Abstract
The importance of RNAs is commonly recognised thanks to protein-coding RNAs, whereas non-coding RNAs (ncRNAs) were conventionally regarded as ‘junk’. In the last decade, ncRNAs’ significance and roles are becoming noticeable in various biological activities, including those in hormonal and metabolic regulation. Among the ncRNAs: microRNA (miRNA) is a small RNA transcript with ~20 nucleotides in length; long non-coding RNA (lncRNA) is an RNA transcript with >200 nucleotides; and circular RNA (circRNA) is derived from back-splicing of pre-mRNA. These ncRNAs can regulate gene expression levels at epigenetic, transcriptional, and post-transcriptional levels through various mechanisms in insects. A better understanding of these crucial regulators is essential to both basic and applied entomology. In this review, we intend to summarise and discuss the current understanding and knowledge of miRNA, lncRNA, and circRNA in the best-studied insect model, the fruit fly Drosophila.
1. Introduction
The fruit fly Drosophila melanogaster is an important model organism used in biological research and is arguably the most extensively studied insect genus. Similar to other holometabolous insects, it has a life cycle enduring multiple morphological transitions—embryos, larvae, pupae, and adults [1]. Accompanying these life stage transitions, alterations in the metabolic system due to hormonal fluctuations result in the turnover of the cell population within the Drosophila’s body. In general, Drosophila is considered to have three instar larval stages determined by molting. During molting, the larvae shed a layer of the cuticle as a fresh layer replaces the previous layer modulated by hormones, a process known as ecdysis. Subsequently, the third instar larvae will transform into pupae and eventually emerge as adults. Two hormonal systems are responsible for modulating the entire process of development and metamorphosis: the sesquiterpenoid and the ecdysteroid systems [1,2,3,4,5]. In this review, we will discuss the established roles of non-coding RNAs (ncRNAs) in Drosophila and how select ncRNAs have been identified that regulate hormonal and metabolic pathways.
2. microRNAs in Drosophila
ncRNAs are a large and diverse class of RNA molecules that do not encode proteins, and their essential roles have only been recognised within the last few decades (Brandenburger et al., 2018 [6]). These include functional ncRNAs such as ribosomal RNA (rRNA) and transfer RNA (tRNA) involved in the protein translation process [7], and others with regulatory effects such as miRNAs and lncRNAs. MiRNAs are processed from larger double-stranded RNA precursors to create RNA molecules of 21–23 nucleotides. In contrast, lncRNAs are longer, more than 200 nucleotides long. With their differences in length, miRNA and lncRNA utilise different mechanisms to regulate the expression of their target genes. Moreover, lncRNA can bind with proteins forming a scaffold of ribonucleoprotein complexes [8] or function as a competing endogenous RNA that serves as a decoy for miRNA to bind to, eventually inhibiting miRNA regulatory effects [9]. In this section, we will first discuss the miRNAs which are summarized in Table 1.
MiRNAs are relatively conserved between bilaterians and cnidarians [10,11,12]. As one of the small ncRNA classes, primary miRNAs are transcribed from DNA and further processed into preliminary miRNAs by enzymes such as Drosha in the canonical miRNA biosynthesis pathway [13]. Preliminary RNAs are hairpin structures cleaved by Dicer and its cofactor Loquacious resulting in the formation of miRNA/miRNA duplexes. The miRNA/miRNA duplexes will be loaded by Argonautes (Ago), and one of the duplex strands will be retained to form a miRNA-induced silencing complex (miRISC). The miRISCs interact with the 3′ untranslated regions (UTRs) of the mRNAs by complementary binding, resulting in the transcriptional repression of the target mRNAs [2,3,14,15,16]. Specific mechanisms of transcriptional repression vary due to the utilization of different Ago proteins in the miRISC. Ago1-RISC represses the translation after 5′cap recognition, and Ago2-RISC blocks the recognition directly [15]. Previous studies also suggested that miRNAs are also capable of binding to open-reading frames (ORF) and 5′UTR [17,18]. Under specific circumstances, miRNA binding can induce mRNA expression by binding to the promoter sites or binding to the 5′TOP motif of rRNA to alleviate translational repressions [19,20,21]. In the Drosophila genome, 258 miRNAs have been annotated (miRBase Release 22.1) [22].
2.1. miRNA in Drosophila Development and Metamorphosis
Some miRNAs, such as miR-34, are maternally inherited in Drosophila and can regulate early embryonic development [23]. In a study knocking out ~130 microRNAs, several miRNAs were found to be essential for survival to adulthood, including bantam, miR-1, the let-7 cluster, miR-8, the miR-309 cluster, miR-263a, and miR-276a [24]. Among the 1065 maternal transcripts, 710 were identified as targets for the SMAUG protein. An additional 92 transcripts were regulated by miR-309 and SMAUG, which cooperatively monitor the degradation of maternal mRNA and the maternal-to-zygotic gene expression transition [25,26,27,28].
During Drosophila embryonic development, miRNAs regulate the apoptotic process [29]. By binding to the 3′UTR of mRNAs expressing reaper (rpr), grim, head involution defective (hid), and sickle (skl), miR-6 and miR-11 are shown to regulate embryonic apoptosis, and a double knock-out of miR-6/miR-11 induces defects in the central nervous system [29]. Another miRNA bantam also functions in the CNS scaling the growth of Class IV dendrite arbors in Drosophila sensory neurons in the late embryonic to early larval stages [30]. In larval development, miR-281, miR-311, miR-79, miR-92, miR-305, miR-131, and miR-31a were predicted to target genes involved in this process [31].
In addition to miRNA involvement in metamorphosis, miRNAs have been suggested to regulate gene expression in forming adult wing structures [32]. In Drosophila, let-7, miR-125, miR-1, miR-2b, miR-2c, miR-9a, and miR-13b control wing development [32,33,34,35], while loss of miR-987 function resulted in indirect flight muscle defects in an age-dependent manner [35]. Another morphogenesis aspect during the adult stage is the abdomen where miR-965 regulates string (stg) and wingless (wg) mRNAs to control its cell proliferation and migration [36], and leg development is controlled by a microRNA cluster miR-6/5/4/286/3/309 [37].

Table 1.
Summarisation of miRNAs reported with their validated targets, function, and hormonal and metabolic regulations according to their references.
Table 1.
Summarisation of miRNAs reported with their validated targets, function, and hormonal and metabolic regulations according to their references.
| miRNA | Validated Targets | Function, Hormonal and Metabolic Regulations | References |
|---|---|---|---|
| Development and Metamorphosis | |||
| miR-6, miR-11 | rpr, grim, hid, skl | Embryonic apoptosis and CNS development | [29] |
| bantam | Akt | Dendrite arbour growth | [30] |
| miR-281, miR-311, miR-79, miR-92, miR-305, miR-131, miR-31a | N/A | Larval development | [31] |
| let-7, miR-125 | N/A | Wing development | [33] |
| miR-9a | dLMO (Beadex) | Wing development | [34] |
| miR-1, miR-2b, miR-2c, miR-13b, miR-987 | N/A | Wing development | [35] |
| miR-965 | stg, wg | Abdomen morphogenesis | [36] |
| miR-6/5/4/286/3/309 | N/A | Leg development | [37] |
| miR-277, miR-304 | mbl | Muscle development | [38] |
| Sesquiterpenoid and Ecdysteroid system | |||
| bantam | JHAMT | JH biosynthesis | [39] |
| miR-252, miR-304 | JHAMT | JH biosynthesis | [39] |
| miR-8, miR-14, miR-34, miR-278 | Met/Gce | JH signalling pathway | [39] |
| miR-927 | Kr-h1 | JH signalling pathway | [40] |
| bantam | phantom, shade, disembodied | Ecdysone biosynthesis | [41] |
| miR-14 | EcR | Ecdysone signalling pathway | [42] |
| let-7, miR-125 | dpt | Regulated by ecdysone; innate immune systems | [43] |
| miR-8 | N/A | Cell growth regulated by ecdysone; induce JH biosynthesis pathway | [44] |
| miR-252 | Abi | Cell division | [45] |
| Insulin pathway and Lipid metabolism | |||
| miR-8 | ush | Insulin signalling pathway; body size | [46] |
| miR-7 | cpa | Insulin secretion pathway | [47] |
| miR-9a | short neuropeptide F receptor 1 | Insulin signalling pathway; body size | [48] |
| miR-14 | sg | Insulin-producing pathway; fat accumulation | [49] |
| miR-305 | Dp53 | TOR regulator signalling in response to nutrient | [50] |
| miR-305 | InR, PI3K, Hairless | Notch and insulin pathways in adaptive homeostasis | [50,51] |
| miR-277 | FAO | Fatty acid metabolism; homeostasis | [52] |
| miR-14 | N/A | Fatty acid metabolism | [53] |
| miR-278 | ex | Insulin sensibility | [54] |
| Sexual Development | |||
| miR-184 | saxophone | Female germline development; nurse cell nutrient support; Egg chamber size | [55] |
| miR-318 | Tramtrack69 | Oogenesis; chorion gene amplification | [56] |
| miR-282 | rut | Egg production; apoptotic activity | [57] |
| miR-989 | N/A | Border cell migration in ovaries | [58] |
| bantam | N/A | Adult germline stem cell formation | [59] |
| bantam | dFmr1 | GSCs maintenance in ovaries | [60] |
| miR-7, miR-309, miR-278 | dap | Cell cycle regulations | [61] |
| let-7 | N/A | Sex-biased ecdysone signalling; aging in testis stem cells and fertility | [62,63] |
| miR-190 | N/A | Sexually dimorphic in male; regulating neuronal activities and lifespan | [64] |
| Lifespan and Aging | |||
| bantam, miR-1, miR-190, miR-279, miR-996 | N/A | Survival; viability | [24] |
| miR-282 | N/A | Increase lifespan | [57] |
| miR-305 | N/A | Aging; locomotor activity; abnormal protein aggregation in muscle; oxidative stress | [65] |
| miR-184, let-7 | N/A | Prolonged lifespan | [66,67] |
| miR-125 | Chinmo | Prolonged lifespan | [68] |
| miR-34 | Pcl, Su(z)12 | Chaperone expressions; healthy brain aging | [69,70] |
| miR-34 | Lst8 | Healthy brain aging | [71] |
| miR-277 | N/A | Branched-chain amino acid catabolism; growth regulation; Reduced lifespan | [72] |
| Circadian Rhythm and Photoperiod | |||
| miR-124 | BMP signalling | Circadian rhythm; rhythmic normality | [73] |
| miR-276a | tim | Circadian rhythm | [74] |
| miR-279 | unpaired | Circadian behaviour; JAK/STAT circuit | [75] |
| miR-959-964 | N/A | Rhythmic feeding; loop of feeding period control | [76] |
| miR-210 | Fasciclin 2 | Photoreceptor function | [77] |
| miR-263a, miR-263b | clk, cwo | Circadian rhythm | [78] |
| let-7 | cwo | Circadian rhythm | [79] |
2.2. miRNA in the Sesquiterpenoid and Ecdysteroid Systems
The entire process of metamorphosis in insects is initiated and regulated by two hormonal systems, namely the sesquiterpenoid and the ecdysteroid hormone systems [2,3,4,80]. The corpus allatum (CA) in the ring gland of Drosophila produces three forms of sesquiterpenoid hormones: juvenile hormone III (JH III), juvenile hormone bisepoxide (JHB3), and methyl farnesoate (MF). Juvenile hormones induce the expression of specific genes such as Krüppel homolog 1 (Kr-h1). The signalling network results in the inhibitory effect of these genes towards the molting process and postpones the initiation of metamorphosis, prolonging the juvenile characteristics of the larvae.
