ConCISE: Consensus Annotation Propagation of Ion Features in Untargeted Tandem Mass Spectrometry Combining Molecular Networking and In Silico Metabolite Structure Prediction
Abstract
1. Introduction
2. Materials and Methods
2.1. Availability
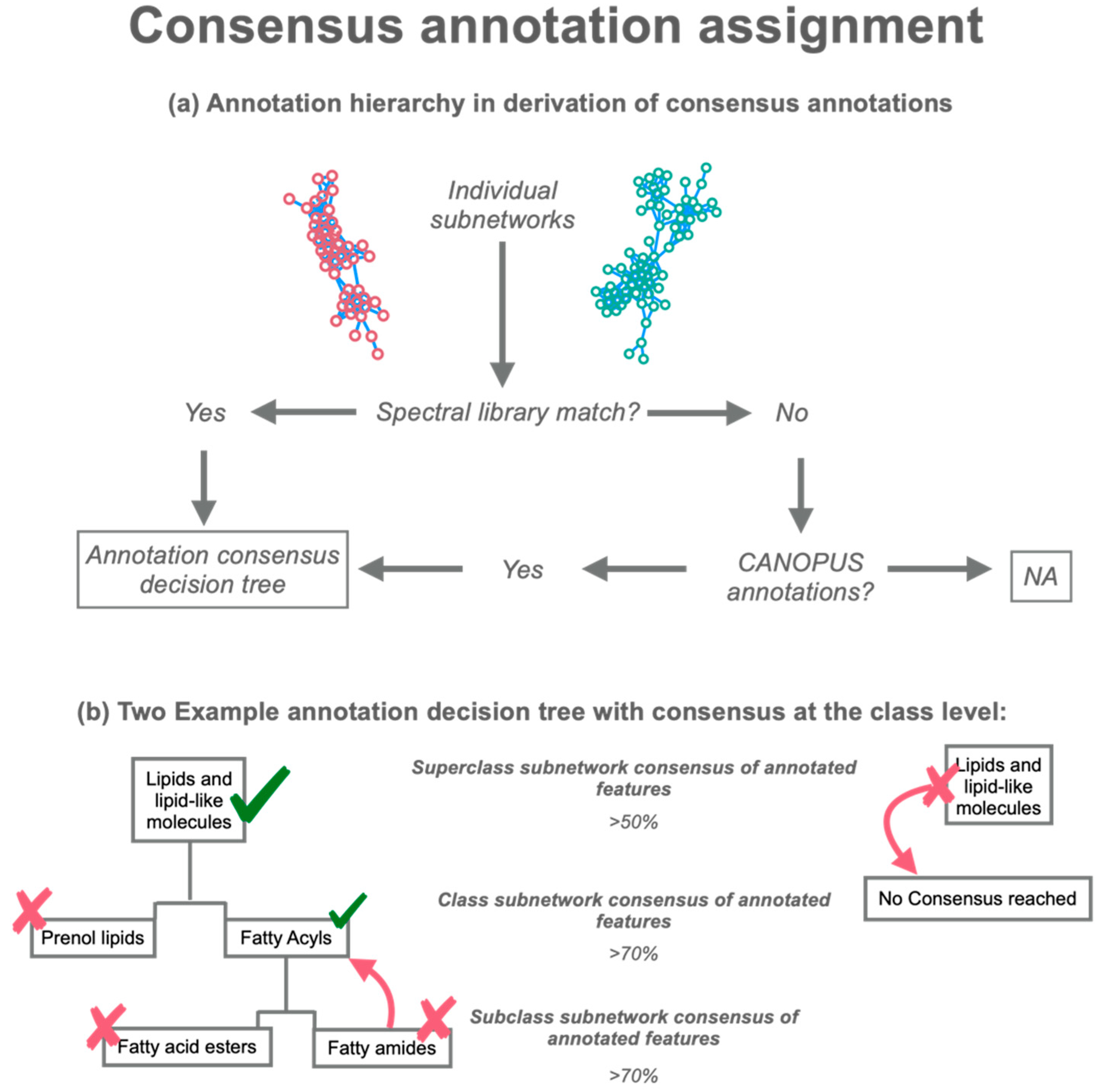
2.2. ConCISE Workflow
2.3. Running ConCISE
2.4. ConCISE Export
3. Results
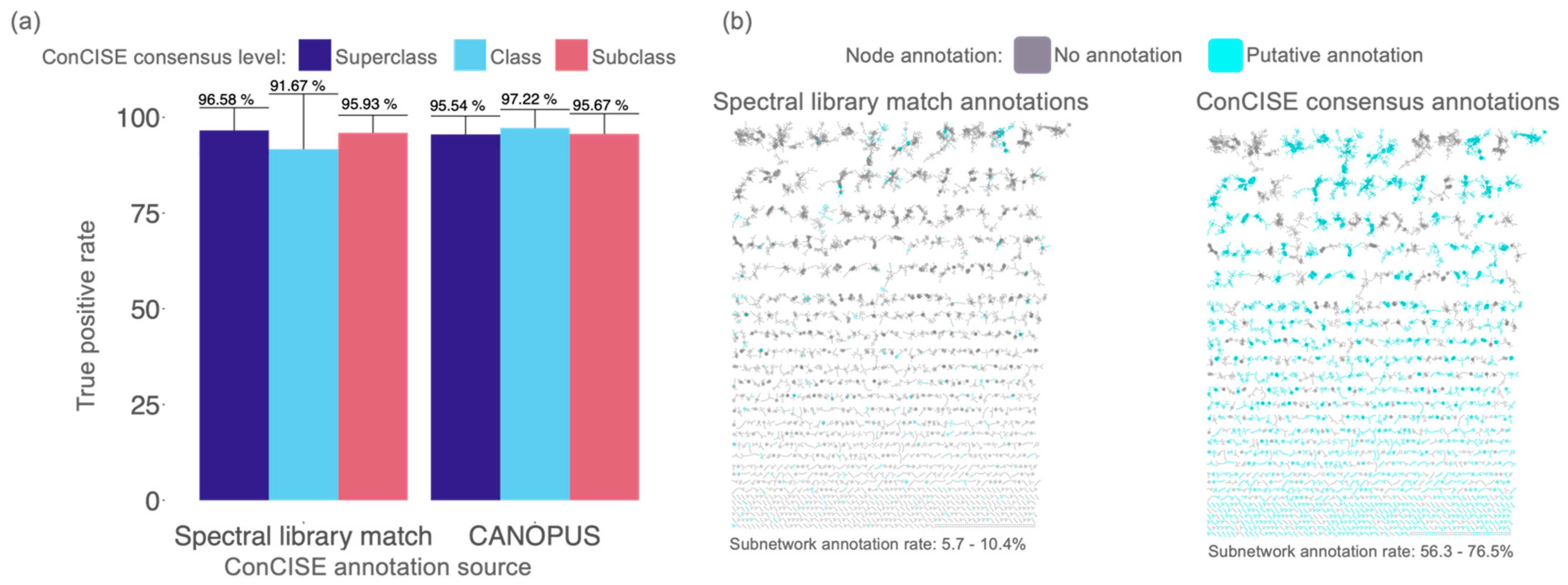
3.1. Manual Validation of ConCISE Consensus Annotations
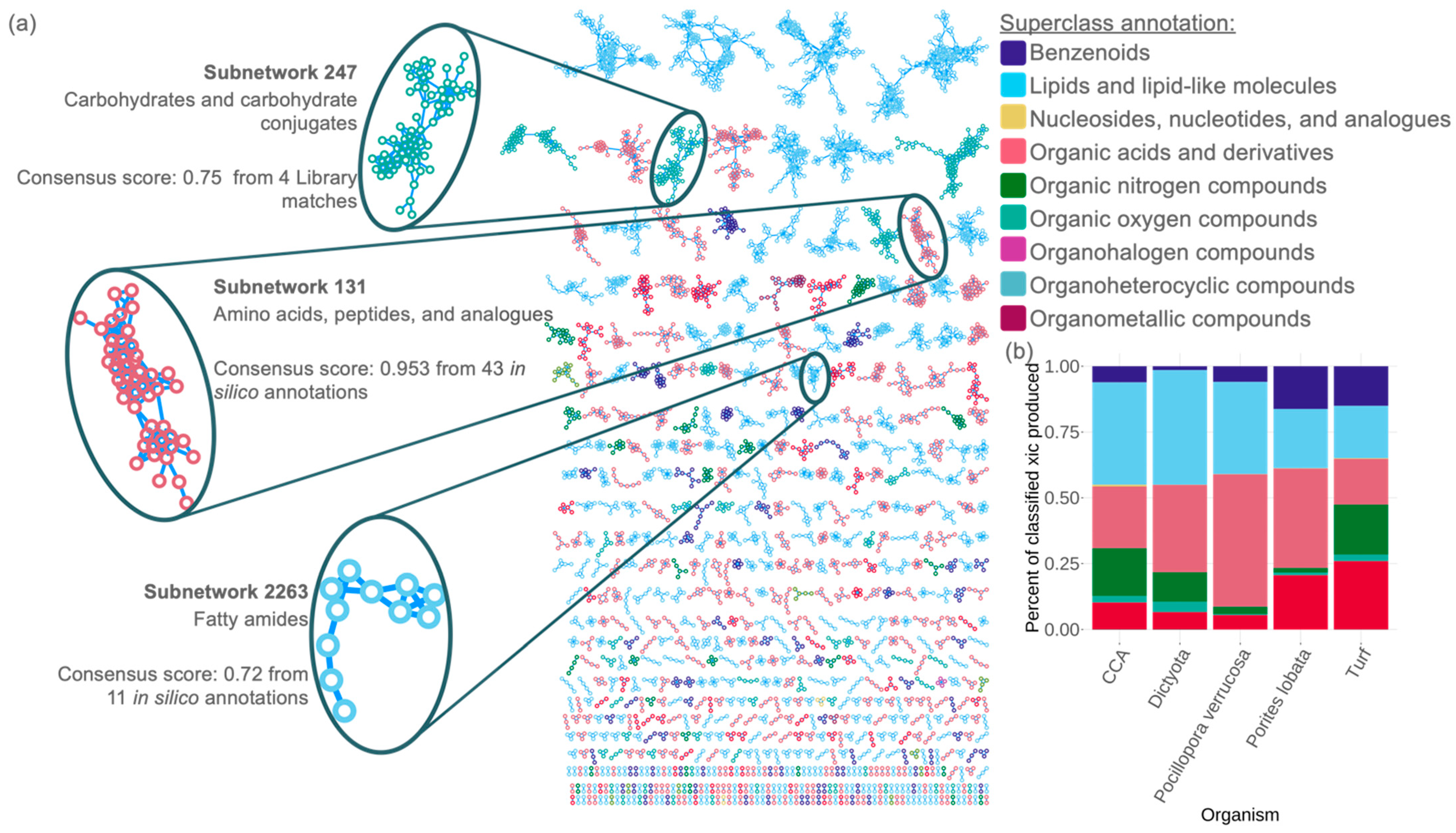
3.2. Case Study 1—Characterization of the Dissolved Organic Matter Pools from Dominant Coral Reef Primary Producers
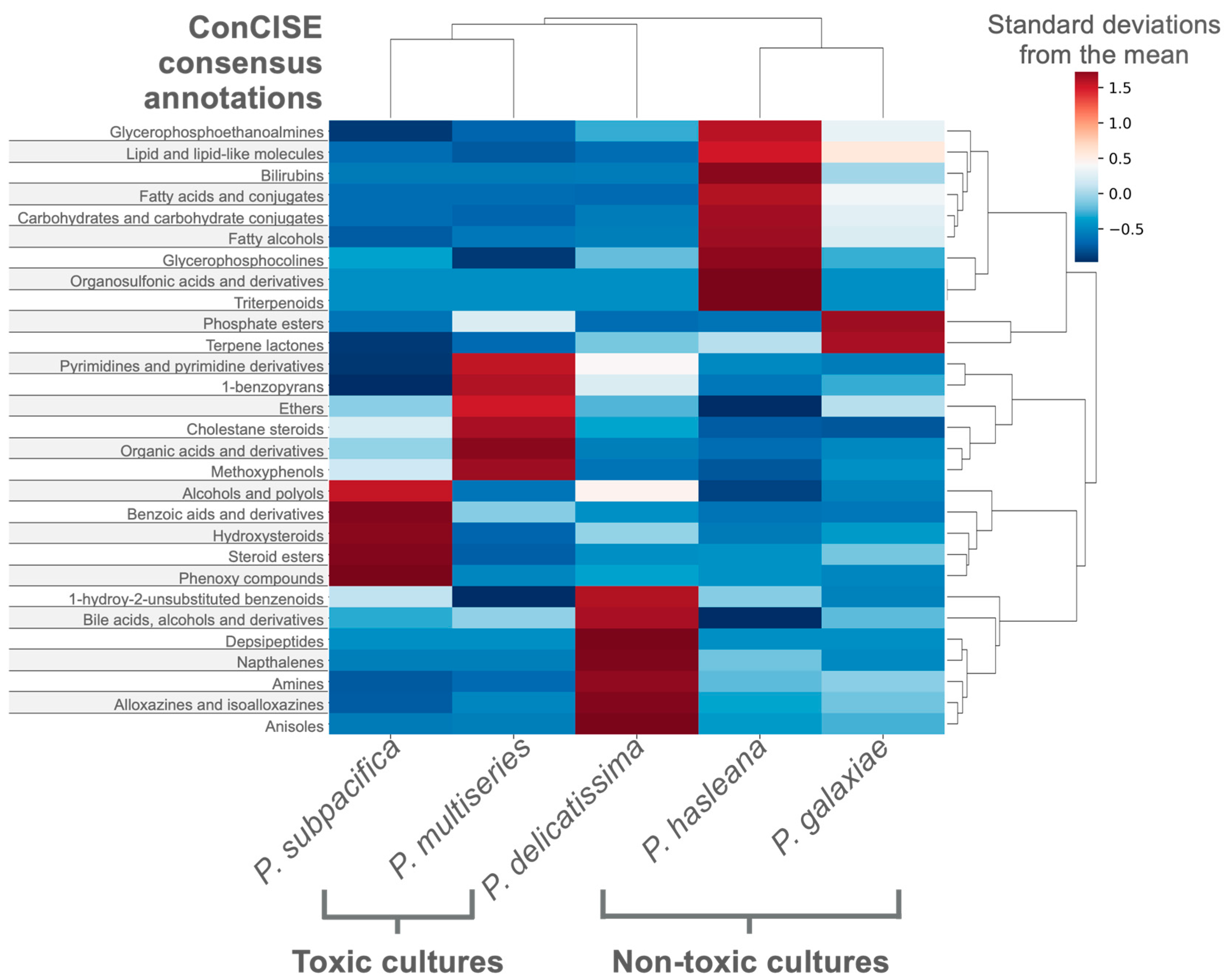
3.3. Case Study 2—Differentiating the Exometabolites of Five Pseudo-nitzschia Species
3.4. Case Study 3—Chemical Surey of Schoenmakerskop in the Eastern Cape of South Africa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wegley Kelly, L.; Nelson, C.E.; Aluwihare, L.I.; Arts, M.G.I.; Dorrestein, P.C.; Koester, I.; Matsuda, S.B.; Petras, D.; Quinlan, Z.A.; Haas, A.F. Molecular Commerce on Coral Reefs: Using Metabolomics to Reveal Biochemical Exchanges Underlying Holobiont Biology and the Ecology of Coastal Ecosystems. Front. Mar. Sci. 2021, 8, 630799. [Google Scholar] [CrossRef]
