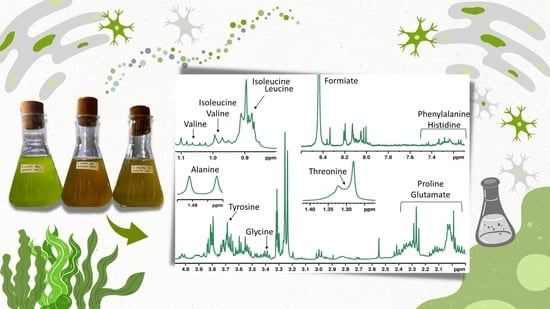
Application of 1H HR-MAS NMR-Based Metabolite Fingerprinting of Marine Microalgae
Abstract
1. Introduction
2. Materials and Methods
2.1. Microalgae Cultivation
2.2. Biomass Harvesting
2.3. Sample Preparation for 1H HR-MAS NMR
2.4. Acquisition of 1H HR-MAS NMR Spectra
2.5. Sample Preparation and Acquisition of 1D and 2D NMR Spectra
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altmann, K.H. Drugs from the oceans: Marine natural products as leads for drug discovery. Chimia 2017, 71, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 34, 235–294. [Google Scholar] [CrossRef]
- Guiry, M.D. How many species of algae are there? J. Phycol. 2012, 48, 1057–1063. [Google Scholar] [CrossRef]
- Hopes, A.; Mock, T. Evolution of Microalgae and Their Adaptations in Different Marine Ecosystems. eLS 2015, 1–9. [Google Scholar] [CrossRef]
- Lauritano, C.; Ferrante, M.I.; Rogato, A. Marine Natural Products from Microalgae: An -Omics Overview. Mar. Drugs 2019, 17, 269. [Google Scholar] [CrossRef]
- Matos, Â.P. The Impact of Microalgae in Food Science and Technology. JAOCS J. Am. Oil Chem. Soc. 2017, 94, 1333–1350. [Google Scholar] [CrossRef]
- Borowitzka, M.A. High-value products from microalgae-their development and commercialization. J. Appl. Phycol. 2013, 25, 743–756. [Google Scholar] [CrossRef]
- Gul, W.; Hamann, M.T. Indole alkaloid marine natural products: An established source of cancer drug leads with considerable promise for the control of parasitic, neurological and other diseases. Life Sci. 2005, 78, 442–453. [Google Scholar] [CrossRef]
- Habbu, P.; Warad, V.; Shastri, R.; Madagundi, S.; Kulkarni, V.H. Antimicrobial metabolites from marine microorganisms. Chin. J. Nat. Med. 2016, 14, 101–116. [Google Scholar] [CrossRef]
- Mayer, A.M.S.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2013, 11, 2510–2573. [Google Scholar] [CrossRef] [PubMed]
- Roselet, F.; Vandamme, D.; Muylaert, K.; Abreu, P.C. Harvesting of microalgae for biomass production. In Microalgae Biotechnology for Development of Biofuel and Wastewater Treatment; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- de Freitas Coêlho, D.; Tundisi, L.L.; Cerqueira, K.S.; da Silva Rodrigues, J.R.; Mazzola, P.G.; Tambourgi, E.B.; de Souza, R.R. Microalgae: Cultivation aspects and bioactive compounds. Braz. Arch. Biol. Technol. 2019, 62, e19180343. [Google Scholar] [CrossRef]
- Thompson, F.; Krüger, R.; Thompson, C.C.; Berlinck, R.G.S.; Coutinho, R.; Landell, M.F.; Pavão, M.; Mourão, P.A.S.; Salles, A.; Negri, N.; et al. Marine Biotechnology in Brazil: Recent Developments and Its Potential for Innovation. Front. Mar. Sci. 2018, 5, 236. [Google Scholar] [CrossRef]