The prothoracic gland (PG) in the ring gland produces ecdysone through the ecdysteroid biosynthesis pathway by enzymes expressed from a set of Halloween genes. Ecdysone is converted into 20-hydroxyecdysone (20E) by the enzyme encoded by shade in the peripheral tissues, such as the fat body [81,82]. Ecdysones are essential for the transition of signalling pathways utilising the transcription factor Chronologically Inappropriate Morphogenesis (Chinmo) to Broad/E93 transcription factors [83]. Williams and Kafatos brought up a theory fifty years ago, suggesting there may be three “master regulatory genes” that can reciprocally inhibit each other, and each will be responsible for regulating a set of “stage-specific genes” in the larval, pupal, and adult stages [84]. Later findings disputed the theory of stage-specific genes since many genes are responsible for metamorphosis and may be expressed in more than one stage. Referencing this theory, Truman and Riddiford found the three “master regulatory” transcription factors Chinmo, Broad, and E93 reciprocally inhibit each other, and each of them induce the expression of different downstream genes throughout metamorphosis due to the induction of JH and 20E [85]. However, the ecdysteroid turnover remains obscure for many insects, including Drosophila. It has been established that the ecdysteroid variations may differ in Drosophila due to diet and development [86]. Regardless of the unknown biosynthesis mechanism, it is well-acknowledged that the two types of hormones collaboratively regulate the proceeding of life stages and control larval molting or onset of metamorphosis in arthropods via binding to their respective receptors [87]. The different levels of antagonistic hormones within the ring gland determine the developmental stages’ transition [88]. Both types of hormones yield the potential as transcription activators and inhibitors, elevating or repressing their target’s transcription level. Some have argued that JH stimulates the Broad-Complex (Br-C) in hemimetabolous insects, and through the evolution to holometabolan insects, JH became inhibitory to the Br-C transcripts [89]. The repression of Br-C by JH in holometabolan insects is well-established and indisputable [90,91,92].
MiRNA can regulate the JH pathway and the ecdysone pathway by binding to the transcripts of protein-coding genes in the process of hormone biosynthesis and the downstream signalling pathways [39,41]. Given that the hormonal pathway of Drosophila regulates the development and onsets of metamorphosis, evaluating miRNA regulation on the hormonal pathways would be critical for understanding insect development.
The miRNA expression levels differ among developmental stages in Drosophila [93,94], suggesting these differentially expressed miRNA may be correlated with development and metamorphosis. Many enzymes catalyze the sesquiterpenoid biosynthetic pathway, and the juvenile hormone acid O-methyltransferase (JHAMT) is responsible for the rate-determining step [3]. The JHAMT expression level determines the JH titre, and DmJHAMT knockdown mutant males have disoriented male genitals [95]. Overexpression of microRNA bantam resulted in similar phenotypes [39]. In addition, in bantam overexpression mutants, JHIII and JHB3 titres were significantly reduced [39]. Moreover, in vitro dual luciferase reporter assay and pull-down assay verify the interaction of bantam and JHAMT mRNA transcript, confirming the regulation of bantam in the sesquiterpenoid pathway. Furthermore, in vitro experiments demonstrate the potential miRNA candidates for regulating the expression of JHAMT mRNA and the JH receptors Methoprene-tolerant (Met) and Germ cell expressed (Gce) mRNA. MiRNA candidates targeting the JHAMT transcript may include bantam, miR-252, and miR-304, and the miRNA candidates targeting the Met/Gce transcript include miR-8, miR-14, miR-34, and miR-278 in Drosophila [39].
Interestingly, bantam inhibits ecdysone production by indirectly regulating some of the Halloween-gene-expressed mRNAs, encoding the enzymes for the ecdysteroid biosynthesis pathway [41]. Boulan et al. suspects the high bantam level was for maintaining lower ecdysone titre and promoting growth, while the lowered bantam level at the third instar larvae stage is for the surge of ecdysteroids for entering the metamorphic transitions [41].
As JH binds to the Met/Gce receptor, the complex will initiate the transcription of Kr-h1. It is worth noting that miR-927 can bind to the transcript of Kr-h1 and down-regulate its expression in Drosophila [40], and its mutants exhibit similar traits to the Kr-h1 mutants [96]. It has been previously established that miR-14 has the potential to regulate the JH system via the Met/Gce receptor. There have also been reports on the regulatory role of miR-14 in ecdysteroid systems. miR-14 knock-out mutants have overexpression of ecdysone receptor (EcR) transcripts and downstream gene transcripts [97]. These two findings suggest that miR-14 may be one of the key regulators in hormonal systems, as shown in Figure 1.

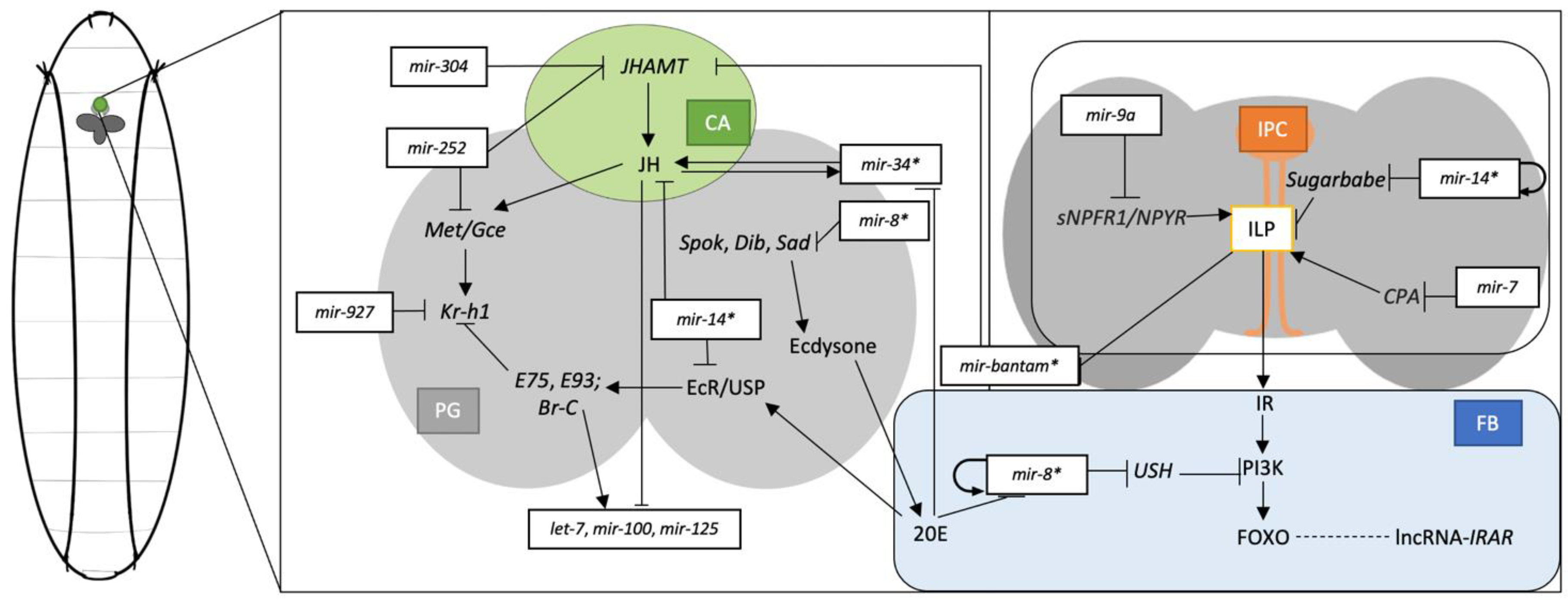
Figure 1.
Drosophila larval hormonal signalling systems and metabolic systems represented with the juvenile hormone (JH), ecdysone, and insulin in the ring gland (RG), insulin-producing cells (IPC) from the brain, and adipose cells from fat body (FB). The RG is represented with corpus allatum (CA) coloured in green, prothoracic gland (PG) in light grey, IPCs in orange, the brain in dark grey, and fat body (FB) in blue. MiRNAs regulation on the specific pathways are represented throughout the pathways. Hormonal crosstalk involves miR-34*, miR-8*, bantam*, and miR-14* as they interplay in multiple systems.
As described, miRNAs may have regulatory effects on the sesquiterpenoid and ecdysteroid systems, and vice versa, since the two systems also induce the expression of miRNAs. Similarly, in Drosophila, let-7 and miR-125 are co-expressed during metamorphosis, and mutants of these miRNAs also experience defects throughout metamorphosis [33]. The expression of let-7, miR-125, miR-100, and miR-34 can also be observed during the Drosophila development stages, each occurring according to specific hormones [40,41]. The let-7, miR-100, and miR-125 cluster expressions are enhanced by the introduction of ecdysone, whereas the expression of miR-34 is strictly inhibited. Meanwhile, the expression level of miR-34 is enhanced by the introduction of JHs, but the let-7, miR-100, and miR-125 are strictly inhibited [41]. Since Qu et al. [39] demonstrated the potential inhibitory effect of miR-34 on Met/Gce, miR-34 can regulate the sesquiterpenoid pathway through feedback inhibition. Br-C knock-out mutants significantly reduce the RNA level of let-7, miR-100, and miR-125. Br-C is crucial for the up-regulation of let-7, miR-100, and miR-125. Eventually, by introducing JH and ecdysones separately, the corresponding miRNA transcript levels increase or decrease accordingly. Thus, JH induces the transcription of miR-34, and ecdysteroid enhances the transcription of let-7, miR-100, and miR-125 via the Br-C [41,42].
Ecdysone-induced miRNA transcription can further regulate other pathways. Let-7 and miR-125 were validated for their regulatory effects on innate immune systems, with let-7 regulating the antimicrobial peptide gene diptericin (dpt) [43]. Conserved ecdysone-induced miR-8 regulates the insulin signalling pathway by targeting the u-shaped (ush) transcript, and the overexpression of miR-8 will result in mutants with larger body sizes [98]. Moreover, enhanced miR-8 has been found to promote cell growth in the CA and elevation in the juvenile hormone biosynthesis pathway independently from the insulin signalling pathway [44]. Apart from the inhibitory effect of miR-252 on JHAMT transcripts, ecdysone-responsive miR-252 controls the cell cycle of Drosophila by targeting the Abelson interacting protein (Abi), suggesting more cell division is occurring through metamorphosis due to the presence of miR-252 [45,94].
Even though many regulatory effects of miRNAs on hormonal systems have been revealed and validated, many potential miRNA-mRNA interactions remain in predictions. The miRNA target predictions are primarily established through the algorithm estimating the stability of annealed complementary pairing of seed regions of miRNAs and mRNAs [99,100]. Certain regions of the transcript sequences may yield a conserved site for miRNA-mRNA interaction. These predictions suggest that some downstream genes of the ecdysteroid system interact with specific miRNAs [31]. Furthermore, the ecdysone-induced miRNA differentially expressed profile with 72 distinct miRNAs was identified with or without 20E [101]. More tools utilising different algorithms are available for more advanced predictions as well. These findings suggest that the current understanding of potential miRNA regulation on the sesquiterpenoid and ecdysteroid pathways is only the tip of the iceberg. The unrevealed modulation will be necessary for a wholesome understanding of the Drosophila endocrine system. Future investigation may also focus on miRNA regulation of hormonal production due to neuropeptide signalling of the ring gland for understanding the crosstalk in the endocrine systems of Drosophila [102].