- Wegley Kelly, L.; Nelson, C.E.; Petras, D.; Koester, I.; Quinlan, Z.A.; Arts, M.G.I.; Nothias, L.-F.; Comstock, J.; White, B.M.; Hopmans, E.C.; et al. Distinguishing the molecular diversity, nutrient content, and energetic potential of exometabolomes produced by macroalgae and reef-building corals. Proc. Natl. Acad. Sci. USA 2022, 119, e2110283119. [Google Scholar] [CrossRef] [PubMed]
- Petras, D.; Koester, I.; Da Silva, R.; Stephens, B.M.; Haas, A.F.; Nelson, C.E.; Kelly, L.W.; Aluwihare, L.I.; Dorrestein, P.C. High-Resolution Liquid Chromatography Tandem Mass Spectrometry Enables Large Scale Molecular Characterization of Dissolved Organic Matter. Front. Mar. Sci. 2017, 4, 405. [Google Scholar] [CrossRef]
- Osterholz, H.; Singer, G.; Wemheuer, B.; Daniel, R.; Simon, M.; Niggemann, J.; Dittmar, T. Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 2016, 10, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.A.; Kujawinski, E.B.; Stubbins, A.; Fatland, R.; Aluwihare, L.I.; Buchan, A.; Crump, B.C.; Dorrestein, P.C.; Dyhrman, S.T.; Hess, N.J.; et al. Deciphering ocean carbon in a changing world. Proc. Natl. Acad. Sci. USA 2016, 113, 3143–3151. [Google Scholar] [CrossRef]
- Da Silva, R.R.; Wang, M.; Nothias, L.-F.; Van Der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLOS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Dittmar, T.; Koch, B.; Hertkorn, N.; Kattner, G. A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 2008, 6, 230–235. [Google Scholar] [CrossRef]
- Forsythe, I.J.; Wishart, D.S. Exploring Human Metabolites Using the Human Metabolome Database. Curr. Protoc. Bioinform. 2009, 25, 14.8.1–14.8.45. [Google Scholar] [CrossRef]
