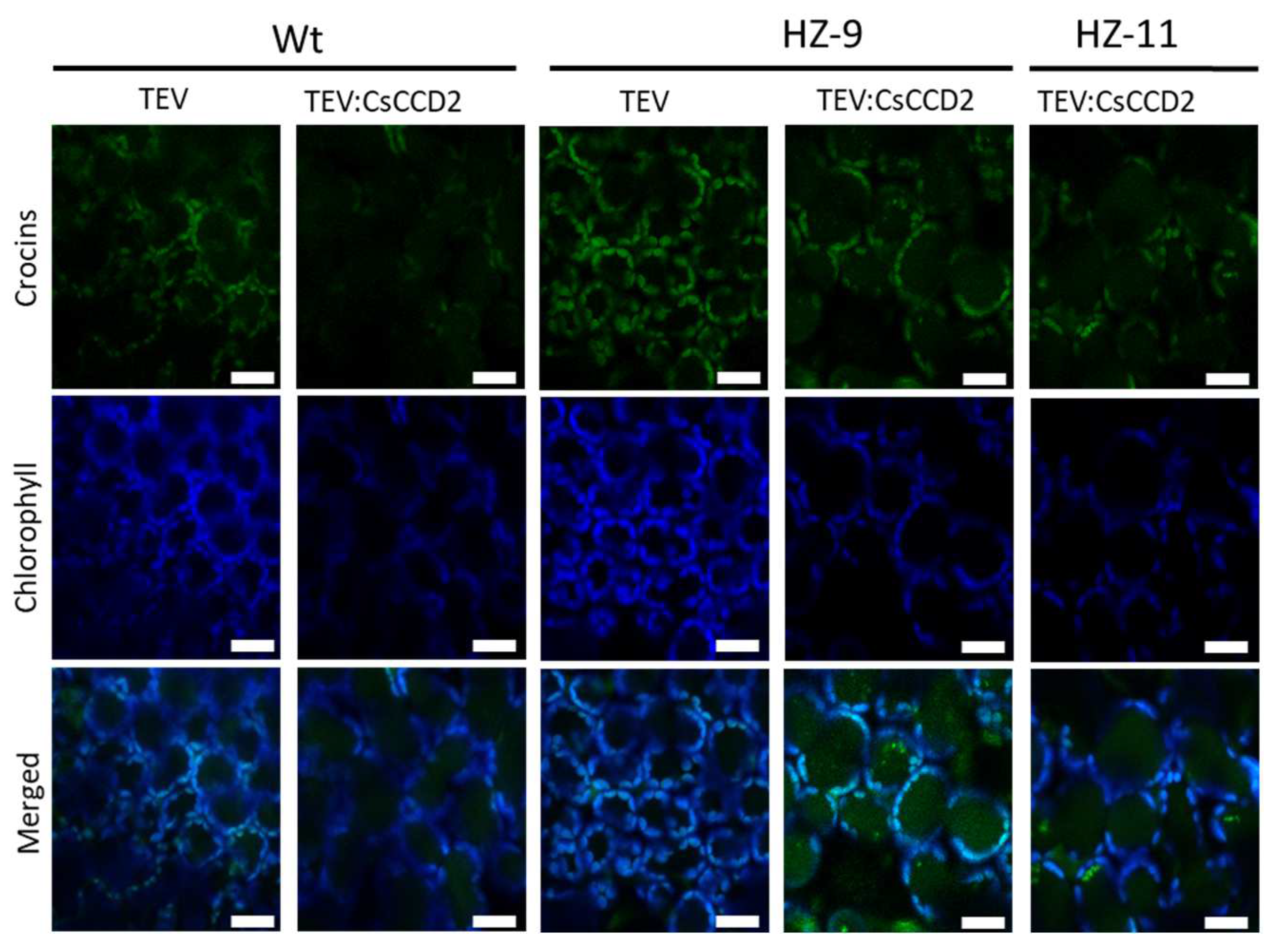
2.7. Confocal Microscopy
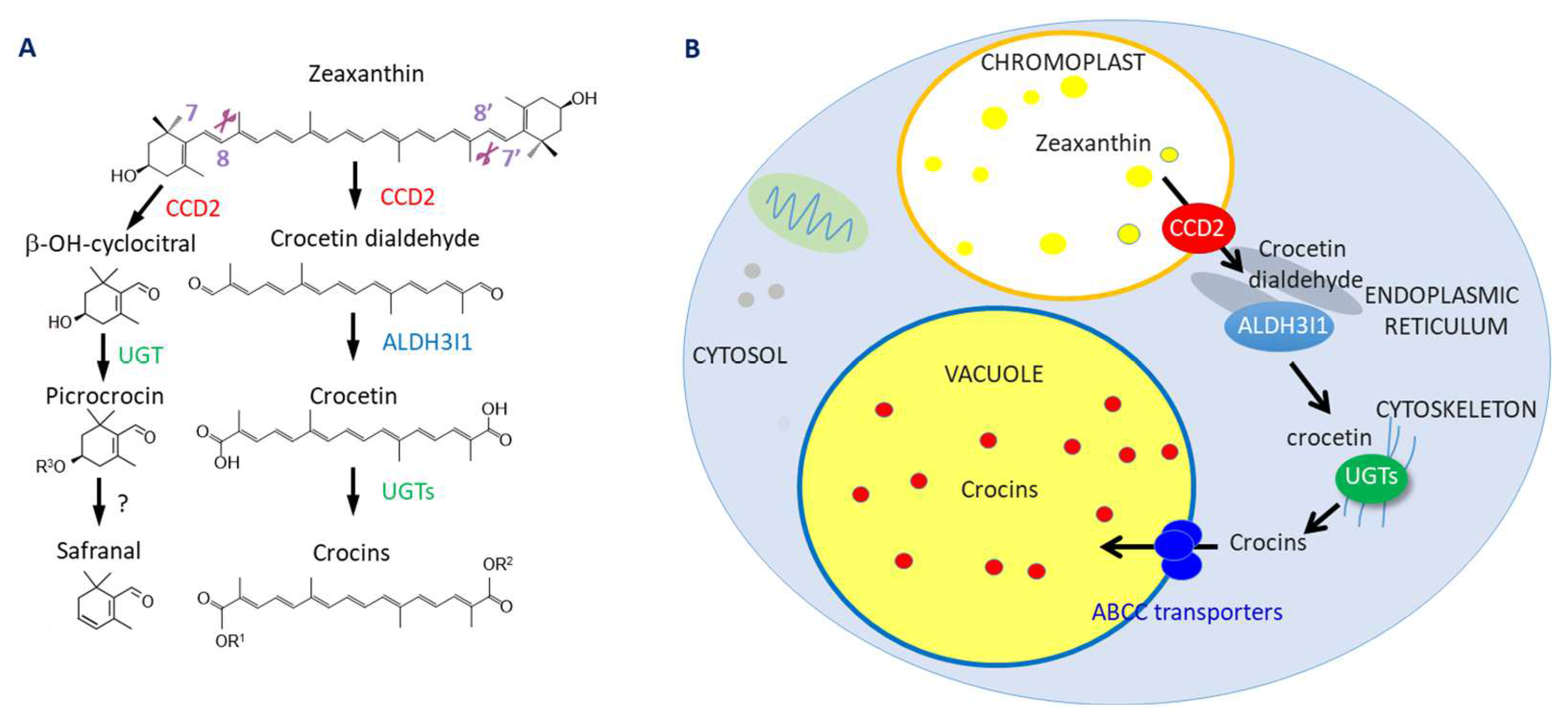
3. Results and Discussion
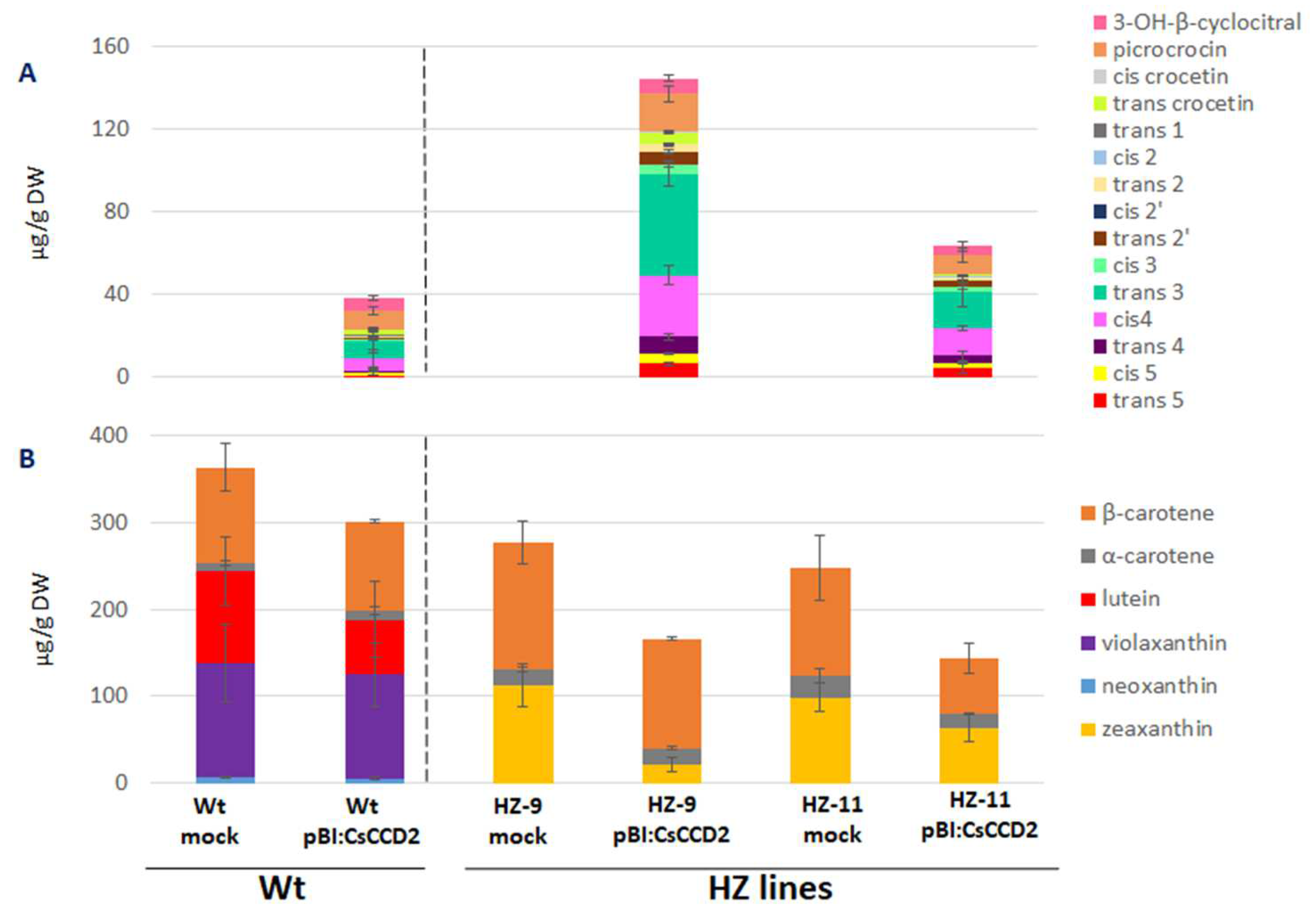
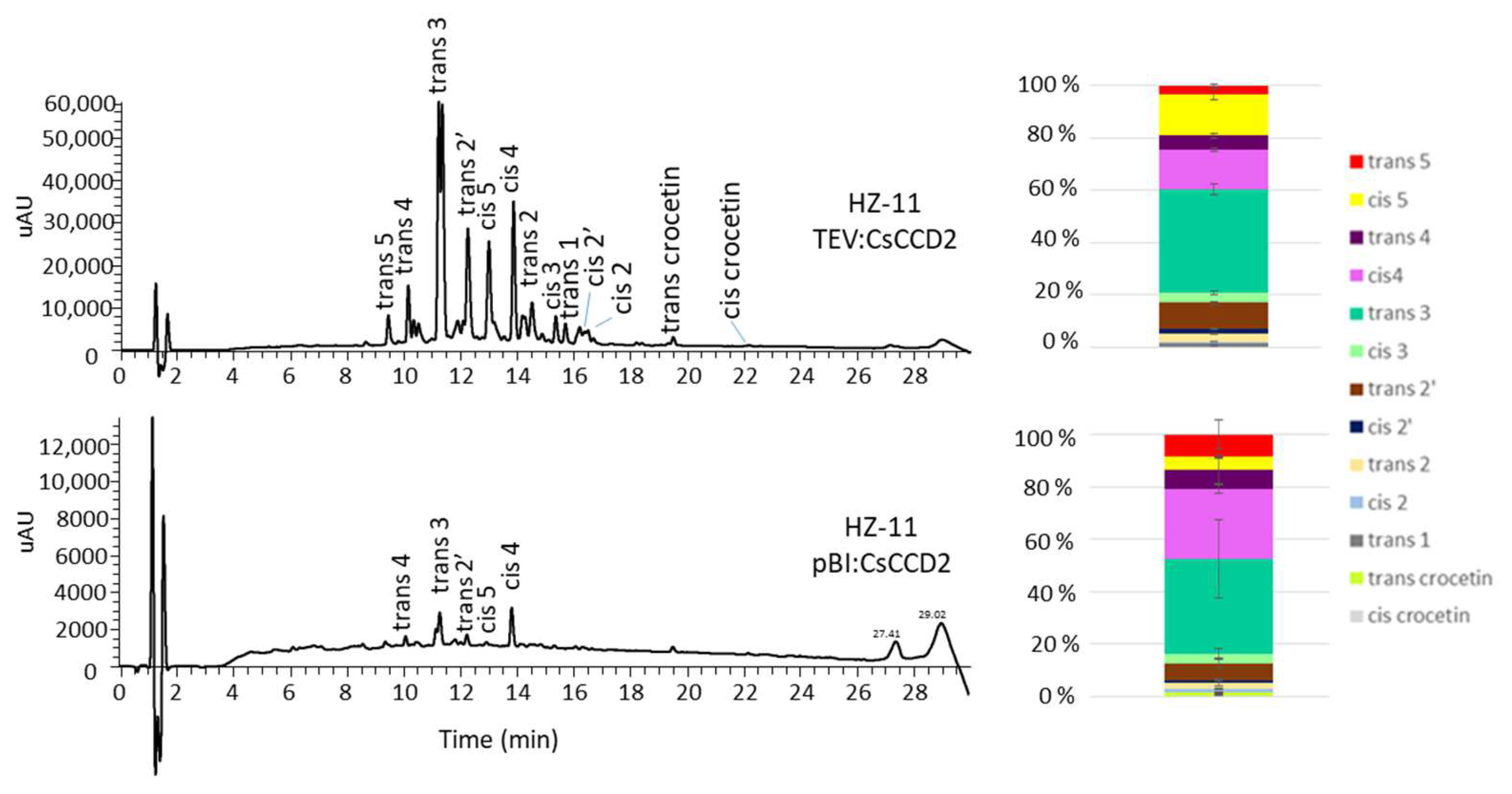
3.1. Selection of Edited Lines as Chassis for the Production of Crocins
3.2. Agroinfiltration
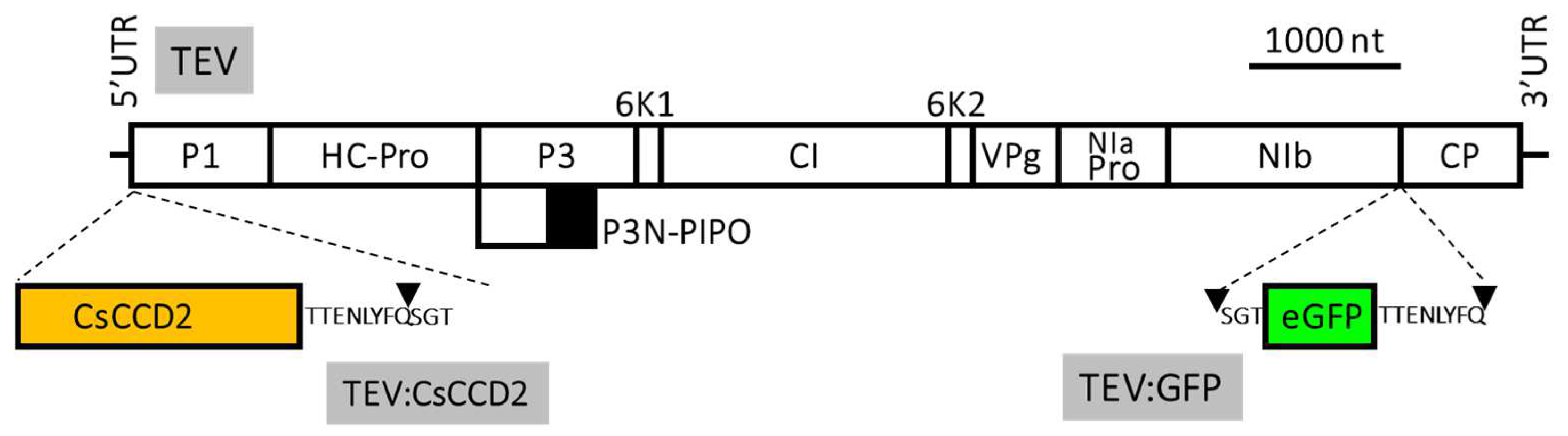
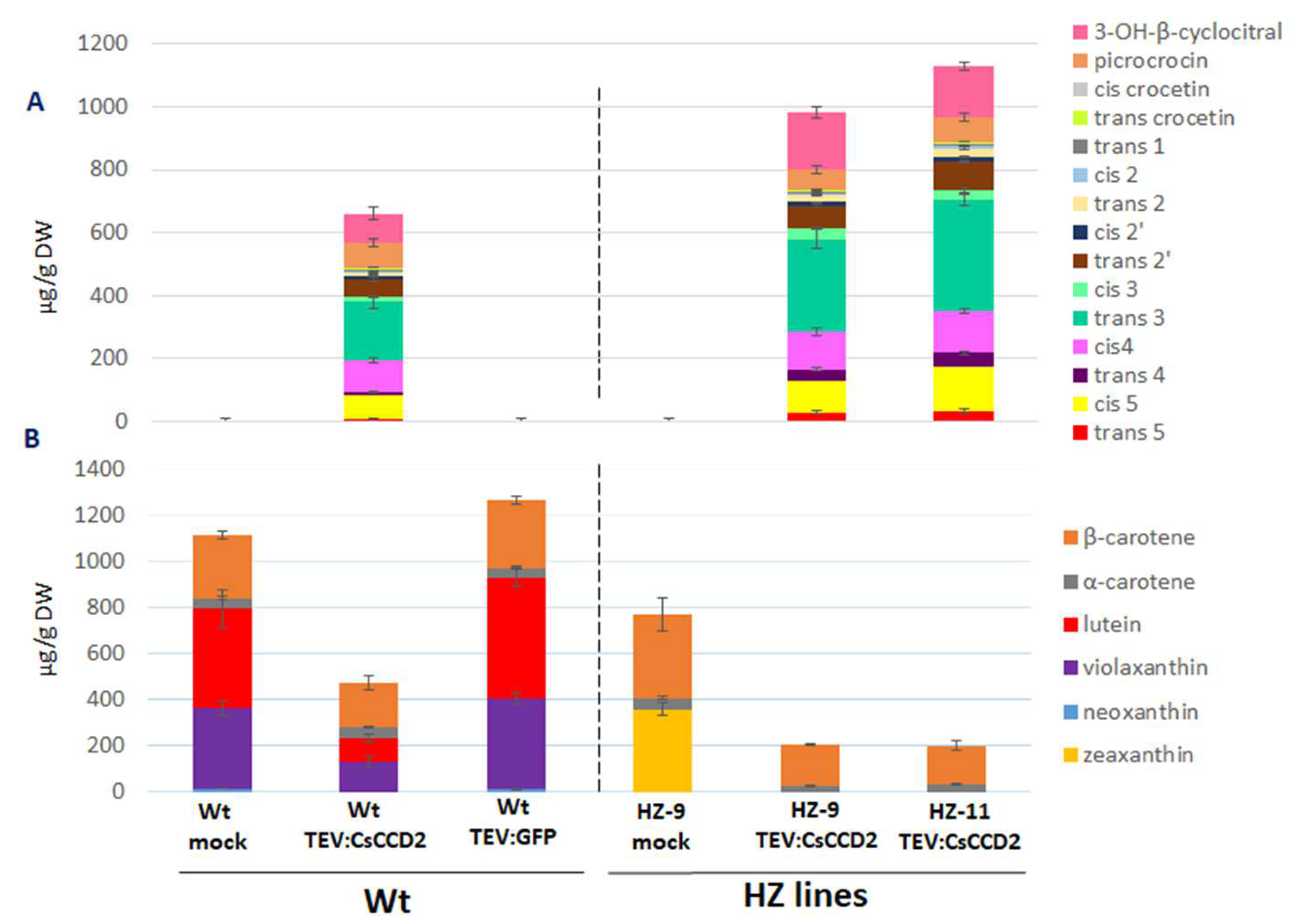
3.3. Viral Expression
3.4. Endogenous Factors and Their Contribution to Crocin Accumulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Rubio Moraga, A.; Ahrazem, O.; Rambla, J.L.; Granell, A.; Gómez Gómez, L. Crocins with high levels of sugar conjugation contribute to the yellow colours of early-spring flowering crocus tepals. PLoS ONE 2013, 8, e71946. [Google Scholar] [CrossRef] [PubMed]
- Frusciante, S.; Diretto, G.; Bruno, M.; Ferrante, P.; Pietrella, M.; Prado-Cabrero, A.; Rubio-Moraga, A.; Beyer, P.; Gomez-Gomez, L.; Al-Babili, S.; et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 12246–12251. [Google Scholar] [CrossRef]
- Demurtas, O.C.; Frusciante, S.; Ferrante, P.; Diretto, G.; Azad, N.H.; Pietrella, M.; Aprea, G.; Taddei, A.R.; Romano, E.; Mi, J.; et al. Candidate Enzymes for Saffron Crocin Biosynthesis Are Localized in Multiple Cellular Compartments. Plant Physiol. 2018, 177, 990–1006. [Google Scholar] [CrossRef] [PubMed]