2.3. Insulin Pathway and Lipid Metabolism
The crosstalk between the hormonal and metabolic pathways are exceptionally sophisticated, as exemplified by the insulin and ecdysteroids signalling pathways. [2]. The previous section mentioned that miR-8 regulates the insulin pathway through ush with the induction of ecdysteroids [98]. More specific to the insulin pathway, the Drosophila insulin-like peptides (dILPs) bind to the insulin receptors (IR) and trigger the downstream phosphorylating cascade involving components such as chico, phosphoinositide-3 kinase (PI3K), and Akt kinase [103,104,105,106]. Since miR-8 regulates the ush transcript in fat cells, miR-8 null mutants suffer from increased brain cell apoptosis and some flies have malformed legs and wings [46]. miR-8 and ush can regulate insulin receptor expression level through the PI3K pathway [46]. miR-7 regulates the secretion of insulin-like peptides (ilp) by targeting the F-actin capping protein alpha (CPA) [47]. On the other hand, insulin signalling represses bantam transcription through dILP regulations, interfering with the ecdysteroid system as well [95].
Additional miRNAs are responsible for insulin pathway regulations and energy metabolism. Insulin-producing cells are regulated by miR-9a and by body sizes [48]. Insulin production from insulin-producing neurosecretory cells is regulated by miR-14 through the regulation of Sugarbabe (sug), which is directly related to the insulin-producing pathway [49]. Overexpression of sug results in fat accumulation and reduces the insulin-like peptide’s mRNA expression level. Unlike sug, miR-14 does not seem to be controlled by nutritional status and may play a part in insulin production and fat metabolism maintenance [49]. The Drosophila homolog of p53 (Dp53) is regulated by miR-305 in a nutrient-dependent matter [50]. The expression level of miR-305 varied between well-fed and nutrient-deprived animals, and the system is induced by TOR regulator signalling [50]. miR-305 was found to regulate the Notch and insulin pathways in intestinal stem cells of Drosophila [51]. Through these two pathways, Foronda et al. also presented the potential of miR-305 in mediating adaptive homeostasis in the Drosophila gut [51]. Similarly, miR-277 regulates fatty acid β-oxidation (FAO) enzymes in intestinal stem cells. The differentially expressed genes are identified in miR-277 mutants [52]. Validations suggested disrupting homeostasis and progenitor cell survival with miR-277 mutants [52]. The alteration of fat metabolism by miR-14 suggested that the triacylglycerol and diacylglycerol levels differ in miR-14 mutants [53].
In the insulin signalling pathway, miR-278 targets expanded (ex) to sustain insulin sensibility [54]. Interestingly, miR-278 is abundantly expressed in wild-type flies, keeping the ex mRNA at lower levels in the adipose tissue, unlike the minute dosage of many other miRNAs [54]. Such abundant expression of miR-278 could be an example of solid modulation of miRNAs on its target transcript, proving the importance of its regulation in a cellular system.
2.4. miRNA in Sexual Development
Larval ovary morphogenesis in Drosophila is closely related to the ecdysone, insulin, Activin, Dpp, and EGFR signalling pathways. It has also been reported that the miRNA pathway monitors Drosophila larval ovary morphogenesis [107]. Sexual development and maturation in Drosophila are monitored and determined by miRNAs. Reports on miR-184 in female germline development reveal the roles it plays [55]. Mutant females are reduced in size and may fail to develop functional nurse cells due to the lack of oocyte nutrient support. This results in the reduced egg chamber size in females [55].
Similarly, miR-318 has multiple roles during oogenesis. Ecdysone signalling pathways induce the expression of miR-318, and miR-318 interferes with the follicle cells’ involvement in chorion gene amplifications to synthesize the eggshell structure [56]. miR-282 is associated with egg production due to elevated apoptotic activity in mutant mir-282 ovaries, possibly due to the increased rutabaga (rut) adenylate cyclase gene targeted by mir-282 [57]. Another study revealed that border cell migration in the ovary requires the modulation of miR-989. Mutants without miR-989 displayed migration of border cells in mutant egg chambers for the somatic cells but not in the germline cells [58].
The Bantam loss-of-function mutant resulted in defects of adult germline stem cells (GSC) [59]. Bantam is also associated with the Drosophila ortholog of Fragile X mental retardation protein (dFmr1) in regulating GSCs maintenance and sustaining GSCs within Drosophila ovaries [60]. Dacapo (dap) is a cyclin-dependent kinase inhibitor that monitors the cell cycle in the GCS. miR-7, miR-309, and miR-278 directly regulate the dap 3′UTR. Among them, miR-7 and miR-278 may affect the cell cycles in GCSs as mir-7 and mir-278 knock-out mutants have reduced division kinetics and a reduced number of progeny cells [61]. However, not only do miR-7, miR-278, miR-309, and bantam serve the sexual development in Drosophila, but the monitoring enzymes of miRNA biosynthesis, Dicer and Pasha, are required for ovary morphogenesis [60]. Yang et al. identified the miRNAs that are differentially expressed in the Drosha mutant [60], including let-7, miR-2, miR-8, miR-14, miR-33, miR-125, miR-184, and miR-277. Nevertheless, the differentially expressed genes in the germline of ovariole morphogenesis have been identified by Tarikere et al. [108]. The target genes of the mentioned miRNAs may be differentially expressed and identified for future studies.
Among miRNAs, let-7 determines sexual identity in male-specific regulation of the X chromosome. There is a sex-biased ecdysone signalling where let-7 plays a key role as modulator [62]. Let-7 also contributes to the regulation of aging in testis stem cells, regulating the fertility of Drosophila [63]. However, miRNAs not only regulate gonadogenesis and the morphological development of the two sexes, but they may also be expressed in a sexual dimorphic manner. Fernandes and Varghese [64] found that miR-190 shows higher expression levels in male flies and maintains neuronal activities and lifespan. This finding suggests that the miRNA expression level differences in males and females may be due to other regulatory differences that are worth further investigation.
2.5. miRNA in Drosophila Lifespan and Aging
Apart from the miRNAs mediating cell death, some miRNAs are associated with lifespan and longevity, and other specific miRNAs result in reduced viability. For instance, bantam, miR-1, miR-190, miR-279, and miR-996 were reported to be essential for survival, and the loss of these miRNAs resulted in reduced viability [24]. As previously mentioned, miR-282 regulated egg production in sexual development. miR-282 knock-out mutants have shortened lifespans by 50% on average [57]. miR-305 regulates the aging effect, and miR-305 expression levels decrease through aging. It has also been shown that miR-305 is correlated with locomotor activity, abnormal protein aggregation in muscle, and oxidative stress [65]. In Drosophila, miR-125, miR-184, and let-7 alter Drosophila lifespan [66,67]. It is first known to have reduced effects on lifespan with let-7 down-regulated mutants, and, following such findings, let-7 overexpression resulted in a prolonged lifespan [66,67]. miR-125 and let-7 expression is elevated in response to dietary restriction, thus extending lifespan. miR-125 regulates the nutrient-dependent transcription factor Chinmo with dietary restriction and restrains the downstream genes of Chinmo from expressing to increase the lifespan in flies [68]. These findings confirm miRNA impacts on increased lifespan and the importance of dietary restrictions.
miR-277 and miR-34 from the same cluster have been reported to interfere with lifespan. miR-277 has been known to control branched-chain amino acid catabolism and further regulated growth regulator TOR kinase. miR-277 expression reduced lifespan, especially on protein-rich diets [72]. On the other hand, miR-34 was associated with healthy brain aging and extended lifespan [69]. miR-34 was proven to regulate Pcl and Su(z)12 mRNA, resulting in higher chaperone expressions [70]. Lst8 is also regulated by miR-34 and has proven to be differentially expressed in mutant animals [71]. Altogether, miR-34 maintains proteostasis progress and results in healthy brain aging.
2.6. miRNA in Drosophila Circadian Rhythm and Photoperiod
Circadian rhythm in animals can be linked to locomotor activity and feeding behavior, sleep/wake patterns, various physiological and metabolic pathways, and even lifespan [109,110]. The circadian rhythm in Drosophila is moderated by the circadian feedback regulatory loop composed of the clock (clk), cycle (cyc), period (per), and timeless (Tim) genes in circadian neurons [111]. Kadener et al. suggested that the 3′UTR can be regulated through miRNAs and the miRNA biosynthesis pathway [112]. As a loss of function in the per mutant will result in an alteration in lipid metabolism and increased susceptibility to starvation with lack of nutrients, it is likely for many metabolic pathways to be altered due to circadian rhythm alterations [113]. miR-276a and miR-124 modulate the circadian rhythm, and mutants exhibit behavioural arrhythmicity [73,74]. miR-279 regulates circadian behaviour through the JAK/STAT circuit for cell signalling [75], while miR-996 regulates rhythmic behaviour [114]. Other than that, the miR-959-964 cluster is determined by rhythmic feeding and forms a loop with feeding period control [76]. Locomotor rhythm is regulated by miR-210 through Fasciclin 2, a cell adhesion molecule possibly associated with photoreceptors [77]. Analysis of the miRNA expression profile due to photoperiod reveals miR-2b, miR-11, miR-34, miR-274, miR-184, and miR-285 [115]. These miRNAs may also be associated with circadian rhythm.
Several miRNAs are differentially regulated when flies are put in constant darkness compared to the light-dark cycle [78]. Through target site predictions, miR-263a and miR-263b are predicted to regulate clk and a circadian rhythm transcription repressor clockwork orange (cwo), as some other miRNAs were identified and predicted to interfere with the circadian rhythm [78]. Let-7 is proven to repress the expression level of cwo. Since circadian prothoracicotropic hormone acts as a transcription factor to initiate ecdysteroid signalling, a regulatory circuit of the circadian cycle consisting of the ecdysteroid system and let-7 is established [79]. It is also worth noting that let-7, miR-375, miR-92a, and bantam are identified as regulating sleep and sleep homeostasis in multiple ways [116,117,118,119,120].
Loss of fragile X mental retardation protein (dFmr) is also associated with altered circadian rhythms and leads to altered expression of miR-1 and miR-281 in fragile X syndrome [110]. It is also suggestive that miR-1 and miR-281 can be associated with circadian rhythm.
3. lncRNA in Drosophila
LncRNAs are RNA molecules with more than 200 nucleotides in length that can function as signals, decoys, guides, and scaffolds [121]. Most lncRNAs are transcribed through the RNA polymerase II (Pol II) with a 5′ cap and 3′ poly-A tail [122]. Even though many lncRNA functions remain unknown, the complexity of lncRNAs in Drosophila is not dismissible for their potential roles in transcriptional activation and inhibition [123]. LncRNAs can interfere with mRNA stability and transcription levels, mRNA splicing, and protein activities, and they act as small RNA precursors or sponges [123]. For instance, the Drosophila long non-coding heat-stress-inducible hsrω transcripts appear to monitor protein synthesis status and are developmentally regulated under non-heat shock conditions [124,125]. The lncRNAs that are discussed in this section are summarised in Table 2. Conservation of lncRNAs across species is still noticeable, with many lncRNA orthologues identified between flies, mice, and humans [126].