- Radakovits, R.; Jinkerson, R.E.; Fuerstenberg, S.I.; Tae, H.; Settlage, R.E.; Boore, J.L.; Posewitz, M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 2012, 3, 686. [Google Scholar] [CrossRef] [PubMed]
- Al-Hoqani, U.; Young, R.; Purton, S. The biotechnological potential of Nannochloropsis. Perspect. Phycol. 2017, 4, 1–15. [Google Scholar] [CrossRef]
- Ma, X.N.; Chen, T.P.; Yang, B.; Liu, J.; Chen, F. Lipid production from Nannochloropsis. Mar. Drugs 2016, 14, 61. [Google Scholar] [CrossRef]
- Ben-Sheleg, A.; Khozin-Godberg, I.; Yaakov, B.; Vonshak, A. Characterization of nannochloropsis oceanica rose bengal mutants sheds light on acclimation mechanisms to high light when grown in low temperature. Plant Cell Physiol. 2021, 62, 1478–1493. [Google Scholar] [CrossRef]
- Guedes, A.C.; Malcata, F. Nutritional Value and Uses of Microalgae in Aquaculture. In Aquaculture; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Serôdio, J.; Lavaud, J. Diatoms and Their Ecological Importance. In Life Below Water. Encyclopedia of the UN Sustainable Development Goals; Leal Filho, W., Azul, A.M., Brandli, L., Lange Salvia, A., Wall, T., Eds.; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Radchenko, I.G.; Il’yash, L.V. Growth and photosynthetic activity of diatom Thalassiosira weissflogii at decreasing salinity. Biol. Bull. 2006, 33, 242–247. [Google Scholar] [CrossRef]
- Halac, S.R.; Villafañe, V.E.; Helbling, E.W. Temperature benefits the photosynthetic performance of the diatoms Chaetoceros gracilis and Thalassiosira weissflogii when exposed to UVR. J. Photochem. Photobiol. B Biol. 2010, 101, 196–205. [Google Scholar] [CrossRef]
- Antal, T.K.; Osipov, V.; Matorin, D.N.; Rubin, A.B. Membrane potential is involved in regulation of photosynthetic reactions in the marine diatom Thalassiosira weissflogii. J. Photochem. Photobiol. B Biol. 2011, 102, 169–173. [Google Scholar] [CrossRef]
- Klein Breteler, W.C.M.; Schogt, N.; Rampen, S. Effect of diatom nutrient limitation on copepod development: Role of essential lipids. Mar. Ecol. Prog. Ser. 2005, 291, 125–133. [Google Scholar] [CrossRef]
- Farooq, H.; Courtier-Murias, D.; Soong, R.; Bermel, W.; Kingery, W.M.; Simpson, A.J. HR-MAS NMR Spectroscopy: A Practical Guide for Natural Samples. Curr. Org. Chem. 2013, 17, 3013–3031. [Google Scholar] [CrossRef]
- Keifer, P.A.; Baltusis, L.; Rice, D.M.; Tymiak, A.A.; Shoolery, J.N. A comparison of NMR spectra obtained for solid-phase-synthesis resins using conventional high-resolution, magic-angle-spinning, and high-resolution magic-angle-spinning probes. J. Magn. Reson. Ser. A 1996, 119, 65–75. [Google Scholar] [CrossRef]