- López-Jimenez, A.J.; Frusciante, S.; Niza, E.; Ahrazem, O.; Rubio-Moraga, Á.; Diretto, G.; Gómez-Gómez, L. A New Glycosyltransferase Enzyme from Family 91, UGT91P3, Is Responsible for the Final Glucosylation Step of Crocins in Saffron (Crocus sativus L.). Int. J. Mol. Sci. 2021, 22, 8815. [Google Scholar] [CrossRef] [PubMed]
- Diretto, G.; Ahrazem, O.; Rubio-Moraga, Á.; Fiore, A.; Sevi, F.; Argandoña, J.; Gómez-Gómez, L. UGT709G1: A novel uridine diphosphate glycosyltransferase involved in the biosynthesis of picrocrocin, the precursor of safranal in saffron (Crocus sativus). New Phytol. 2019, 224, 725–740. [Google Scholar] [CrossRef]
- Demurtas, O.C.; de Brito Francisco, R.; Diretto, G.; Ferrante, P.; Frusciante, S.; Pietrella, M.; Aprea, G.; Borghi, L.; Feeney, M.; Frigerio, L.; et al. ABCC Transporters Mediate the Vacuolar Accumulation of Crocins in Saffron Stigmas. Plant Cell 2019, 31, 2789–2804. [Google Scholar] [CrossRef]
- Xu, Z.; Pu, X.; Gao, R.; Demurtas, O.C.; Fleck, S.J.; Richter, M.; He, C.; Ji, A.; Sun, W.; Kong, J.; et al. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020, 18, 63. [Google Scholar] [CrossRef]
- Ahrazem, O.; Diretto, G.; Argandoña, J.; Rubio-Moraga, Á.; Julve, J.M.; Orzáez, D.; Granell, A.; Gómez-Gómez, L. Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii. J. Exp. Bot. 2017, 68, 4663–4677. [Google Scholar] [CrossRef]
