Widely Targeted Volatilomics and Metabolomics Analysis Reveal the Metabolic Composition and Diversity of Zingiberaceae Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Chemicals
2.3. Sample Preparation
2.4. GC-MS Analysis
2.5. LC-MS Analysis
2.6. Qualitative and Quantitative Analysis of Metabolomics Data
2.7. Statistical Data Analysis
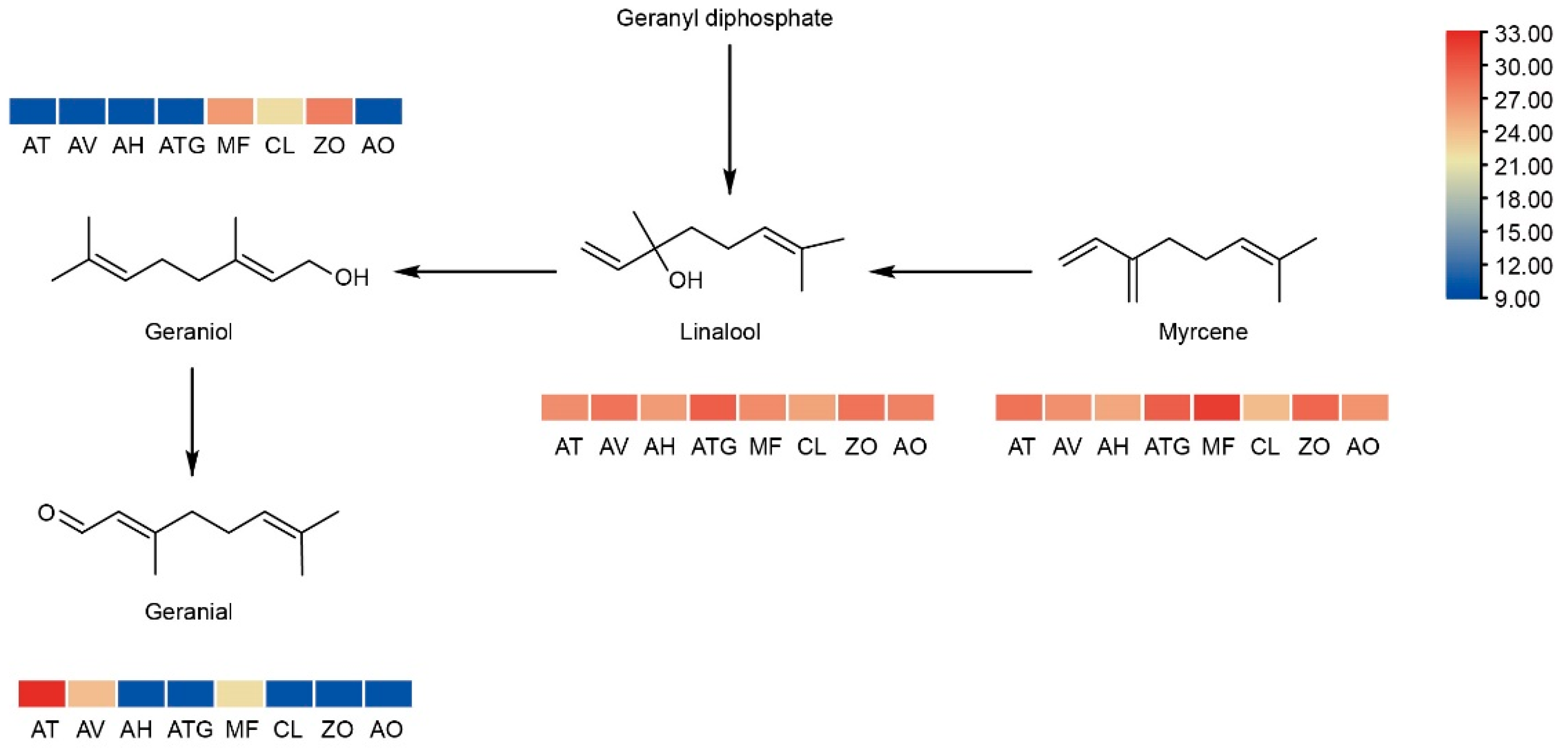
3. Results
3.1. Phylogeny Analysis of Zingiberaceae Plants
3.2. Volatilome and Metabolome Profiling of Zingiberaceae Plants
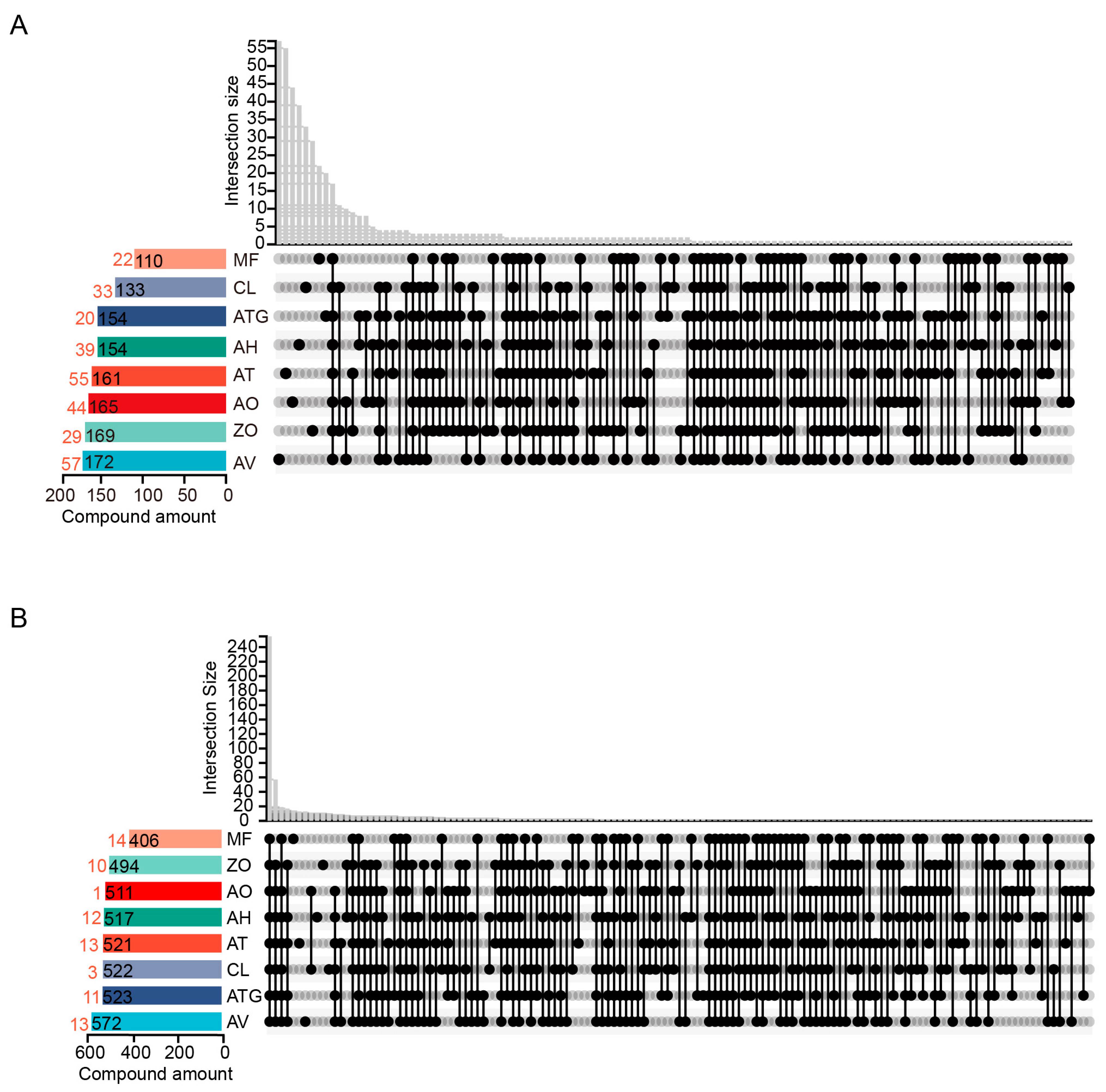
3.3. Venn Diagram Analysis
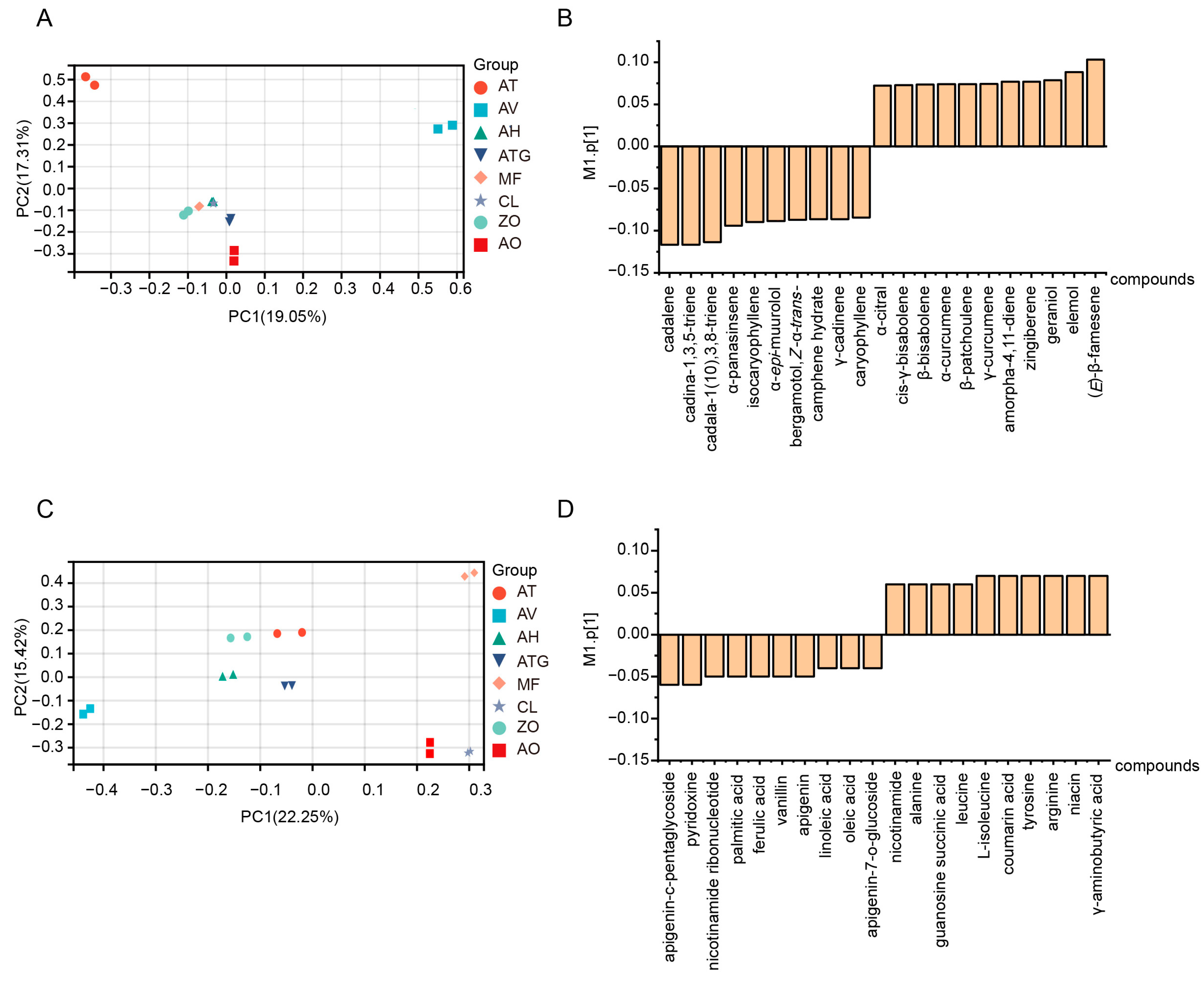
3.4. Principal Component Analysis
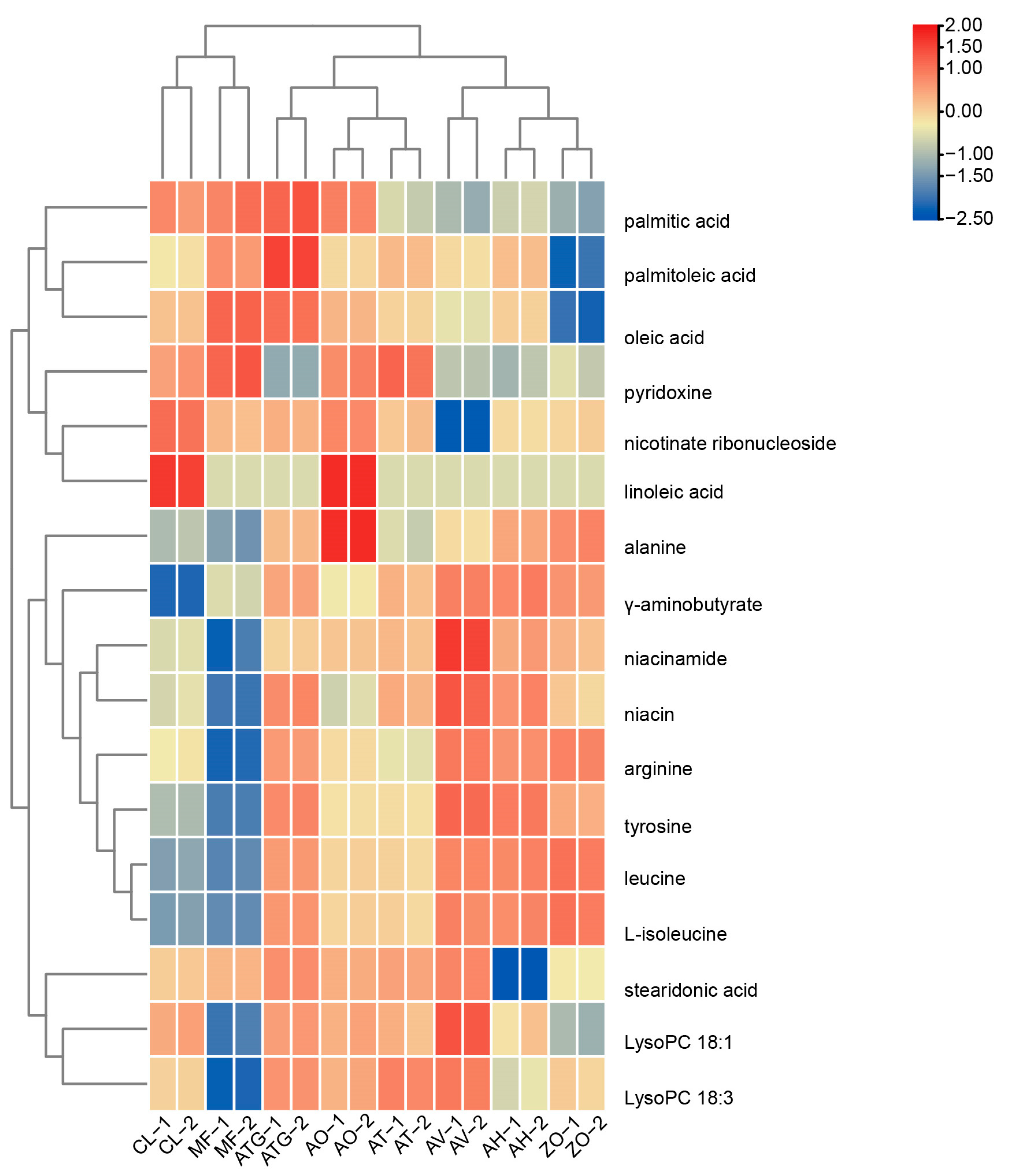
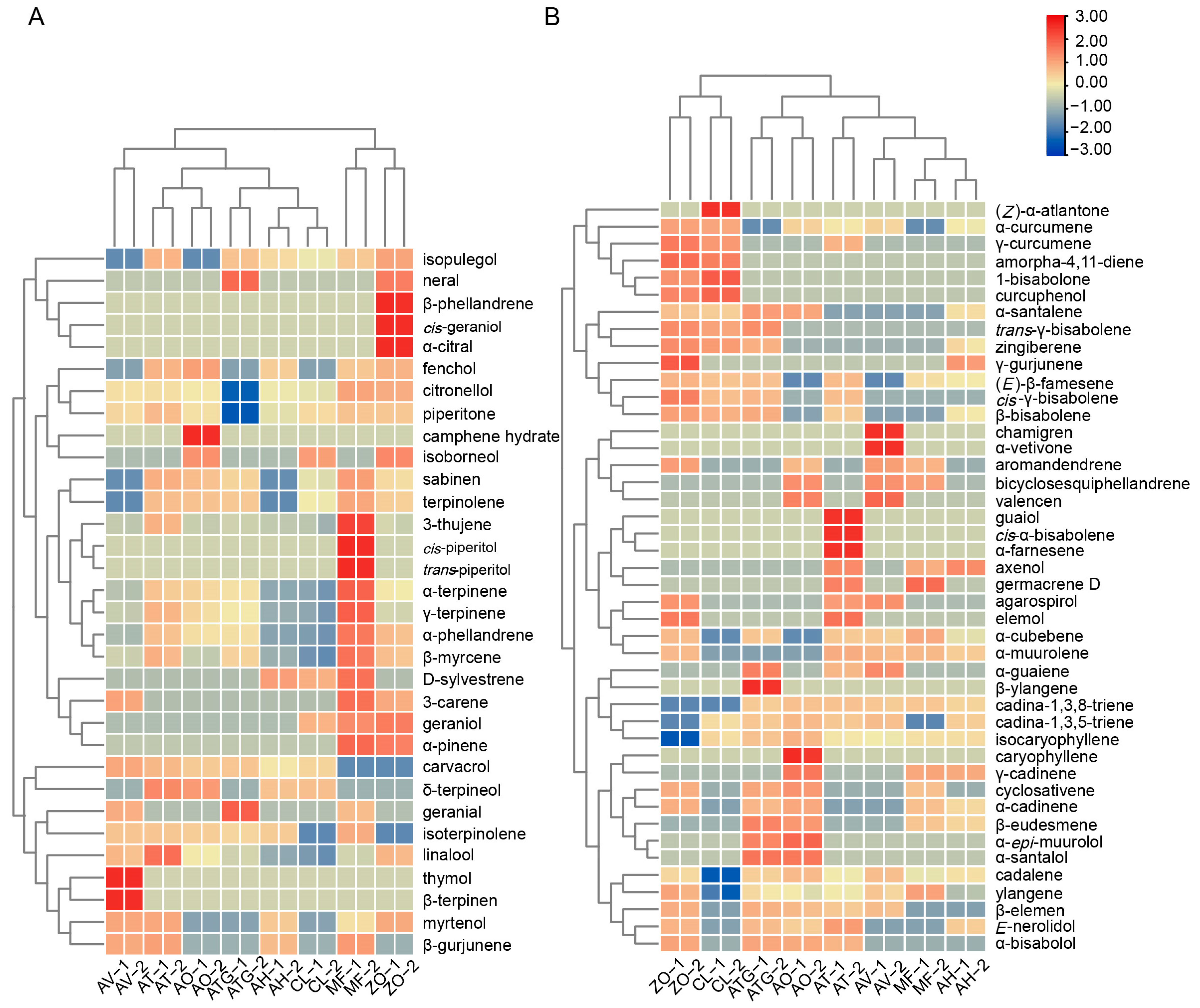
3.5. Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbosa, G.B.; Jayasinghe, N.S.; Natera, S.H.A.; Inutan, E.D.; Peteros, N.P.; Roessner, U. From common to rare Zingiberaceae plants—A metabolomics study using GC-MS. Phytochemistry 2017, 140, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Liu, A.Z.; Newman, M.; Li, Q.J. The molecular phylogeny of Alpinia (Zingiberaceae): A complex and polyphyletic genus of gingers. Am. J. Bot. 2005, 92, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Ao, H.; Wang, J.; Chen, L.; Li, S.; Dai, C. Comparison of Volatile Oil between the Fruits of Amomum villosum Lour. and Amomum villosum Lour. var. xanthioides T. L. Wu et Senjen Based on GC-MS and Chemometric Techniques. Molecules 2019, 24, 1663. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.W.; Israf, D.A.; Tham, C.L. Major Bioactive Compounds in Essential Oils Extracted from the Rhizomes of Zingiber zerumbet (L.) Smith: A Mini-Review on the Anti-allergic and Immunomodulatory Properties. Front. Pharmacol. 2018, 9, 652. [Google Scholar] [CrossRef] [PubMed]