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T.; et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Nothias, L.-F.; Dührkop, K.; Koester, I.; Fleischauer, M.; Hoffmann, M.A.; Petras, D.; Vargas, F.; Morsy, M.; Aluwihare, L.; et al. Database-independent molecular formula annotation using Gibbs sampling through ZODIAC. Nat. Mach. Intell. 2020, 2, 629–641. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminformatics 2016, 8, 3. [Google Scholar] [CrossRef]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef]
- Jupyter, P.; Bussonnier, M.; Forde, J.; Freeman, J.; Granger, B.; Head, T.; Holdgraf, C.; Kelley, K.; Nalvarte, G.; Osheroff, A.; et al. Binder 2.0—Reproducible, interactive, sharable environments for science at scale. In Proceedings of the 17th python in science conference, Austin, Texas, 9–15 July 2018. [Google Scholar]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Heller, S.R.; McNaught, A.; Pletnev, I.; Stein, S.; Tchekhovskoi, D. InChI, the IUPAC International Chemical Identifier. J. Cheminform. 2015, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, Texas, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, N.; Brainard, R.E.; Fisher, R.; Moews, M.; Plaisance, L.; Caley, M.J. Coral reef biodiversity. Life World’s Ocean. Divers. Distrib. Abundance 2010, 4, 65–74. [Google Scholar]