- Diretto, G.; López-Jiménez, A.J.; Ahrazem, O.; Frusciante, S.; Song, J.; Rubio-Moraga, Á.; Gómez-Gómez, L. Identification and characterization of apocarotenoid modifiers and carotenogenic enzymes for biosynthesis of crocins in Buddleja davidii flowers. J. Exp. Bot. 2021, 72, 3200–3218. [Google Scholar] [CrossRef]
- Frusciante, S.; Demurtas, O.C.; Sulli, M.; Mini, P.; Aprea, G.; Diretto, G.; Karcher, D.; Bock, R.; Giuliano, G. Heterologous expression of Bixa orellana cleavage dioxygenase 4-3 drives crocin but not bixin biosynthesis. Plant Physiol. 2022, 188, 1469–1482. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gómez-Gómez, L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 650–663. [Google Scholar] [CrossRef]
- Zheng, X.; Mi, J.; Balakrishna, A.; Liew, K.X.; Ablazov, A.; Sougrat, R.; Al-Babili, S. Gardenia carotenoid cleavage dioxygenase 4a is an efficient tool for biotechnological production of crocins in green and non-green plant tissues. Plant Biotechnol. J. 2022, 20, 2202–2216. [Google Scholar] [CrossRef]
- Martí, M.; Diretto, G.; Aragonés, V.; Frusciante, S.; Ahrazem, O.; Gómez-Gómez, L.; Daròs, J.A. Efficient production of saffron crocins and picrocrocin in Nicotiana benthamiana using a virus-driven system. Metab. Eng. 2020, 61, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Zhu, C.; Huang, X.; Rubio-Moraga, A.; Capell, T.; Christou, P.; Gómez-Gómez, L. Metabolic Engineering of Crocin Biosynthesis in Nicotiana Species. Front. Plant Sci. 2022, 13, 861140. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Morote, L.; Zhu, C.; Ahrazem, O.; Capell, T.; Christou, P.; Gómez-Gómez, L. The Biosynthesis of Non-Endogenous Apocarotenoids in Transgenic Nicotiana glauca. Metabolites 2022, 12, 575. [Google Scholar] [CrossRef]
- Ahrazem, O.; Diretto, G.; Rambla, J.L.; Rubio-Moraga, Á.; Lobato-Gómez, M.; Frusciante, S.; Argandoña, J.; Presa, S.; Granell, A.; Gómez-Gómez, L. Engineering high levels of saffron apocarotenoids in tomato. Hortic. Res. 2022, 9, uhac074. [Google Scholar] [CrossRef]
- Bally, J.; Nakasugi, K.; Jia, F.; Jung, H.; Ho, S.Y.; Wong, M.; Paul, C.M.; Naim, F.; Wood, C.C.; Crowhurst, R.N.; et al. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 2015, 1, 15165. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [PubMed]
- Bedoya, L.; Martínez, F.; Rubio, L.; Daròs, J.A. Simultaneous equimolar expression of multiple proteins in plants from a disarmed potyvirus vector. J. Biotechnol. 