- Mahanta, B.P.; Sarma, N.; Kemprai, P.; Begum, T.; Saikia, L.; Lal, M.; Haldar, S. Hydrodistillation based multifaceted value addition to Kaempferia galanga L. leaves, an agricultural residue. Ind. Crops Prod. 2020, 154, 112642. [Google Scholar] [CrossRef]
- Zhao, T.; Specht, C.D.; Dong, Z.; Ye, Y.; Liu, H.; Liao, J. Transcriptome analyses provide insights into development of the Zingiber zerumbet flower, revealing potential genes related to floral organ formation and patterning. Plant. Growth Regul. 2020, 90, 331–345. [Google Scholar] [CrossRef]
- Mohamad, H.; Lajis, N.H.; Abas, F.; Ali, A.M.; Sukari, M.A.; Kikuzaki, H.; Nakatani, N. Antioxidative Constituents of Etlingera elatior. J. Nat. Prod. 2005, 68, 285–288. [Google Scholar] [CrossRef]
- Cai, R.; Yue, X.; Wang, Y.; Yang, Y.; Sun, D.; Li, H.; Chen, L. Chemistry and bioactivity of plants from the genus Amomum. J. Ethnopharmacol. 2021, 281, 114563. [Google Scholar] [CrossRef]
- Cao, Y.; Xie, L.; Liu, K.; Liang, Y.; Dai, X.; Wang, X.; Lu, J.; Zhang, X.; Li, X. The antihypertensive potential of flavonoids from Chinese Herbal Medicine: A review. Pharmacol. Res. 2021, 174, 105919. [Google Scholar] [CrossRef]
- Chan, E.W.C.; Lim, Y.Y.; Wong, S.K.; Lim, K.K.; Tan, S.P.; Lianto, F.S.; Yong, M.Y. Effects of different drying methods on the antioxidant properties of leaves and tea of ginger species. Food Chem. 2009, 113, 166–172. [Google Scholar] [CrossRef]