Table 2.
Summarisation of lncRNAs reported with their validated targets, function, and hormonal and metabolic regulations according to their references.
3.1. lncRNA in Development, Metamorphosis, and Ecdysteroid Hormone Systems
In Drosophila embryogenesis, intraabdominal (iab) is known for regulation of the homeotic (Hox) transcription factors. Ultrabithorax (Ubx), Abdominal-A (Abd-A), and Abdominal-B (Abd-B) are known for the spatiotemporal expression pattern in development and its mutants will have disrupted abdominal segments [127]. However, for lncRNA:PS4, despite having the same expression profile as Ubx, promoter deletion in the lncRNA:PS4 mutant shows no direct expression difference in Ubx [140]. Another lncRNA, acal, is shown to be responsible for dorsal closure through the JNK signalling pathway, and mutations in acal are lethal [128].
LncRNAs that are differentially expressed throughout metamorphosis have been identified previously. Among all, 21% and 42% lncRNAs were significantly up-regulated at late-embryonic and late-larval stages, respectively, suggesting a drastic change in transcriptome throughout different life stages [141]. For example, leg development in Drosophila is dependent on lncRNA:CR33938 and a mutant of this lncRNA results in alteration of expression level in leg development genes [129]. Relevant to the metamorphosis process that is regulated by the ecdysone signalling pathways, a study on the hormone response network for ecdysone reveals four ecdysone-induced lncRNAs: CR43432, CR43626, CR45391, and CR45424, without any further analysis [142].
3.2. lncRNA in Nutrient Metabolism and Aging
Metabolism can be altered due to many reasons, and one is infection-induced alteration. The lncRNA IBIN is overexpressed in response to bacterial infection [130]. The induction of IBIN results in the up-regulation of carbohydrate metabolism gene cluster expression and the down-regulation of the amino acid metabolism gene cluster [130]. Such findings suggest that transcriptomic regulation of the immune system and the modulation of pathway preferences in energy metabolism can be regulated by lncRNAs. Another study suggests that with changes in environmental nutrition, lncRNA-IRAR mutants of overexpression are more sensitive to the changes and CRISPR-Cas9 mutants are less sensitive [131]. The underlying mechanism has been identified as the insulin receptor transcript expression level due to the similar expression pattern through FOXO binding in the insulin pathway [131].
The environmental nutrient affects the aging process through dietary restriction. The expression profile of flies with dietary restrictions and fully fed flies identifies 102 differentially expressed lncRNAs and 1406 differentially expressed mRNAs, suggesting the potential roles of these transcripts may be differentially expressed due to dietary restrictions. The differentially expressed transcripts are annotated with aging-related signalling pathways in the GO and KEGG databases [143].
3.3. lncRNA in Sexual Development
Many annotated lncRNAs have been identified as playing roles in sexual development. Integrated data with RNA-seq and ChIP-seq identified the expressed mRNA and lncRNA throughout the Drosophila melanogaster lifespan revealing a high level of lncRNAs in pupal stages and adult males in comparison to adult females, suggesting the expression profile differences in sexes for lncRNA expression profiles [132]. The conservation of lncRNAs in the Drosophila genus reveals that trends observed in Drosophila melanogaster may not be followed by Drosophila pseudoobscura. Both species have shown sex differences in lncRNA expression profiles to be more abundant in males for male development. However, the pupal lncRNA expression profile elevation was not observed in D. pseudoobscura [144].
The lncRNA expression profile shows excess lncRNA expression in adult males within days of emergence, potentially associated with adult male sexual development [132]. Male-specific abdominal (msa) lncRNA is required for accessory gland development in males. The MSA transcript overlaps with lncRNA iab-8, both embedded with miR-iab-8. The lncRNA iab-8 and the miRNA embedded with miR-iab-8 have a regulatory effect on Hox genes, whereas the lncRNA msa regulates secondary cell morphology and male fertility [133]. The two different lncRNAs, even though embedding the same miRNA, have different regulatory effects on different tissues [133]. This difference is caused by the micro-peptides encoded within the small ORF within the lncRNA msa transcript, demonstrating the potential for micro-peptide encoding lncRNAs [145].
The lncRNAs roX1 and roX2 are associated with the male-specific lethal (MSL) protein complex, and double roX gene knock-out mutants are lethal in males, suggesting the essential role of the roX genes [134]. The formation of the MSL protein complex and roX genes will increase X-linked gene transcription, resulting in a hyperactive X chromosome in males [135]. Since females have two pairs of X chromosomes, the dosage compensation effect is not required. Therefore, roX gene expression is only detected at the initial blastoderm stage in embryos [146]. Furthermore, the processes of spermatogenesis and oogenesis have been reported to be regulated by lncRNAs. Systematic identification of testes-associated lncRNAs reveals that around 30% of lncRNAs are essential for male fertility and late spermatogenesis, with one severe case of CR44455/6 knock-out mutant, suggesting the potential regulatory effects of lncRNAs [136]. However, CR45362, even though not mentioned in the systematic identification, has been reported to regulate spermatogenesis by interacting with α-Spectrin for spermatid nuclear bundling, as knock-out mutants exhibit disrupted spermatid nuclear bundling and α-Spectrin interactions [143]. An interesting case is demonstrated with Oskar mRNA in female oogenesis. Oskar is identified as a protein-coding gene, but Jenny et al. showed that it can regulate early oogenesis progress solely with the mRNA 3′UTR rescue; therefore, functioning as a lncRNA [137].
3.4. lncRNA in Drosophila Circadian Rhythm
The lncRNA yellow-achaete intergenic (yar) is reported to have a regulatory effect on circadian rhythm, affecting sleep behavior in Drosophila [139]. The conservation of yar is established among Drosophila species, and transcriptional regulation is also conserved. Mutants with a deletion in yar show no morphological differences, but behavioral changes in sleep regulation have been observed. Fragmented and reduced nighttime sleep is caused by yar mutants and results in sleep deprivation [139]. Unlike many other lncRNAs retained in the nucleus, yar is cytoplasmic and, therefore, may be suggestive of possible post-translational regulation for future investigation [139].
3.5. lncRNA and miRNA Interactions
In the previous section, we mentioned that lncRNA msa, lncRNA iab-8, and miR-iab-8 were encoded within the same region, demonstrating some potential connections of miRNA-lncRNA interplay [133]. However, a recent study on target-directed miRNA degradation revealed that miR-310 family degradation can be triggered by the CR43432 named Marge for proper cuticle development [147]. Moreover, a study on differentially expressed mRNA and lncRNA during dietary restrictions summarized a competing endogenous RNA network in Drosophila aging pathways [143]. The authors analyzed all the differentially expressed genes and transcripts of flies aged 7 days to 42 days, and, eventually, identified several pivotal regulatory axes and validated each through qRT-PCR [143]. The lncRNA and miRNA interactions have been reviewed for immune regulation in insect–pathogen interactions, but only miRNAs are identified in Drosophila, and no miRNA-lncRNA regulatory axis has been identified [148]. Altogether, these studies can bring fresh perspectives into the current understanding of the non-coding RNA regulatory axis within Drosophila.
4. circRNA in Drosophila
Another type of non-coding RNA, namely circular RNAs or circRNAs, can also have multiple diverse functions in regulating gene expression [149,150,151,152]. The circRNAs in Drosophila discussed in this section are summarised in Table 3. In general, circRNAs are stable due to their covalently closed loop structures [153]. The biogenesis of different types of circRNAs vary depending on their composition as introns or exons [153,154,155,156,157,158]. To date, circRNAs have been reported in fungi, plants, protists, and animals [159,160].
In some cases, circRNAs and related regulatory pathways are also highly conserved in animals such as between Drosophila and mammals [161,162,163,164]. For instance, in human and mouse cell lines, knockdown of the RNA-editing enzyme ADAR (adenosine deaminase acting on RNA) induces RNA circularisation processes. Decreases of ADAR levels in Drosophila due to high temperature will also lead to increased RNA circularisation [161]. Different to the linear mRNAs with the lack of 5′ and 3′ UTRs, the GW182-mediated circRNA degradation and the pathway controlling circRNA export from the nucleus are also conserved between Drosophila and human [165,166,167,168].
In Drosophila, circRNA transcripts can be identified in the embryonic stages as circular stable intronic sequences RNAs (sisRNAs), which are maternally inherited and can repress gene expression during embryogenesis [169], including sisR-4 from deadpan locus triggered deadpan expression, forming a positive feedback loop [170].
Another circRNA known as circMbl, derived from the muscleblind (mbl) transcript, is constantly expressed throughout, except during embryogenesis and early adulthood [171]. Mbl, regulated by miR-277 and miR-304, is necessary for photoreceptor differentiation in Drosophila eyes and muscle development [38,172]. Protein MBLs may initiate the expression of their pre-mRNA [164]. With circMbl having multiple binding sites for MBL, the interaction between MBL, mbl pre-mRNA, and circMbl resembles a regulatory circuit when excess MBLs are translated as MBL promotes the biogenesis of circMbl, which acts as a protein sponge to MBLs [164]. Moreover, circMbl encoding-proteins in fly head extracts are enriched in synaptosomes and modulated by starvation and FOXO, whereas the MBL, circMbl, and mbl isoforms are expressed in a tissue-specific manner [173]. Knockdown of circMbl results in abnormal developmental phenotypes, such as high lethality and muscular defects [174].
During neuronal development in Drosophila, circRNA Ect4-derived immune suppressor (Edis) has been found to be essential, as Edis depletion shows wiring defects of the neuromuscular junctions and mushroom body neuronal development [175,176]. The circRNA encoded peptide Edis-p can also block Relish from activating the immune deficiency (IMD) pathway of the immune system, whereas Relish binds to the zinc finger transcriptional factor castor which promotes and up-regulates castor expression that functions in central nervous system development [175,176]. Furthermore, previous studies have also identified that circRNAs were abundantly localised in the Drosophila brain and central nervous system and would accumulate with aging [177,178,179].
A circRNA circSfl encoded by the sulfateless gene also produces peptides, and the overexpression of circSfl was found to extend the lifespan of around 15% of female flies [180]. Since there were less overall circRNAs accumulated in the brain of insulin dilp mutants through aging, and circSfl up-regulation in median-neurosecretory-cell-ablated flies was dFOXO-dependent, a relationship between insulin signalling pathways, circSfl, and aging has been proposed [180].

Table 3.
Summarisation of circRNAs reported with their validated targets, function, and hormonal and metabolic regulations according to their references.
Table 3.
Summarisation of circRNAs reported with their validated targets, function, and hormonal and metabolic regulations according to their references.
| circRNA | Validated Targets | Function, Hormonal and Metabolic Regulations | References |
|---|---|---|---|
| sisR-4 | deadpan | Embryogenesis; positive feedback loop formation | [168,169] |
| circMbl | muscleblind | Eye and muscle development; negative feedback loop formation | [163,173] |
| Edis | castor | Neuromuscular junctions; mushroom body neuronal development; IMD pathway | [175,176] |
| circSfl | N/A | Increased female lifespan; insulin pathway; aging | [180] |
| circBoule | Hsc4, Hsp60C | Male fertility due to heat-stress | [181] |
Last but not least, in mature adults, circBoule is produced from the conserved reproductive gene Boule and can regulate heat-shock proteins to gradually decrease protein levels once exposed to a higher temperature. Loss of circBoule resulted in reduced male fertility in flies under a heat-stress environment [181], and such stress-induced fertility decline phenomena can also be found in humans. The above examples highlighted the importance of functional roles of circRNAs in Drosophila.