2010, 150, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Majer, E.; Navarro, J.A.; Daròs, J.A. A potyvirus vector efficiently targets recombinant proteins to chloroplasts, mitochondria and nuclei in plant cells when expressed at the amino terminus of the polyprotein. Biotechnol. J. 2015, 10, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Cordero, T.; Cerdán, L.; Carbonell, A.; Katsarou, K.; Kalantidis, K.; Daròs, J.A. Dicer-Like 4 Is Involved in Restricting the Systemic Movement of Zucchini yellow mosaic virus in Nicotiana benthamiana. Mol. Plant-Microbe Interact. MPMI 2017, 30, 63–71. [Google Scholar] [CrossRef]
- Thole, V.; Worland, B.; Snape, J.W.; Vain, P. The pCLEAN dual binary vector system for Agrobacterium-mediated plant transformation. Plant Physiol. 2007, 145, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.; Christensen, L.P. Simple saponification method for the quantitative determination of carotenoids in green vegetables. J. Agric. Food Chem. 2005, 53, 6598–6602. [Google Scholar] [CrossRef]
- Sulli, M.; Mandolino, G.; Sturaro, M.; Onofri, C.; Diretto, G.; Parisi, B.; Giuliano, G. Molecular and biochemical characterization of a potato collection with contrasting tuber carotenoid content. PLoS ONE 2017, 12, e0184143. [Google Scholar] [CrossRef]