- Mustafa, I.; Chin, N.L. Antioxidant Properties of Dried Ginger (Zingiber officinale Roscoe) var. Bentong. Foods 2023, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Ammon, H.P.T.; Wahl, M.A. Pharmacology of Curcuma longa. Planta Med. 1991, 57, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Cao, J.; Wang, D.; Qiu, J.; Kong, F. Identification of Ginger (Zingiber officinale Roscoe) Volatiles and Localization of Aroma-Active Constituents by GC–Olfactometry. J. Agric. Food Chem. 2017, 65, 4140–4145. [Google Scholar] [CrossRef] [PubMed]
- Chatzinasiou, L.; Booker, A.; MacLennan, E.; Mackonochie, M.; Heinrich, M. Turmeric (Curcuma longa L.) products: What quality differences exist? J. Herb. Med. 2019, 17–18, 100281. [Google Scholar] [CrossRef]
- Singh, C.; Manglembi, N.; Swapana, N.; Chanu, S. Ethnobotany, Phytochemistry and Pharmacology of Zingiber cassumunar Roxb. (Zingiberaceae). J. Pharmacogn. Phytochem. 2015, 4, 1–6. [Google Scholar]
- Chan, E.; Lim, Y.; Omar, M. Antioxidant and antibacterial activity of leaves of Etlingera species (Zingiberaceae) in Peninsular Malaysia. Food Chem. 2007, 104, 1586–1593. [Google Scholar] [CrossRef]
- Giang, P.M.; Son, P.T.; Matsunami, K.; Otsuka, H. New Diarylheptanoids from Amomum muricarpum LMER. Chem. Pharm. Bull. 2006, 54, 139–140. [Google Scholar] [CrossRef]
- Peng, W.; Li, P.; Ling, R.; Wang, Z.; Feng, X.; Liu, J.; Yang, Q.; Yan, J. Diversity of Volatile Compounds in Ten Varieties of Zingiberaceae. Molecules 2022, 27, 565. [Google Scholar] [CrossRef]
- Ivanović, M.; Makoter, K.; Islamčević Razboršek, M. Comparative Study of Chemical Composition and Antioxidant Activity of Essential Oils and Crude Extracts of Four Characteristic Zingiberaceae Herbs. Plants 2021, 10, 501. [Google Scholar] [CrossRef]