- Lindon, J.C.; Beckonert, O.P.; Holmes, E.; Nicholson, J.K. High-resolution magic angle spinning NMR spectroscopy: Application to biomedical studies. Prog. Nucl. Magn. Reson. Spectrosc. 2009, 55, 79–100. [Google Scholar] [CrossRef]
- Dutra, L.M.; da Conceição Santos, A.D.; Lourenço, A.V.F.; Nagata, N.; Heiden, G.; Campos, F.R.; Barison, A. 1H HR-MAS NMR and chemometric methods for discrimination and classification of Baccharis (Asteraceae): A proposal for quality control of Baccharis trimera. J. Pharm. Biomed. Anal. 2020, 184, 113200. [Google Scholar] [CrossRef]
- Ali, S.; Rech, K.S.; Badshah, G.; Soares, F.L.F.; Barison, A. 1H HR-MAS NMR-Based Metabolomic Fingerprinting to Distinguish Morphological Similarities and Metabolic Profiles of Maytenus ilicifolia, a Brazilian Medicinal Plant. J. Nat. Prod. 2021, 84, 1707–1714. [Google Scholar] [CrossRef]
- da Conceição Santos, A.D.; Fonseca, F.A.; Dutra, L.M.; de Fátima Costa Santos, M.; Menezes, L.R.A.; Campos, F.R.; Nagata, N.; Ayub, R.; Barison, A. 1H HR-MAS NMR-based metabolomics study of different persimmon cultivars (Diospyros kaki) during fruit development. Food Chem. 2018, 239, 511–519. [Google Scholar] [CrossRef]
- Chauton, M.S.; Størseth, T.R.; Krane, J. High-resolution magic angle spinning NMR analysis of whole cells of Chaetoceros muelleri (Bacillariophyceae) and comparison with 13C-NMR and distortionless enhancement by polarization transfer 13C-NMR analysis of lipophilic extracts. J. Phycol. 2004, 40, 611–618. [Google Scholar] [CrossRef]
- Couto, C.; Hernández, C.P.; Alves Sobrinho, R.C.M.; Mendes, C.R.B.; Roselet, F.; Abreu, P.C. Optimization of a low-cost fertilizer-based medium for large-scale cultivation of the coastal diatom Conticribra weissflogii using response surface methodology and its effects on biomass composition. J. Appl. Phycol. 2021, 33, 2767–2781. [Google Scholar] [CrossRef]
- Kubelka, B.G.; Roselet, F.; Pinto, W.T.; Abreu, P.C. The action of small bubbles in increasing light exposure and production of the marine microalga Nannochloropsis oceanica in massive culture systems. Algal Res. 2018, 35, 569–576. [Google Scholar] [CrossRef]
- Roselet, F.; Vandamme, D.; Roselet, M.; Muylaert, K.; Abreu, P.C. Effects of pH, Salinity, Biomass Concentration, and Algal Organic Matter on Flocculant Efficiency of Synthetic Versus Natural Polymers for Harvesting Microalgae Biomass. Bioenergy Res. 2017, 10, 427–437. [Google Scholar] [CrossRef]
- Martínez-Bisbal, M.C.; Carbó Mestre, N.; Martínez-Máñez, R.; Bauzá, J.; Alcañiz Fillol, M. Microalgae degradation follow up by voltammetric electronic tongue, impedance spectroscopy and NMR spectroscopy. Sens. Actuators B Chem. 2019, 281, 44–52. [Google Scholar] [CrossRef]