- Fraser, P.D.; Pinto, M.E.; Holloway, D.E.; Bramley, P.M. Technical advance: Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. Cell Mol. Biol. 2000, 24, 551–558. [Google Scholar] [CrossRef]
- Fantini, E.; Falcone, G.; Frusciante, S.; Giliberto, L.; Giuliano, G. Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol. 2013, 163, 986–998. [Google Scholar] [CrossRef]
- Rodriguez-Amaya, D.B. A Guide to Carotenoid Analysis in Foods; ILSI Press: Washington, WA, USA, 1999. [Google Scholar]
- Butnariu, M.; Rodino, S.; Petrache, P.; Negoescu, C.; Butu, M. Determination and quantification of maize zeaxanthin stability. Dig. J. Nanomater. Biostructures 2014, 9, 745–755. [Google Scholar]
- Miller, N.J.; Sampson, J.; Candeias, L.P.; Bramley, P.M.; Rice-Evans, C.A. Antioxidant activities of carotenes and xanthophylls. FEBS Lett. 1996, 384, 240–242. [Google Scholar] [CrossRef] [PubMed]
- Ordoudi, S.A.; Tsimidou, M.Z. Crocin bleaching assay (CBA) in structure-radical scavenging activity studies of selected phenolic compounds. J. Agric. Food Chem. 2006, 54, 9347–9356. [Google Scholar] [CrossRef]
- Sarfarazi, M.; Jafari, S.M.; Rajabzadeh, G.; Feizi, J. Development of an environmentally-friendly solvent-free extraction of saffron bioactives using subcritical water. LWT 2019, 114, 108428. [Google Scholar] [CrossRef]