- Luca, S.V.; Trifan, A.; Zengin, G.; Sinan, K.I.; Uba, A.I.; Korona-Glowniak, I.; Skalicka-Woźniak, K. Evaluating the phyto-complexity and poly-pharmacology of spices: The case of Aframomum melegueta K. Schum (Zingiberaceae). Food Biosci. 2022, 49, 10929. [Google Scholar] [CrossRef]
- Asamenew, G.; Kim, H.-W.; Lee, M.-K.; Lee, S.-H.; Kim, Y.J.; Cha, Y.-S.; Yoo, S.M.; Kim, J.-B. Characterization of phenolic compounds from normal ginger (Zingiber officinale Rosc.) and black ginger (Kaempferia parviflora Wall.) using UPLC–DAD–QToF–MS. Eur. Food Res. Technol. 2019, 245, 653–665. [Google Scholar] [CrossRef]
- Yue, W.; Sun, W.; Rao, R.S.P.; Ye, N.; Yang, Z.; Chen, M. Non-targeted metabolomics reveals distinct chemical compositions among different grades of Bai Mudan white tea. Food Chem. 2019, 277, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef]
- Shen, S.; Zhan, C.; Yang, C.; Fernie, A.R.; Luo, J. Metabolomics-centered mining of plant metabolic diversity and function: Past decade and future perspectives. Mol. Plant. 2023, 16, 43–63. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, K.; Zhou, X.; Fang, C. Integrative Analysis of Metabolome and Transcriptome Reveals the Role of Strigolactones in Wounding-Induced Rice Metabolic Re-Programming. Metabolites 2022, 12, 789. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Guo, Y.; Wang, Y.; Li, M.; Li, K.; Liu, X.; Fang, C.; Luo, J. Metabolomic Analysis Reveals Nutritional Diversity among Three Staple Crops and Three Fruits. Foods 2022, 11, 550. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Cao, G.; Hou, X.; Huang, M.; Du, P.; Tan, T.; Zhang, Y.; Zhou, H.; Liu, X.; Liu, L.; et al. Development of a widely targeted volatilomics method for profiling volatilomes in plants. Mol. Plant. 2022, 15, 189–202. [Google Scholar] [CrossRef]