- Sathasivam, R.; Radhakrishnan, R.; Hashem, A.; Abd_Allah, E.F. Microalgae metabolites: A rich source for food and medicine. Saudi J. Biol. Sci. 2019, 26, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Zanella, L.; Vianello, F. Microalgae of the genus Nannochloropsis: Chemical composition and functional implications for human nutrition. J. Funct. Foods 2020, 68, 103919. [Google Scholar] [CrossRef]
- Hughes, A.H.; Magot, F.; Tawfike, A.F.; Rad-Menéndez, C.; Thomas, N.; Young, L.C.; Stucchi, L.; Carettoni, D.; Stanley, M.S.; Edrada-Ebel, R.; et al. Exploring the chemical space of macro-and micro-algae using comparative metabolomics. Microorganisms 2021, 9, 311. [Google Scholar] [CrossRef]
- Park, Y.H.; Han, S.-I.; Oh, B.; Kim, H.S.; Jeon, M.S.; Kim, S.; Choi, Y.-E. Microalgal secondary metabolite productions as a component of biorefinery: A review. Biooresour. Technol. 2022, 344 Pt A, 126206. [Google Scholar] [CrossRef]
- Abreu, A.C.; Molina-Miras, A.; Aguilera-Saéz, L.M.; López-Rosales, L.; Cerón-Garciá, M.D.C.; Sánchez-Mirón, A.; Olmo-Garciá, L.; Carrasco-Pancorbo, A.; Garciá-Camacho, F.; Molina-Grima, E.; et al. Production of Amphidinols and Other Bioproducts of Interest by the Marine Microalga Amphidinium carterae Unraveled by Nuclear Magnetic Resonance Metabolomics Approach Coupled to Multivariate Data Analysis. J. Agric. Food Chem. 2019, 67, 9667–9682. [Google Scholar] [CrossRef]
- Ma, N.L.; Aziz, A.; Teh, K.Y.; Lam, S.S.; Cha, T.S. Metabolites Re-programming and Physiological Changes Induced in Scenedesmus regularis under Nitrate Treatment. Sci. Rep. 2018, 8, 9746. [Google Scholar] [CrossRef]
- Azizan, A.; Bustamam, M.S.A.; Maulidiani, M.; Shaari, K.; Ismail, I.S.; Nagao, N.; Abas, F. Metabolite profiling of the microalgal diatom chaetoceros calcitrans and correlation with antioxidant and nitric oxide inhibitory Activities via 1H NMR-Based Metabolomics. Mar. Drugs 2018, 16, 154. [Google Scholar] [CrossRef]
- Crable, B.R.; Plugge, C.M.; McInerney, M.J.; Stams, A.J.M. Formate formation and formate conversion in biological fuels production. Enzym. Res. 2011, 2011, 532536. [Google Scholar] [CrossRef]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Eadsforth, T.C.; Gardiner, M.; Maluf, F.V.; McElroy, S.; James, D.; Frearson, J.; Gray, D.; Hunter, W.N. Assessment of Pseudomonas aeruginosa N5,N10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase as a potential antibacterial drug target. PLoS ONE 2012, 7, e35973. [Google Scholar] [CrossRef] [PubMed]