- Franke, A.A.; Lai, J.F.; Halm, B.M. Absorption, distribution, metabolism, and excretion of isoflavonoids after soy intake. Arch. Biochem. Biophys. 2014, 559, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Ghag, S.B.; Adki, V.S.; Ganapathi, T.R.; Bapat, V.A. Plant Platforms for Efficient Heterologous Protein Production. Biotechnol. Bioprocess Eng. BBE 2021, 26, 546–567. [Google Scholar] [CrossRef]
- Wei, X.; Su, X.; Cao, P.; Liu, X.; Chang, W.; Li, M.; Zhang, X.; Liu, Z. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 2016, 534, 69–74. [Google Scholar] [CrossRef]
- Qin, X.; Suga, M.; Kuang, T.; Shen, J.R. Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 2015, 348, 989–995. [Google Scholar] [CrossRef]
- Demurtas, O.C.; Nicolia, A.; Diretto, G. Terpenoid Transport in Plants: How Far from the Final Picture? Plants 2023, 12, 634. [Google Scholar] [CrossRef]
- Gaillard, C.; Dufaud, A.; Tommasini, R.; Kreuz, K.; Amrhein, N.; Martinoia, E. A herbicide antidote (safener) induces the activity of both the herbicide detoxifying enzyme and of a vacuolar transporter for the detoxified herbicide. FEBS Lett. 1994, 352, 219–221. [Google Scholar] [CrossRef]
- Molina-Hidalgo, F.J.; Vazquez-Vilar, M.; D’Andrea, L.; Demurtas, O.C.; Fraser, P.; Giuliano, G.; Bock, R.; Orzáez, D.; Goossens, A. Engineering Metabolism in Nicotiana Species: A Promising Future. Trends Biotechnol. 