- Eyob, S.; Appelgren, M.; Rohloff, J.; Tsegaye, A.; Messele, G. Chemical Composition of Essential Oils from Fresh Plant Parts of Korarima (Aframomum corrorima) Cultivated in the Highland of Southern Ethiopia. J. Essent. Oil Res. 2007, 19, 372–375. [Google Scholar] [CrossRef]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.; Zhou, J.; Lu, D.; Wang, S.; et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1975. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Zaid, A.; Haw, X.R.; Alkatib, H.H.; Sasidharan, S.; Marriott, P.J.; Wong, Y.F. Phytochemical Constituents and Antiproliferative Activities of Essential Oils from Four Varieties of Malaysian Zingiber officinale Roscoe against Human Cervical Cancer Cell Line. Plants 2022, 11, 1280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Hou, D.; Zou, W.; Wang, J.; Luo, R.; Wang, M.; Yu, H. Comparison of Widely Targeted Metabolomics and Untargeted Metabolomics of Wild Ophiocordyceps sinensis. Molecules 2022, 27, 3645. [Google Scholar] [CrossRef]
- Wu, Z.; Guo, Z.; Wang, K.; Wang, R.; Fang, C. Comparative Metabolomic Analysis Reveals the Role of OsHPL1 in the Cold-Induced Metabolic Changes in Rice. Plants 2023, 12, 2032. [Google Scholar] [CrossRef]
- Pereira, I.; Severino, P.; Santos, A.C.; Silva, A.M.; Souto, E.B. Linalool bioactive properties and potential applicability in drug delivery systems. Colloids Surf. B 2018, 171, 566–578. [Google Scholar] [CrossRef]
- Silva, G.; Marques, J.N.J.; Linhares, E.P.M.; Bonora, C.M.; Costa, E.T.; Saraiva, M.F. Review of anticancer activity of monoterpenoids: Geraniol, nerol, geranial and neral. Chem. Biol. Interact. 2022, 362, 109994. [Google Scholar] [CrossRef]