- Ramos, D.F.; Bartolomeu Halicki, P.C.; da Silva Canielles Caprara, C.; Borges, P.; da D’Oca, C.R.M.; de Santos, M.F.C.; D’Oca, M.G.M.; Roselet, F.; Almeida da Silva, P.E.; Abreu, P.C. Chemical Profile and Antimicrobial Activity of the Marine Diatom Chaetoceros muelleri. Chem. Biodivers. 2022, 19, e202100846. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, E.C.; Syit, D.A.; Grossart, H.P.; Ullrich, M.S. Chemotaxis of Marinobacter adhaerens and its impact on attachmentto the diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 2012, 78, 6900–6907. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, H.; Wang, S.; Xu, Y.; Liang, H.; Liu, H.; Sonnenschein, E.C. The Draft Genome of the Centric Diatom Conticribra weissflogii (Coscinodiscophyceae, Ochrophyta). Protist 2021, 172, 125845. [Google Scholar] [CrossRef] [PubMed]
- Stachura-Suchoples, K.; Williams, D.M. Description of Conticribra tricircularis, a new genus and species of Thalassiosirales, with a discussion on its relationship to other continuous cribra species of Thalassiosira Cleve (Bacillariophyta) and its freshwater origin. Eur. J. Phycol. 2009, 44, 477–486. [Google Scholar] [CrossRef]
- Ribalet, F.; Intertaglia, L.; Lebaron, P.; Casotti, R. Differential effect of three polyunsaturated aldehydes on marine bacterial isolates. Aquat. Toxicol. 2008, 86, 249–255. [Google Scholar] [CrossRef]
- Rizzi, J.; Moro, T.R.; Winnischofer, S.M.B.; Colusse, G.A.; Tamiello, C.S.; Trombetta-Lima, M.; Noleto, G.R.; Dolga, A.M.; Duarte, M.E.R.; Noseda, M.D. Chemical structure and biological activity of the (1 → 3)-linked β-D-glucan isolated from marine diatom Conticribra weissflogii. Int. J. Biol. Macromol. 2022, 224, 584–593. [Google Scholar] [CrossRef]
- Barrera-Alba, J.J.; Abreu, P.C.; Tenenbaum, D.R. Seasonal and inter-annual variability in phytoplankton over a 22-year period in a tropical coastal region in the southwestern Atlantic Ocean. Cont. Shelf Res. 2019, 176, 51–63. [Google Scholar] [CrossRef]


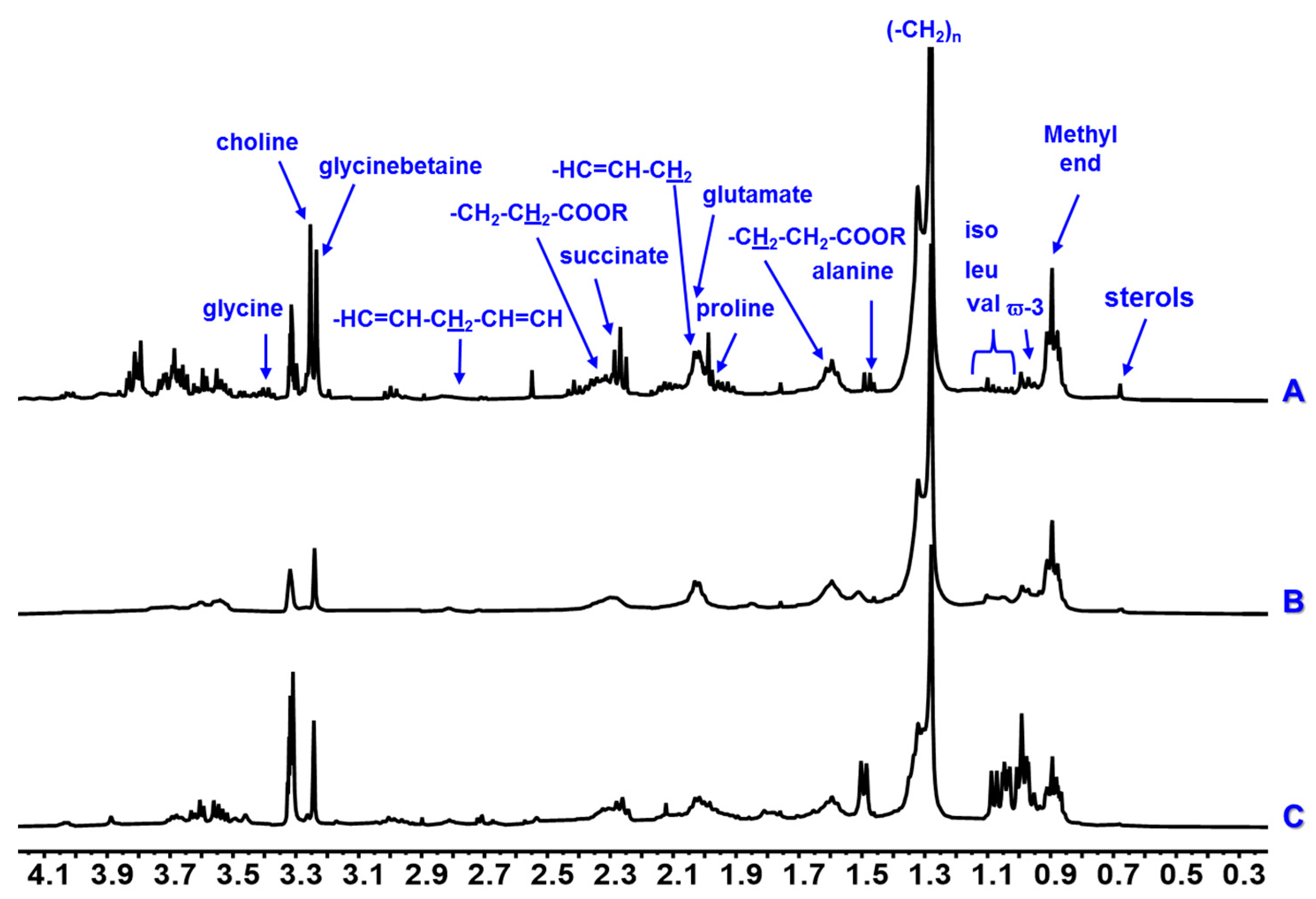
| Class | Compound | δH mult. (J in Hz) | CDCl3 | D2O | CD3OD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N. O | C. M | C. W | N. O | C. M | C. W | N. O | C. M | C. W | |||
| Amino Acids | Isoleucine | 0.95 t (7.0), 1.02 d (7.0) | - | - | - | x | x | x | x | x | x |
| Leucine | 0.99 m | - | - | - | x | x | x | x | x | x | |
| Valine | 1.02 d (7.0), 1.07 d (7.0) | - | - | - | x | x | x | x | x | x | |
| Alanine | 1.48 d (7.0) | - | - | - | x | x | x | x | x | x | |
| Proline | 1.97–2.07 m | - | - | - | - | - | - | x | x | x | |
| Glutamate | 2.08 m, 2.14 m, 2.35 m | - | - | - | - | - | - | x | x | x | |