2021, 39, 901–913. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demurtas, O.C.; Sulli, M.; Ferrante, P.; Mini, P.; Martí, M.; Aragonés, V.; Daròs, J.-A.; Giuliano, G. Production of Saffron Apocarotenoids in Nicotiana benthamiana Plants Genome-Edited to Accumulate Zeaxanthin Precursor. Metabolites 2023, 13, 729. https://doi.org/10.3390/metabo13060729
Demurtas OC, Sulli M, Ferrante P, Mini P, Martí M, Aragonés V, Daròs J-A, Giuliano G. Production of Saffron Apocarotenoids in Nicotiana benthamiana Plants Genome-Edited to Accumulate Zeaxanthin Precursor. Metabolites. 2023; 13(6):729. https://doi.org/10.3390/metabo13060729
Chicago/Turabian StyleDemurtas, Olivia Costantina, Maria Sulli, Paola Ferrante, Paola Mini, Maricarmen Martí, Verónica Aragonés, José-Antonio Daròs, and Giovanni Giuliano. 2023. "Production of Saffron Apocarotenoids in Nicotiana benthamiana Plants Genome-Edited to Accumulate Zeaxanthin Precursor" Metabolites 13, no. 6: 729. https://doi.org/10.3390/metabo13060729
APA StyleDemurtas, O. C., Sulli, M., Ferrante, P., Mini, P., Martí, M., Aragonés, V., Daròs, J.-A., & Giuliano, G. (2023). Production of Saffron Apocarotenoids in Nicotiana benthamiana Plants Genome-Edited to Accumulate Zeaxanthin Precursor. Metabolites, 13(6), 729. https://doi.org/10.3390/metabo13060729