- Surendran, S.; Qassadi, F.; Surendran, G.; Lilley, D.; Heinrich, M. Myrcene-What Are the Potential Health Benefits of This Flavouring and Aroma Agent? Front. Nutr. 2021, 8, 699666. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, J.; Chen, J.; Xiao, L.; Zhang, Y.; Wang, F.; Li, X. Efficient Biosynthesis of R-(-)-Linalool through Adjusting the Expression Strategy and Increasing GPP Supply in Escherichia coli. J. Agric. Food Chem. 2020, 68, 8381–8390. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, J.; Yao, G.; Bao, S.; Wan, X.; Wang, F.; Wang, K.; Song, T.; Han, P.; Liu, T.; et al. A novel, genetically encoded whole-cell biosensor for directed evolution of myrcene synthase in Escherichia coli. Biosens. Bioelectron. 2023, 228, 115176. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, W.; Jiang, L.; Pu, X.; Yang, Y.; Zhang, G.; Luo, Y. Functional characterization of a geraniol synthase-encoding gene from Camptotheca acuminata and its application in production of geraniol in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2016, 43, 1281–1292. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, Y.; Deng, J.; Gao, G.; Ding, C.; Zhang, L.; Yang, R. The Complete Chloroplast Genome Sequences of 14 Curcuma Species: Insights Into Genome Evolution and Phylogenetic Relationships within Zingiberales. Front. Genet. 2020, 11, 802. [Google Scholar] [CrossRef]