- White, A.T.; Ross, M.; Flores, M. Benefits and costs of coral reef and wetland management, Olango Island, Philippines. Collect. Essays Econ. Coral Reefs 2000, 215–227. [Google Scholar]
- Cesar, H.S. Coral reefs: Their functions, threats and economic value. In Collected Essays on the Economics of Coral Reefs; Cesar, H.S.J., Ed.; CORDIO, Department of Biology and Environmental Sciences, Kalmar University: Kalmar, Sweden, 2000; pp. 14–39. [Google Scholar]
- Mathieu, L.F.; Langford, I.H.; Kenyon, W. Valuing marine parks in a developing country: A case study of the Seychelles. Environ. Dev. Econ. 2003, 8, 373–390. [Google Scholar] [CrossRef]
- Ahmed, M.; Umali, G.M.; Chong, C.K.; Rull, M.F.; Garcia, M.C. Valuing recreational and conservation benefits of coral reefs—The case of Bolinao, Philippines. Ocean. Coast. Manag. 2007, 50, 103–118. [Google Scholar] [CrossRef]
- Hughes, T.P.; Barnes, M.L.; Bellwood, D.R.; Cinner, J.E.; Cumming, G.S.; Jackson, J.B.C.; Kleypas, J.; Van De Leemput, I.A.; Lough, J.M.; Morrison, T.H.; et al. Coral reefs in the Anthropocene. Nature 2017, 546, 82–90. [Google Scholar] [CrossRef] [PubMed]
- McDole, T.; Nulton, J.; Barott, K.L.; Felts, B.; Hand, C.; Hatay, M.; Lee, H.; Nadon, M.O.; Nosrat, B.; Salamon, P.; et al. Assessing Coral Reefs on a Pacific-Wide Scale Using the Microbialization Score. PLoS ONE 2012, 7, e43233. [Google Scholar] [CrossRef]