5. Conclusions and Future Perspectives
In this review, we have discussed miRNA, lncRNA, and circRNA regulation on development and metamorphosis, ecdysteroid and sesquiterpenoid systems, nutrition metabolism, sexual development, lifespan and viability, and circadian rhythm. Among all the pathways, some miRNAs have played multiple roles and served as crosstalk key components between pathways (Figure 1). Many studies start with the identification of expression profiles, lead with the miRNA target predictions, and result in mutant validations. Such comprehensive analysis can support the findings with a detailed understanding of the underlying mechanisms. Even with the global identification of miRNA-mRNA and the genome-wide lncRNA and circRNA identifications [99,141], validations of all the in silico predictions are still required to further understand the underlying mechanisms in ncRNA regulation, especially for their crosstalk in hormonal and metabolic pathways in Drosophila.
Another aspect is the potential of micro-peptide encoding ncRNAs. As mentioned previously, pri-miR-8 has been reported to encode for a small ORF that encodes for a miPEP-8, which can regulate the expression of hundreds more genes and will form a regulatory loop with miR-8 [182]. Similar ORFs have also been reported by other ncRNAs, such as lncRNA-MSA, miR-14, and many more micro-peptides encoded in lncRNAs and circRNAs [145,183,184]. How miPEPs may have regulatory effects on the hormonal and metabolic systems and whether these miPEPs may also be regulatory targets for considerations of pathway disturbances such as insecticides all require further investigation in the future.
Funding
This work was supported by Hong Kong Research Grant General Research Fund GRF (14100420) and CUHK Direct Grant (4053489, 4053547). KKC was supported by a postgraduate studentship provided by The Chinese University of Hong Kong.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Truman, J.W. The Evolution of Insect Metamorphosis. Curr. Biol. 2019, 29, R1252–R1268. [Google Scholar] [CrossRef]
- Cao, J.Q.; Tong, W.S.; Yu, H.Y.; Tobe, S.S.; Bendena, W.G.; Hui, J.H.L. The Role of MicroRNAs in Drosophila Regulation of Insulin-Like Peptides and Ecdysteroid Signalling: Where Are We Now? In Advances in Insect Physiology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 53, pp. 55–85. [Google Scholar] [CrossRef]
- Qu, Z.; Bendena, W.G.; Tobe, S.S.; Hui, J.H.L. Juvenile Hormone and Sesquiterpenoids in Arthropods: Biosynthesis, Signaling, and Role of MicroRNA. J. Steroid Biochem. Mol. Biol. 2018, 184, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.S.K.; Law, S.T.S.; Li, C.; Qu, Z.; Bendena, W.G.; Tobe, S.S.; Hui, J.H.L. Diversity of Insect Sesquiterpenoid Regulation. Front. Genet. 2020, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Truman, J.W.; Riddiford, L.M. The Evolution of Insect Metamorphosis: A Developmental and Endocrine View. Phil. Trans. R. Soc. B 2019, 374, 20190070. [Google Scholar] [CrossRef]
- Brandenburger, T.; Salgado Somoza, A.; Devaux, Y.; Lorenzen, J.M. Noncoding RNAs in Acute Kidney Injury. Kidney Int. 2018, 94, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Briata, P.; Gherzi, R. Long Non-Coding RNA-Ribonucleoprotein Networks in the Post-Transcriptional Control of Gene Expression. ncRNA 2020, 6, 40. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Yuva-Aydemir, Y.; Simkin, A.; Gascon, E.; Gao, F.-B. MicroRNA-9: Functional Evolution of a Conserved Small Regulatory RNA. RNA Biol. 2011, 8, 557–564. [Google Scholar] [CrossRef]
- Nong, W.; Cao, J.; Li, Y.; Qu, Z.; Sun, J.; Swale, T.; Yip, H.Y.; Qian, P.Y.; Qiu, J.-W.; Kwan, H.S.; et al. Jellyfish Genomes Reveal Distinct Homeobox Gene Clusters and Conservation of Small RNA Processing. Nat. Commun. 2020, 11, 3051. [Google Scholar] [CrossRef]
- Li, Y.; Hui, J.H.L. Small RNAs in Cnidaria: A Review. Evol. Appl. 2022. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell. Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Kawamata, T.; Tomari, Y. Drosophila Argonaute1 and Argonaute2 Employ Distinct Mechanisms for Translational Repression. Mol. Cell 2009, 34, 58–67. [Google Scholar] [CrossRef]
- Jo, M.H.; Shin, S.; Jung, S.-R.; Kim, E.; Song, J.-J.; Hohng, S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell 2015, 59, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, W.; Liu, Y.; Liu, T.; Li, C.; Wang, L. Oncogenic Role of MicroRNA-532-5p in Human Colorectal Cancer via Targeting of the 5’UTR of RUNX3. Oncol. Lett. 2018, 15, 7215–7220. [Google Scholar] [CrossRef]
- Gu, W.; Xu, Y.; Xie, X.; Wang, T.; Ko, J.-H.; Zhou, T. The Role of RNA Structure at 5′ Untranslated Region in MicroRNA-Mediated Gene Regulation. RNA 2014, 20, 1369–1375. [Google Scholar] [CrossRef]
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA MiR-324-3p Induces Promoter-Mediated Expression of RelA Gene. PLoS ONE 2013, 8, e79467. [Google Scholar] [CrossRef]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 Induces Expression of Genes with Complementary Promoter Sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein MRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Soni, K.; Choudhary, A.; Patowary, A.; Singh, A.R.; Bhatia, S.; Sivasubbu, S.; Chandrasekaran, S.; Pillai, B. miR-34 Is Maternally Inherited in Drosophila Melanogaster and Danio Rerio. Nucleic Acids Res. 2013, 41, 4470–4480. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Song, S.; Weng, R.; Verma, P.; Kugler, J.M.; Buescher, M.; Rouam, S.; Cohen, S.M. Systematic Study of Drosophila MicroRNA Functions Using a Collection of Targeted Knockout Mutations. Dev. Cell 2014, 31, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Tadros, W.; Goldman, A.L.; Babak, T.; Menzies, F.; Vardy, L.; Orr-Weaver, T.; Hughes, T.R.; Westwood, J.T.; Smibert, C.A.; Lipshitz, H.D. SMAUG Is a Major Regulator of Maternal mRNA Destabilization in Drosophila and Its Translation Is Activated by the PAN GU Kinase. Dev. Cell 2007, 12, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Stark, A.; Brennecke, J.; Cohen, S.M. Temporal Reciprocity of MiRNAs and Their Targets during the Maternal-to-Zygotic Transition in Drosophila. Curr. Biol. 2008, 18, 501–506. [Google Scholar] [CrossRef]
- Iwasaki, Y.W.; Siomi, H. miRNA Regulatory Ecosystem in Early Development. Mol. Cell 2014, 56, 615–616. [Google Scholar] [CrossRef][Green Version]
- Fu, S.; Nien, C.Y.; Liang, H.L.; Rushlow, C. Co-Activation of MicroRNAs by Zelda Is Essential for Early Drosophila Development. Development 2014, 141, 2108–2118. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Chen, Y.W.; Weng, R.; Lim, S.F.; Buescher, M.; Zhang, R.; Cohen, S.M. Overlapping Functions of MicroRNAs in Control of Apoptosis during Drosophila Embryogenesis. Cell Death Differ. 2012, 19, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Parrish, J.Z.; Xu, P.; Kim, C.C.; Jan, L.Y.; Jan, Y.N. The MicroRNA Bantam Functions in Epithelial Cells to Regulate Scaling Growth of Dendrite Arbors in Drosophila Sensory Neurons. Neuron 2009, 63, 788–802. [Google Scholar] [CrossRef]
- Enright, A.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D. MicroRNA Targets in Drosophila. Genome Biol. 2003, 4, P8. [Google Scholar] [CrossRef]
- Belles, X. MicroRNAs and the Evolution of Insect Metamorphosis. Annu. Rev. Entomol. 2017, 62, 111–125. [Google Scholar] [CrossRef]
- Caygill, E.E.; Johnston, L.A. Temporal Regulation of Metamorphic Processes in Drosophila by the Let-7 and MiR-125 Heterochronic MicroRNAs. Curr. Biol. 2008, 18, 943–950. [Google Scholar] [CrossRef]
- Biryukova, I.; Asmar, J.; Abdesselem, H.; Heitzler, P. Drosophila Mir-9a Regulates Wing Development via Fine-Tuning Expression of the LIM Only Factor, DLMO. Dev. Biol. 2009, 327, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Fulga, T.A.; McNeill, E.M.; Binari, R.; Yelick, J.; Blanche, A.; Booker, M.; Steinkraus, B.R.; Schnall-Levin, M.; Zhao, Y.; DeLuca, T.; et al. A Transgenic Resource for Conditional Competitive Inhibition of Conserved Drosophila MicroRNAs. Nat. Commun. 2015, 6, 7279. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Cohen, S.M. miR-965 Controls Cell Proliferation and Migration during Tissue Morphogenesis in the Drosophila Abdomen. eLife 2015, 4, e07389. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Yiu, W.C.; Yip, H.Y.; Nong, W.; Yu, C.W.C.; Lee, I.H.T.; Wong, A.Y.P.; Wong, N.W.Y.; Cheung, F.K.M.; Chan, T.F.; et al. Micro-RNA Clusters Integrate Evolutionary Constraints on Expression and Target Affinities: The MiR-6/5/4/286/3/309 Cluster in Drosophila. Mol. Biol. Evol. 2020, 37, 2955–2965. [Google Scholar] [CrossRef]
- Cerro-Herreros, E.; Fernandez-Costa, J.M.; Sabater-Arcis, M.; Llamusi, B.; Artero, R. Derepressing Muscleblind Expression by MiRNA Sponges Ameliorates Myotonic Dystrophy-like Phenotypes in Drosophila. Sci. Rep. 2016, 6, 36230. [Google Scholar] [CrossRef]
- Qu, Z.; Bendena, W.G.; Nong, W.; Siggens, K.W.; Noriega, F.G.; Kai, Z.; Zang, Y.; Koon, A.C.; Chan, H.Y.E.; Chan, T.F.; et al. MicroRNAs Regulate the Sesquiterpenoid Hormonal Pathway in Drosophila and Other Arthropods. Proc. R. Soc. B 2017, 284, 20171827. [Google Scholar] [CrossRef]