- Liu, K.; Abdullah, A.A.; Huang, M.; Nishioka, T.; Altaf-Ul-Amin, M.; Kanaya, S. Novel Approach to Classify Plants Based on Metabolite-Content Similarity. Biomed. Res. Int. 2017, 2017, 5296729. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Guo, P.; Tsui, S.W.; Chen, H.; Zhao, Z. An ethnobotanical survey of medicinal spices used in Chinese hotpot. Food Res. Int. 2012, 48, 226–232. [Google Scholar] [CrossRef]
- Meng, L.; Song, W.; Chen, S.; Hu, F.; Pang, B.; Cheng, J.; He, B.; Sun, F. Widely targeted metabolomics analysis reveals the mechanism of quality improvement of flue-cured tobacco. Front. Plant. Sci. 2022, 13, 1074029. [Google Scholar] [CrossRef]
- Borgonetti, V.; Governa, P.; Manetti, F.; Galeotti, N. Zingiberene, a non-zinc-binding class I HDAC inhibitor: A novel strategy for the management of neuropathic pain. Phytomedicine 2023, 111, 154670. [Google Scholar] [CrossRef]
- Salas-Oropeza, J.; Jimenez-Estrada, M.; Perez-Torres, A.; Castell-Rodriguez, A.E.; Becerril-Millan, R.; Rodriguez-Monroy, M.A.; Jarquin-Yanez, K.; Canales-Martinez, M.M. Wound Healing Activity of alpha-Pinene and alpha-Phellandrene. Molecules 2021, 26, 2488. [Google Scholar] [CrossRef]
- Eddin, L.B.; Jha, N.K.; Goyal, S.N.; Agrawal, Y.O.; Subramanya, S.B.; Bastaki, S.M.A.; Ojha, S. Health Benefits, Pharmacological Effects, Molecular Mechanisms, and Therapeutic Potential of alpha-Bisabolol. Nutrients 2022, 14, 1370. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro-Neto, F.R.; Lopes, E.M.; Acha, B.T.; Gomes, L.D.S.; Dias, W.A.; Reis Filho, A.C.D.; Leal, B.S.; Rodrigues, D.; Silva, J.D.N.; Dittz, D.; et al. alpha-Phellandrene exhibits antinociceptive and tumor-reducing effects in a mouse model of oncologic pain. Toxicol. Appl. Pharmacol. 2021, 418, 115497. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shang, K. Isoborneol as a natural sporulation quenching agent to control Aspergillus flavus. Nat. Prod. Res. 2022, 19, 1–5. [Google Scholar] [CrossRef]