- Nelson, C.E.; Goldberg, S.J.; Kelly, L.W.; Haas, A.F.; Smith, J.E.; Rohwer, F.; Carlson, C.A. Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 2013, 7, 962–979. [Google Scholar] [CrossRef] [PubMed]
- Haas, A.F.; Fairoz, M.F.M.; Kelly, L.W.; Nelson, C.E.; Dinsdale, E.A.; Edwards, R.A.; Giles, S.; Hatay, M.; Hisakawa, N.; Knowles, B.; et al. Global microbialization of coral reefs. Nat. Microbiol. 2016, 42, 16042. [Google Scholar] [CrossRef]
- Barott, K.L.; Rohwer, F.L. Unseen players shape benthic competition on coral reefs. Trends Microbiol. 2012, 20, 621–628. [Google Scholar] [CrossRef]
- Guannel, M.L.; Horner-Devine, M.C.; Rocap, G. Bacterial community composition differs with species and toxigenicity of the diatom Pseudo-nitzschia. Aquat. Microb. Ecol. 2011, 64, 117–133. [Google Scholar] [CrossRef]
- Sison-Mangus, M.P.; Jiang, S.; Tran, K.N.; Kudela, R.M. Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J. 2014, 8, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Hmelo, L.R.; Van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Koester, I.; Quinlan, Z.A.; Nothias, L.F.; White, M.E.; Rabines, A.; Petras, D.; Brunson, J.K.; Dührkop, K.; Ludwig, M.; Böcker, S. Illuminating the dark metabolome of Pseudo-nitzschia-microbiome associations. Environ. Microbiol. 2022, 24, 5408–5424. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quinlan, Z.A.; Koester, I.; Aron, A.T.; Petras, D.; Aluwihare, L.I.; Dorrestein, P.C.; Nelson, C.E.; Wegley Kelly, L. ConCISE: Consensus Annotation Propagation of Ion Features in Untargeted Tandem Mass Spectrometry Combining Molecular Networking and In Silico Metabolite Structure Prediction. Metabolites 2022, 12, 1275. https://doi.org/10.3390/metabo12121275
Quinlan ZA, Koester I, Aron AT, Petras D, Aluwihare LI, Dorrestein PC, Nelson CE, Wegley Kelly L. ConCISE: Consensus Annotation Propagation of Ion Features in Untargeted Tandem Mass Spectrometry Combining Molecular Networking and In Silico Metabolite Structure Prediction. Metabolites. 2022; 12(12):1275. https://doi.org/10.3390/metabo12121275
Chicago/Turabian StyleQuinlan, Zachary A., Irina Koester, Allegra T. Aron, Daniel Petras, Lihini I. Aluwihare, Pieter C. Dorrestein, Craig E. Nelson, and Linda Wegley Kelly. 2022. "ConCISE: Consensus Annotation Propagation of Ion Features in Untargeted Tandem Mass Spectrometry Combining Molecular Networking and In Silico Metabolite Structure Prediction" Metabolites 12, no. 12: 1275. https://doi.org/10.3390/metabo12121275
APA StyleQuinlan, Z. A., Koester, I., Aron, A. T., Petras, D., Aluwihare, L. I., Dorrestein, P. C., Nelson, C. E., & Wegley Kelly, L. (2022). ConCISE: Consensus Annotation Propagation of Ion Features in Untargeted Tandem Mass Spectrometry Combining Molecular Networking and In Silico Metabolite Structure Prediction. Metabolites, 12(12), 1275. https://doi.org/10.3390/metabo12121275