- He, Q.; Zhang, Y.; Dong, W. MicroRNA miR-927 Targets the Juvenile Hormone Primary Response Gene Krüppel Homolog1 to Control Drosophila Developmental Growth. Insect. Mol. Biol. 2020, 29, 545–554. [Google Scholar] [CrossRef]
- Boulan, L.; Martín, D.; Milán, M. Bantam MiRNA Promotes Systemic Growth by Connecting Insulin Signaling and Ecdysone Production. Curr. Biol. 2013, 23, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Chawla, G.; Sokol, N.S. Hormonal Activation of Let-7-C MicroRNAs via EcR Is Required for Adult Drosophila Melanogaster Morphology and Function. Development 2012, 139, 1788–1797. [Google Scholar] [CrossRef]
- Garbuzov, A.; Tatar, M. Hormonal Regulation of Drosophila MicroRNA Let-7 and MiR-125 That Target Innate Immunity. Fly 2010, 4, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wen, D.; Li, E.Y.; Palli, S.R.; Li, S.; Wang, J.; Liu, S. MicroRNA MiR-8 Promotes Cell Growth of Corpus Allatum and Juvenile Hormone Biosynthesis Independent of Insulin/IGF Signaling in Drosophila Melanogaster. Insect Biochem. Mol. Biol. 2021, 136, 103611. [Google Scholar] [CrossRef]
- Lim, D.; Lee, S.; Yun Han, J.; Choi, M.; Hong, J.; Seong, Y.; Kwon, Y.; Sik Lee, Y. Ecdysone-responsive MicroRNA-252-5p Controls the Cell Cycle by Targeting Abi in Drosophila. FASEB J. 2018, 32, 4519–4533. [Google Scholar] [CrossRef] [PubMed]
- Hyun, S.; Lee, J.H.; Jin, H.; Nam, J.; Namkoong, B.; Lee, G.; Chung, J.; Kim, V.N. Conserved MicroRNA MiR-8/MiR-200 and Its Target USH/FOG2 Control Growth by Regulating PI3K. Cell 2009, 139, 1096–1108. [Google Scholar] [CrossRef]
- Agbu, P.; Cassidy, J.J.; Braverman, J.; Jacobson, A.; Carthew, R.W. MicroRNA MiR-7 Regulates Secretion of Insulin-Like Peptides. Endocrinology 2020, 161, bqz040. [Google Scholar] [CrossRef]
- Suh, Y.S.; Bhat, S.; Hong, S.-H.; Shin, M.; Bahk, S.; Cho, K.S.; Kim, S.-W.; Lee, K.-S.; Kim, Y.-J.; Jones, W.D.; et al. Genome-Wide MicroRNA Screening Reveals That the Evolutionary Conserved MiR-9a Regulates Body Growth by Targeting SNPFR1/NPYR. Nat. Commun. 2015, 6, 7693. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.; Lim, S.F.; Cohen, S.M. Drosophila MiR-14 Regulates Insulin Production and Metabolism through Its Target, Sugarbabe. Genes Dev. 2010, 24, 2748–2753. [Google Scholar] [CrossRef]
- Barrio, L.; Dekanty, A.; Milán, M. MicroRNA-Mediated Regulation of Dp53 in the Drosophila Fat Body Contributes to Metabolic Adaptation to Nutrient Deprivation. Cell Rep. 2014, 8, 528–541. [Google Scholar] [CrossRef]
- Foronda, D.; Weng, R.; Verma, P.; Chen, Y.-W.; Cohen, S.M. Coordination of Insulin and Notch Pathway Activities by MicroRNA MiR-305 Mediates Adaptive Homeostasis in the Intestinal Stem Cells of the Drosophila Gut. Genes Dev. 2014, 28, 2421–2431. [Google Scholar] [CrossRef]
- Zipper, L.; Batchu, S.; Kaya, N.H.; Antonello, Z.A.; Reiff, T. The MicroRNA MiR-277 Controls Physiology and Pathology of the Adult Drosophila Midgut by Regulating the Expression of Fatty Acid β-Oxidation-Related Genes in Intestinal Stem Cells. Metabolites 2022, 12, 315. [Google Scholar] [CrossRef]
- Xu, P.; Vernooy, S.Y.; Guo, M.; Hay, B.A. The Drosophila MicroRNA Mir-14 Suppresses Cell Death and Is Required for Normal Fat Metabolism. Curr. Biol. 2003, 13, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Teleman, A.A.; Cohen, S.M. Drosophila Lacking MicroRNA MiR-278 Are Defective in Energy Homeostasis. Genes Dev. 2006, 20, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Iovino, N.; Pane, A.; Gaul, U. miR-184 Has Multiple Roles in Drosophila Female Germline Development. Dev. Cell 2009, 17, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Deng, Q.; Guo, T.; Hong, X.; Kugler, J.-M.; Yang, X.; Cohen, S.M. Regulation of Pattern Formation and Gene Amplification During Drosophila Oogenesis by the MiR-318 MicroRNA. Genetics 2015, 200, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Vilmos, P.; Bujna, Á.; Szuperák, M.; Havelda, Z.; Várallyay, É.; Szabad, J.; Kucerova, L.; Somogyi, K.; Kristó, I.; Lukácsovich, T.; et al. Viability, Longevity, and Egg Production of Drosophila Melanogaster Are Regulated by the MiR-282 MicroRNA. Genetics 2013, 195, 469–480. [Google Scholar] [CrossRef]
- Kugler, J.M.; Chen, Y.W.; Weng, R.; Cohen, S.M. miR-989 Is Required for Border Cell Migration in the Drosophila Ovary. PLoS ONE 2013, 8, e67075. [Google Scholar] [CrossRef]
- Shcherbata, H.R.; Ward, E.J.; Fischer, K.A.; Yu, J.-Y.; Reynolds, S.H.; Chen, C.-H.; Xu, P.; Hay, B.A.; Ruohola-Baker, H. Stage-Specific Differences in the Requirements for Germline Stem Cell Maintenance in the Drosophila Ovary. Cell Stem Cell 2007, 1, 698–709. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, S.; Xia, L.; Wang, J.; Wen, S.; Jin, P.; Chen, D. The Bantam MicroRNA Is Associated with Drosophila Fragile X Mental Retardation Protein and Regulates the Fate of Germline Stem Cells. PLoS Genet. 2009, 5, e1000444. [Google Scholar] [CrossRef]
- Yu, J.Y.; Reynolds, S.H.; Hatfield, S.D.; Shcherbata, H.R.; Fischer, K.A.; Ward, E.J.; Long, D.; Ding, Y.; Ruohola-Baker, H. Dicer-1-Dependent Dacapo Suppression Acts Downstream of Insulin Receptor in Regulating Cell Division of Drosophila Germline Stem Cells. Development 2009, 136, 1497–1507. [Google Scholar] [CrossRef]
- Fagegaltier, D.; König, A.; Gordon, A.; Lai, E.C.; Gingeras, T.R.; Hannon, G.J.; Shcherbata, H.R. A Genome-Wide Survey of Sexually Dimorphic Expression of Drosophila MiRNAs Identifies the Steroid Hormone-Induced MiRNA Let-7 as a Regulator of Sexual Identity. Genetics 2014, 198, 647–668. [Google Scholar] [CrossRef]
- Toledano, H.; D’Alterio, C.; Czech, B.; Levine, E.; Jones, D.L. The Let-7–Imp Axis Regulates Ageing of the Drosophila Testis Stem-Cell Niche. Nature 2012, 485, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.; Varghese, J. Sexually Dimorphic MicroRNA MiR-190 Regulates Lifespan in Male Drosophila. RNA Biol. 2022, 19, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Sato, T.; Ohkawa, Y.; Inoue, Y.H. Identification of MiR-305, a MicroRNA That Promotes Aging, and Its Target MRNAs in Drosophila. Genes Cells 2018, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Chawla, G.; Deosthale, P.; Childress, S.; Wu, Y.; Sokol, N.S. A Let-7-to-MiR-125 MicroRNA Switch Regulates Neuronal Integrity and Lifespan in Drosophila. PLoS Genet. 2016, 12, e1006247. [Google Scholar] [CrossRef] [PubMed]
- Gendron, C.M.; Pletcher, S.D. Micro RNA s Mir-184 and Let-7 Alter Drosophila Metabolism and Longevity. Aging Cell 2017, 16, 1434–1438. [Google Scholar] [CrossRef]
- Pandey, M.; Bansal, S.; Bar, S.; Yadav, A.K.; Sokol, N.S.; Tennessen, J.M.; Kapahi, P.; Chawla, G. miR-125-Chinmo Pathway Regulates Dietary Restriction-Dependent Enhancement of Lifespan in Drosophila. eLife 2021, 10, e62621. [Google Scholar] [CrossRef]
- Liu, N.; Landreh, M.; Cao, K.; Abe, M.; Hendriks, G.-J.; Kennerdell, J.R.; Zhu, Y.; Wang, L.-S.; Bonini, N.M. The MicroRNA MiR-34 Modulates Ageing and Neurodegeneration in Drosophila. Nature 2012, 482, 519–523. [Google Scholar] [CrossRef]
- Kennerdell, J.R.; Liu, N.; Bonini, N.M. miR-34 Inhibits Polycomb Repressive Complex 2 to Modulate Chaperone Expression and Promote Healthy Brain Aging. Nat. Commun. 2018, 9, 4188. [Google Scholar] [CrossRef]
- Srinivasan, A.R.; Tran, T.T.; Bonini, N.M. Loss of MiR-34 in Drosophila Dysregulates Protein Translation and Protein Turnover in the Aging Brain. Aging Cell 2022, 21, e13559. [Google Scholar] [CrossRef]
- Esslinger, S.M.; Schwalb, B.; Helfer, S.; Michalik, K.M.; Witte, H.; Maier, K.C.; Martin, D.; Michalke, B.; Tresch, A.; Cramer, P.; et al. Drosophila MiR-277 Controls Branched-Chain Amino Acid Catabolism and Affects Lifespan. RNA Biol. 2013, 10, 1042–1056. [Google Scholar] [CrossRef] [PubMed]
- Garaulet, D.L.; Sun, K.; Li, W.; Wen, J.; Panzarino, A.M.; O’Neil, J.L.; Hiesinger, P.R.; Young, M.W.; Lai, E.C. miR-124 Regulates Diverse Aspects of Rhythmic Behavior in Drosophila. J. Neurosci. 2016, 36, 3414–3421. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Rosbash, M. miR-276a Strengthens Drosophila Circadian Rhythms by Regulating Timeless Expression. Proc. Natl. Acad. Sci. USA 2016, 113, E2965–E2972. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Sehgal, A. Regulation of Circadian Behavioral Output via a MicroRNA-JAK/STAT Circuit. Cell 2012, 148, 765–779. [Google Scholar] [CrossRef]
- Vodala, S.; Pescatore, S.; Rodriguez, J.; Buescher, M.; Chen, Y.W.; Weng, R.; Cohen, S.M.; Rosbash, M. The Oscillating MiRNA 959–964 Cluster Impacts Drosophila Feeding Time and Other Circadian Outputs. Cell Metab. 2012, 16, 601–612. [Google Scholar] [CrossRef]
- Niu, Y.; Liu, Z.; Nian, X.; Xu, X.; Zhang, Y. miR-210 Controls the Evening Phase of Circadian Locomotor Rhythms through Repression of Fasciclin 2. PLoS Genet. 2019, 15, e1007655. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lee, J.-E.; Padgett, R.W.; Edery, I. Circadian Regulation of a Limited Set of Conserved MicroRNAs in Drosophila. BMC Genom. 2008, 9, 83. [Google Scholar] [CrossRef]
- Chen, W.; Liu, Z.; Li, T.; Zhang, R.; Xue, Y.; Zhong, Y.; Bai, W.; Zhou, D.; Zhao, Z. Regulation of Drosophila Circadian Rhythms by MiRNA Let-7 Is Mediated by a Regulatory Cycle. Nat. Commun. 2014, 5, 5549. [Google Scholar] [CrossRef]
- Cheong, S.P.S.; Huang, J.; Bendena, W.G.; Tobe, S.S.; Hui, J.H.L. Evolution of Ecdysis and Metamorphosis in Arthropods: The Rise of Regulation of Juvenile Hormone. Integr. Comp. Biol. 2015, 55, 878–890. [Google Scholar] [CrossRef]