- Fidyt, K.; Fiedorowicz, A.; Strzadala, L.; Szumny, A. beta-caryophyllene and beta-caryophyllene oxide-natural compounds of anticancer and analgesic properties. Cancer Med. 2016, 5, 3007–3017. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, J.; Khondkar, W.; Morelli, M.B.; Wang, X.; Santulli, G.; Trimarco, V. Arginine and Endothelial Function. Biomedicines 2020, 8, 277. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Su, R.; Yuan, H.; Zhou, H.; Jiangfang, Y.; Liu, X.; Luo, J. Widely Targeted Volatilomics and Metabolomics Analysis Reveal the Metabolic Composition and Diversity of Zingiberaceae Plants. Metabolites 2023, 13, 700. https://doi.org/10.3390/metabo13060700
Zhang Y, Su R, Yuan H, Zhou H, Jiangfang Y, Liu X, Luo J. Widely Targeted Volatilomics and Metabolomics Analysis Reveal the Metabolic Composition and Diversity of Zingiberaceae Plants. Metabolites. 2023; 13(6):700. https://doi.org/10.3390/metabo13060700
Chicago/Turabian StyleZhang, Youjin, Rongxiu Su, Honglun Yuan, Haihong Zhou, Yiding Jiangfang, Xianqing Liu, and Jie Luo. 2023. "Widely Targeted Volatilomics and Metabolomics Analysis Reveal the Metabolic Composition and Diversity of Zingiberaceae Plants" Metabolites 13, no. 6: 700. https://doi.org/10.3390/metabo13060700
APA StyleZhang, Y., Su, R., Yuan, H., Zhou, H., Jiangfang, Y., Liu, X., & Luo, J. (2023). Widely Targeted Volatilomics and Metabolomics Analysis Reveal the Metabolic Composition and Diversity of Zingiberaceae Plants. Metabolites, 13(6), 700. https://doi.org/10.3390/metabo13060700