| Choline | 3.20 s | - | - | - | x | x | x | x | x | x | |
| Glycinebetaine | 3.26 s | - | - | - | x | x | x | x | x | x | |
| Glycine | 3.59 s | - | - | - | x | x | x | x | x | x | |
| Phenylalanine | 7.24 m, 7.30 m, 7.34 m | - | - | - | x | x | x | x | - | x | |
| Histidine | 7.24 m, 8.13 s | - | - | - | - | - | - | x | - | x | |
| Tyrosine | 6.89 d (8.1), 7.19 d (8.1) | - | - | - | x | x | x | x | x | x | |
| Glutatione | 8.23 s | - | - | - | x | x | x | - | - | - | |
| Organic acids | Lactate | 1.33 d (6.7) | - | - | - | x | x | x | - | - | - |
| Succinate | 2.30 s | - | - | - | x | - | x | x | x | x | |
| Formate | 8.47 s | - | - | - | x | - | - | x | - | - | |
| Sterols | 0.73 m | x | x | x | - | - | - | x | x | x | |
| Monounsaturated Fatty acids | Methylic | 0.89 m | x | x | x | - | - | - | x | x | x |
| Methylenic | 1.35–1.25 m | x | x | x | - | - | - | x | x | x | |
| –CH2–CH2–COOR | 1.59 m | x | x | x | - | - | - | x | x | x | |
| –CH2–CH2–COOR | 2.34 m | x | x | x | - | - | - | x | x | x | |
| PUFAs | Methylic ϖ-3 | 0.98 t (7.0) | x | - | - | - | - | - | x | - | - |
| –CH2–CH2–COOR | 1.62 m | x | x | x | - | - | - | x | x | x | |
| Allylic | 2.06 m | x | x | x | - | - | - | x | x | x | |
| –CH2–CH2–COOR | 2.34 m | x | x | x | - | - | - | x | x | x | |
| bis-allylic | 2.81 m | x | x | x | - | - | - | x | x | x | |
| Vinylic | 5.40–5.30 m | x | x | x | - | - | - | x | x | x | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caprara, C.d.S.C.; Mathias, T.K.; Santos, M.d.F.C.; D’Oca, M.G.M.; D’Oca, C.D.R.M.; Roselet, F.; Abreu, P.C.; Ramos, D.F. Application of 1H HR-MAS NMR-Based Metabolite Fingerprinting of Marine Microalgae. Metabolites 2023, 13, 202. https://doi.org/10.3390/metabo13020202
Caprara CdSC, Mathias TK, Santos MdFC, D’Oca MGM, D’Oca CDRM, Roselet F, Abreu PC, Ramos DF. Application of 1H HR-MAS NMR-Based Metabolite Fingerprinting of Marine Microalgae. Metabolites. 2023; 13(2):202. https://doi.org/10.3390/metabo13020202
Chicago/Turabian StyleCaprara, Carolina da Silva Canielles, Tatiane Ksyvickas Mathias, Maria de Fátima C. Santos, Marcelo G. M. D’Oca, Caroline Da R. M. D’Oca, Fabio Roselet, Paulo Cesar Abreu, and Daniela Fernandes Ramos. 2023. "Application of 1H HR-MAS NMR-Based Metabolite Fingerprinting of Marine Microalgae" Metabolites 13, no. 2: 202. https://doi.org/10.3390/metabo13020202
APA StyleCaprara, C. d. S. C., Mathias, T. K., Santos, M. d. F. C., D’Oca, M. G. M., D’Oca, C. D. R. M., Roselet, F., Abreu, P. C., & Ramos, D. F. (2023). Application of 1H HR-MAS NMR-Based Metabolite Fingerprinting of Marine Microalgae. Metabolites, 13(2), 202. https://doi.org/10.3390/metabo13020202