- Niwa, Y.S.; Niwa, R. Neural Control of Steroid Hormone Biosynthesis during Development in the Fruit Fly Drosophila Melanogaster. Genes Genet. Syst. 2014, 89, 27–34. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Sonobe, H. 20-Hydroxyecdysone. In Handbook of Hormones; Elsevier: Amsterdam, The Netherlands, 2016; pp. 560–563. [Google Scholar] [CrossRef]
- Syed, M.H.; Mark, B.; Doe, C.Q. Steroid Hormone Induction of Temporal Gene Expression in Drosophila Brain Neuroblasts Generates Neuronal and Glial Diversity. eLife 2017, 6, e26287. [Google Scholar] [CrossRef]
- Williams, C.M.; Kafatos, F.C. Theoretical Aspects of the Action of Juvenile Hormone. In Insect Juvenile Hormones. Chemistry and Action; Academic Press: New York, NY, USA, 1972; pp. 29–44. [Google Scholar]
- Truman, J.W.; Riddiford, L.M. Chinmo Is the Larval Member of the Molecular Trinity That Directs Drosophila Metamorphosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2201071119. [Google Scholar] [CrossRef] [PubMed]
- Lavrynenko, O.; Rodenfels, J.; Carvalho, M.; Dye, N.A.; Lafont, R.; Eaton, S.; Shevchenko, A. The Ecdysteroidome of Drosophila: Influence of Diet and Development. Development 2015, 142, dev.124982. [Google Scholar] [CrossRef]
- Ono, H. Ecdysone Differentially Regulates Metamorphic Timing Relative to 20-Hydroxyecdysone by Antagonizing Juvenile Hormone in Drosophila Melanogaster. Dev. Biol. 2014, 391, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, K.; Gao, Y.; Liu, X.; Chen, W.; Ge, W.; Feng, Q.; Palli, S.R.; Li, S. Antagonistic Actions of Juvenile Hormone and 20-Hydroxyecdysone within the Ring Gland Determine Developmental Transitions in Drosophila. Proc. Natl. Acad. Sci. USA 2017, 115, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Belles, X.; Santos, C.G. The MEKRE93 (Methoprene Tolerant-Krüppel Homolog 1-E93) Pathway in the Regulation of Insect Metamorphosis, and the Homology of the Pupal Stage. Insect Biochem. Mol. Biol. 2014, 52, 60–68. [Google Scholar] [CrossRef]
- Huang, J.; Tian, L.; Peng, C.; Abdou, M.; Wen, D.; Wang, Y.; Li, S.; Wang, J. DPP-Mediated TGFβ Signaling Regulates Juvenile Hormone Biosynthesis by Activating the Expression of Juvenile Hormone Acid Methyltransferase. Development 2011, 138, 2283–2291. [Google Scholar] [CrossRef]
- Kayukawa, T.; Jouraku, A.; Ito, Y.; Shinoda, T. Molecular Mechanism Underlying Juvenile Hormone-Mediated Repression of Precocious Larval–Adult Metamorphosis. Proc. Natl. Acad. Sci. USA 2017, 114, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Nagamine, K.; Ito, Y.; Nishita, Y.; Ishikawa, Y.; Shinoda, T. Krüppel Homolog 1 Inhibits Insect Metamorphosis via Direct Transcriptional Repression of Broad-Complex, a Pupal Specifier Gene. J. Biol. Chem. 2016, 291, 1751–1762. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The Small RNA Profile during Drosophila Melanogaster Development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef]
- Sempere, L.F.; Sokol, N.S.; Dubrovsky, E.B.; Berger, E.M.; Ambros, V. Temporal Regulation of MicroRNA Expression in Drosophila Melanogaster Mediated by Hormonal Signals and Broad-Complex Gene Activity. Dev. Biol. 2003, 259, 9–18. [Google Scholar] [CrossRef]
- Niwa, R.; Niimi, T.; Honda, N.; Yoshiyama, M.; Itoyama, K.; Kataoka, H.; Shinoda, T. Juvenile Hormone Acid O-Methyltransferase in Drosophila Melanogaster. Insect Biochem. Mol. Biol. 2008, 38, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Pecasse, F.; Beck, Y.; Ruiz, C.; Richards, G. Krüppel-Homolog, a Stage-Specific Modulator of the Prepupal Ecdysone Response, Is Essential for Drosophila Metamorphosis. Dev. Biol. 2000, 221, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.; Cohen, S.M. MicroRNA MiR-14 Acts to Modulate a Positive Autoregulatory Loop Controlling Steroid Hormone Signaling in Drosophila. Genes Dev. 2007, 21, 2277–2282. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Kim, V.N.; Hyun, S. Conserved MicroRNA MiR-8 Controls Body Size in Response to Steroid Signaling in Drosophila. Genes Dev. 2012, 26, 1427–1432. [Google Scholar] [CrossRef]
- Wessels, H.-H.; Lebedeva, S.; Hirsekorn, A.; Wurmus, R.; Akalin, A.; Mukherjee, N.; Ohler, U. Global Identification of Functional MicroRNA-MRNA Interactions in Drosophila. Nat. Commun. 2019, 10, 1626. [Google Scholar] [CrossRef]
- Agarwal, V.; Subtelny, A.O.; Thiru, P.; Ulitsky, I.; Bartel, D.P. Predicting MicroRNA Targeting Efficacy in Drosophila. Genome Biol. 2018, 19, 152. [Google Scholar] [CrossRef]
- Lim, D.; Lee, S.; Choi, M.; Han, J.Y.; Seong, Y.; Na, D.; Kwon, Y.; Lee, Y.S. The Conserved MicroRNA MiR-8-3p Coordinates the Expression of V-ATPase Subunits to Regulate Ecdysone Biosynthesis for Drosophila Metamorphosis. FASEB J. 2020, 34, 6449–6465. [Google Scholar] [CrossRef]
- Bendena, W.G.; Hui, J.H.L.; Chin-Sang, I.; Tobe, S.S. Neuropeptide and MicroRNA Regulators of Juvenile Hormone Production. Gen. Comp. Endocrinol. 2020, 295, 113507. [Google Scholar] [CrossRef]
- Edgar, B.A. How Flies Get Their Size: Genetics Meets Physiology. Nat. Rev. Genet. 2006, 7, 907–916. [Google Scholar] [CrossRef]
- Böhni, R.; Riesgo-Escovar, J.; Oldham, S.; Brogiolo, W.; Stocker, H.; Andruss, B.F.; Beckingham, K.; Hafen, E. Autonomous Control of Cell and Organ Size by CHICO, a Drosophila Homolog of Vertebrate IRS1–4. Cell 1999, 97, 865–875. [Google Scholar] [CrossRef]
- Radimerski, T.; Montagne, J.; Rintelen, F.; Stocker, H.; van der Kaay, J.; Downes, C.P.; Hafen, E.; Thomas, G. DS6K-Regulated Cell Growth Is DPKB/DPI(3)K-Independent, but Requires DPDK1. Nat. Cell Biol. 2002, 4, 251–255. [Google Scholar] [CrossRef]
- Das, R.; Sebo, Z.; Pence, L.; Dobens, L.L. Drosophila Tribbles Antagonizes Insulin Signaling-Mediated Growth and Metabolism via Interactions with Akt Kinase. PLoS ONE 2014, 9, e109530. [Google Scholar] [CrossRef]
- Yang, H.; Li, M.; Hu, X.; Xin, T.; Zhang, S.; Zhao, G.; Xuan, T.; Li, M. MicroRNA-Dependent Roles of Drosha and Pasha in the Drosophila Larval Ovary Morphogenesis. Dev. Biol. 2016, 416, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Tarikere, S.; Ylla, G.; Extavour, C.G. Distinct Gene Expression Dynamics in Germ Line and Somatic Tissue during Ovariole Morphogenesis in Drosophila Melanogaster. G3-Genes Genomes Genet. 2022, 12, jkab305. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Poidevin, M.; Han, E.; Bi, J.; Jin, P. Circadian Rhythm-Dependent Alterations of Gene Expression in Drosophila Brain Lacking Fragile X Mental Retardation Protein. PLoS ONE 2012, 7, e37937. [Google Scholar] [CrossRef] [PubMed]
- Giebultowicz, J.M. Circadian Regulation of Metabolism and Healthspan in Drosophila. Free Radic. Biol. Med. 2018, 119, 62–68. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, Y. Emerging Roles for MicroRNA in the Regulation of Drosophila Circadian Clock. BMC Neurosci. 2018, 19, 1. [Google Scholar] [CrossRef]
- Kadener, S.; Menet, J.S.; Sugino, K.; Horwich, M.D.; Weissbein, U.; Nawathean, P.; Vagin, V.V.; Zamore, P.D.; Nelson, S.B.; Rosbash, M. A Role for MicroRNAs in the Drosophila Circadian Clock. Genes Dev. 2009, 23, 2179–2191. [Google Scholar] [CrossRef]
- Schäbler, S.; Amatobi, K.M.; Horn, M.; Rieger, D.; Helfrich-Förster, C.; Mueller, M.J.; Wegener, C.; Fekete, A. Loss of Function in the Drosophila Clock Gene Period Results in Altered Intermediary Lipid Metabolism and Increased Susceptibility to Starvation. Cell. Mol. Life Sci. 2020, 77, 4939–4956. [Google Scholar] [CrossRef]
- Sun, K.; Jee, D.; de Navas, L.F.; Duan, H.; Lai, E.C. Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of Drosophila Mir-279 and Mir-996. PLoS Genet. 2015, 11, e1005245. [Google Scholar] [CrossRef]
- Pegoraro, M.; Fishman, B.; Zonato, V.; Zouganelis, G.; Francis, A.; Kyriacou, C.P.; Tauber, E. Photoperiod-Dependent Expression of MicroRNA in Drosophila. IJMS 2022, 23, 4935. [Google Scholar] [CrossRef]
- Goodwin, P.R.; Meng, A.; Moore, J.; Hobin, M.; Fulga, T.A.; Van Vactor, D.; Griffith, L.C. MicroRNAs Regulate Sleep and Sleep Homeostasis in Drosophila. Cell Rep. 2018, 23, 3776–3786. [Google Scholar] [CrossRef]
- You, S.; Fulga, T.A.; Van Vactor, D.; Jackson, F.R. Regulation of Circadian Behavior by Astroglial MicroRNAs in Drosophila. Genetics 2018, 208, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Fu, X.; Du, J.; Wu, B.; Zhao, X.; Zhu, J.; Zhao, Z. Regulation of Circadian Rhythm and Sleep by MiR-375-timeless Interaction in Drosophila. FASEB J. 2020, 34, 16536–16551. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Rosbash, M. MicroRNA-92a Is a Circadian Modulator of Neuronal Excitability in Drosophila. Nat. Commun. 2017, 8, 14707. [Google Scholar] [CrossRef] [PubMed]
- Hobin, M.; Dorfman, K.; Adel, M.; Rivera-Rodriguez, E.J.; Kuklin, E.A.; Ma, D.; Griffith, L.C. The Drosophila MicroRNA Bantam Regulates Excitability in Adult Mushroom Body Output Neurons to Promote Early Night Sleep. iScience 2022, 25, 104874. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. R. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Lan, W.; Li, M.; Zhao, K.; Liu, J.; Wu, F.-X.; Pan, Y.; Wang, J. LDAP: A Web Server for LncRNA-Disease Association Prediction. Bioinformatics 2016, 33, 458–460. [Google Scholar] [CrossRef]
- Li, K.; Tian, Y.; Yuan, Y.; Fan, X.; Yang, M.; He, Z.; Yang, D. Insights into the Functions of LncRNAs in Drosophila. IJMS 2019, 20, 4646. [Google Scholar] [CrossRef]
- Bendena, W.G.; Garbe, J.C.; Traverse, K.L.; Lakhotia, S.C.; Pardue, M.L. Multiple Inducers of the Drosophila Heat Shock Locus 93D (Hsr Omega): Inducer-Specific Patterns of the Three Transcripts. J. Cell Biol. 1989, 108, 2017–2028. [Google Scholar] [CrossRef]
- Pardue, M.L.; Bendena, W.G.; Fini, M.E.; Garbe, J.C.; Hogan, N.C.; Traverse, K.L. Hsr-Omega, A Novel Gene Encoded by a Drosophila Heat Shock Puff. Biol. Bull. 1990, 179, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Camilleri-Robles, C.; Amador, R.; Klein, C.C.; Guigó, R.; Corominas, M.; Ruiz-Romero, M. Genomic and Functional Conservation of LncRNAs: Lessons from Flies. Mamm. Genome 2022, 33, 328–342. [Google Scholar] [CrossRef]
- Bae, E.; Calhoun, V.C.; Levine, M.; Lewis, E.B.; Drewell, R.A. Characterization of the Intergenic RNA Profile at Abdominal-A and Abdominal-B in the Drosophila Bithorax Complex. Proc. Natl. Acad. Sci. USA 2002, 99, 16847–16852. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Barrera, L.D.; Gutiérrez-Pérez, I.; Domínguez, M.; Riesgo-Escovar, J.R. Acal Is a Long Non-Coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure. PLoS Genet. 2015, 11, e1004927. [Google Scholar] [CrossRef] [PubMed]
- Tse, J.; Li, T.H.; Zhang, J.; Lee, A.C.K.; Lee, I.; Qu, Z.; Lin, X.; Hui, J.; Chan, T.F. Single-Cell Atlas of the Drosophila Leg Disc Identifies a Long Non-Coding RNA in Late Development. IJMS 2022, 23, 6796. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Salminen, T.S.; Järvelä-Stölting, M.; Vesala, L.; Rämet, M. Immune-Inducible Non-Coding RNA Molecule LincRNA-IBIN Connects Immunity and Metabolism in Drosophila Melanogaster. PLoS Pathog. 2019, 15, e1007504. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Y.; Qi, G. LncRNA-IRAR-mediated Regulation of Insulin Receptor Transcripts in Drosophila Melanogaster during Nutritional Stress. Insect Mol. Biol. 2022, 31, 261–272. [Google Scholar] [CrossRef]
- Chen, M.J.M.; Chen, L.K.; Lai, Y.S.; Lin, Y.Y.; Wu, D.C.; Tung, Y.A.; Liu, K.Y.; Shih, H.T.; Chen, Y.J.; Lin, Y.L.; et al. Integrating RNA-Seq and ChIP-Seq Data to Characterize Long Non-Coding RNAs in Drosophila Melanogaster. BMC Genom. 2016, 17, 220. [Google Scholar] [CrossRef]
- Maeda, R.K.; Sitnik, J.L.; Frei, Y.; Prince, E.; Gligorov, D.; Wolfner, M.F.; Karch, F. The LncRNA Male-Specific Abdominal Plays a Critical Role in Drosophila Accessory Gland Development and Male Fertility. PLoS Genet. 2018, 14, e1007519. [Google Scholar] [CrossRef]
- Meller, V.H.; Rattner, B.P. The RoX Genes Encode Redundant Male-Specific Lethal Transcripts Required for Targeting of the MSL Complex. EMBO J. 2002, 21, 1084–1091. [Google Scholar] [CrossRef]
- Deng, X.; Meller, V.H. RoX RNAs Are Required for Increased Expression of X-Linked Genes in Drosophila Melanogaster Males. Genetics 2006, 174, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.; Yang, L.; Xiong, T.; Di, C.; Ma, D.; Wu, M.; Xue, Z.; Zhang, X.; Long, L.; Zhang, W.; et al. Critical Roles of Long Noncoding RNAs in Drosophila Spermatogenesis. Genome Res. 2016, 26, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Bouska, M.J.; Bai, H. Long Noncoding RNA Regulation of Spermatogenesis via the Spectrin Cytoskeleton in Drosophila. G3-Genes Genomes Genet. 2021, 11, jkab080. [Google Scholar] [CrossRef] [PubMed]
- Jenny, A.; Hachet, O.; Závorszky, P.; Cyrklaff, A.; Weston, M.D.J.; Johnston, D.S.; Erdélyi, M.; Ephrussi, A. A Translation-Independent Role of Oskar RNA in Early Drosophila Oogenesis. Development 2006, 133, 2827–2833. [Google Scholar] [CrossRef] [PubMed]
- Soshnev, A.A.; Ishimoto, H.; McAllister, B.F.; Li, X.; Wehling, M.D.; Kitamoto, T.; Geyer, P.K. A Conserved Long Noncoding RNA Affects Sleep Behavior in Drosophila. Genetics 2011, 189, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Kosman, D.; McGinnis, W.; Tour, E. The Expression of Drosophila Melanogaster Hox Gene Ultrabithorax Is Not Overtly Regulated by the Intronic Long Noncoding RNA LncRNA:PS4 in a Wild-Type Genetic Background. G3-Genes Genomes Genet. 2022, 12, jkab374. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, Y.; Zhang, X.; Jia, S.; Chen, S.; Kang, L. Genome-Wide Identification and Developmental Expression Profiling of Long Noncoding RNAs during Drosophila Metamorphosis. Sci. Rep. 2016, 6, 23330. [Google Scholar] [CrossRef]
- Stoiber, M.; Celniker, S.; Cherbas, L.; Brown, B.; Cherbas, P. Diverse Hormone Response Networks in 41 Independent Drosophila Cell Lines. G3-Genes Genomes Genet. 2016, 6, 683–694. [Google Scholar] [CrossRef]
- Yang, D.; Lian, T.; Tu, J.; Gaur, U.; Mao, X.; Fan, X.; Li, D.; Li, Y.; Yang, M. LncRNA Mediated Regulation of Aging Pathways in Drosophila Melanogaster during Dietary Restriction. Aging 2016, 8, 2182–2203. [Google Scholar] [CrossRef]
- Nyberg, K.G.; Machado, C.A. Comparative Expression Dynamics of Intergenic Long Noncoding RNAs in the Genus Drosophila. Genome Biol. Evol. 2016, 8, 1839–1858. [Google Scholar] [CrossRef]
- Immarigeon, C.; Frei, Y.; Delbare, S.Y.N.; Gligorov, D.; Machado Almeida, P.; Grey, J.; Fabbro, L.; Nagoshi, E.; Billeter, J.-C.; Wolfner, M.F.; et al. Identification of a Micropeptide and Multiple Secondary Cell Genes That Modulate Drosophila Male Reproductive Success. Proc. Natl. Acad. Sci. USA 2021, 118, e2001897118. [Google Scholar] [CrossRef] [PubMed]
- Meller, V.H.; Wu, K.H.; Roman, G.; Kuroda, M.I.; Davis, R.L. RoX1 RNA Paints the X Chromosome of Male Drosophila and Is Regulated by the Dosage Compensation System. Cell 1997, 88, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Kingston, E.R.; Blodgett, L.W.; Bartel, D.P. Endogenous Transcripts Direct MicroRNA Degradation in Drosophila, and This Targeted Degradation Is Required for Proper Embryonic Development. Mol. Cell 2022, 82, 3872–3884.e9. [Google Scholar] [CrossRef]
- Moure, U.A.E.; Tan, T.; Sha, L.; Lu, X.; Shao, Z.; Yang, G.; Wang, Y.; Cui, H. Advances in the Immune Regulatory Role of Non-Coding RNAs (MiRNAs and LncRNAs) in Insect-Pathogen Interactions. Front. Immunol. 2022, 13, 856457. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z. Efficient Backsplicing Produces Translatable Circular MRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Li, Y.; Niringiyumukiza, J.D.; Su, P.; Xiang, W. Circular RNA Involvement in Aging: An Emerging Player with Great Potential. Mech. Ageing Dev. 2019, 178, 16–24. [Google Scholar] [CrossRef]
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef]
- Wang, S.; Xiao, F.; Li, J.; Fan, X.; He, Z.; Yan, T.; Yang, M.; Yang, D. Circular RNAs Involved in the Regulation of the Age-Related Pathways. IJMS 2022, 23, 10443. [Google Scholar] [CrossRef]
- Meng, X.; Li, X.; Zhang, P.; Wang, J.; Zhou, Y.; Chen, M. Circular RNA: An Emerging Key Player in RNA World. Brief. Bioinform. 2016, 18, bbw045. [Google Scholar] [CrossRef]
- Guria, A.; Sharma, P.; Natesan, S.; Pandi, G. Circular RNAs—The Road Less Traveled. Front. Mol. Biosci. 2020, 6, 146. [Google Scholar] [CrossRef]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, Present, and Future of CircRNA s. EMBO J 2019, 38, e100836. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xiao, M.-S.; Li, Z.; Huang, C. New Progresses of Circular RNA Biology: From Nuclear Export to Degradation. RNA Biol. 2021, 18, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E. Circular RNAs: Unexpected Outputs of Many Protein-Coding Genes. RNA Biol. 2016, 14, 1007–1017. [Google Scholar] [CrossRef]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-Protein Interactions: Functions, Mechanisms, and Identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Bao, Y.; Yee, M.-C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse Alternative Back-Splicing and Alternative Splicing Landscape of Circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial Control of Drosophila Circular RNA Expression by Intronic Repeats, HnRNPs, and SR Proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis Competes with Pre-MRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Jia, R.; Xiao, M.-S.; Li, Z.; Shan, G.; Huang, C. Defining an Evolutionarily Conserved Role of GW182 in Circular RNA Degradation. Cell Discov. 2019, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A Length-Dependent Evolutionarily Conserved Pathway Controls Nuclear Export of Circular RNAs. Genes Dev. 2018, 32, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Y.; Lin, J.; Song, Z.; Wang, Q.; Zhao, W.; Wang, Y.; Xiu, X.; Deng, Y.; Li, X.; et al. Exportin 4 Depletion Leads to Nuclear Accumulation of a Subset of Circular RNAs. Nat. Commun. 2022, 13, 5769. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kearse, M.G.; Huang, C. The Nuclear Export of Circular RNAs Is Primarily Defined by Their Length. RNA Biol. 2019, 16, 1–4. [Google Scholar] [CrossRef]
- Pek, J.W.; Osman, I.; Tay, M.L.-I.; Zheng, R.T. Stable Intronic Sequence RNAs Have Possible Regulatory Roles in Drosophila Melanogaster. J. Cell Biol. 2015, 211, 243–251. [Google Scholar] [CrossRef]
- Tay, M.L.I.; Pek, J.W. Maternally Inherited Stable Intronic Sequence RNA Triggers a Self-Reinforcing Feedback Loop during Development. Curr. Biol. 2017, 27, 1062–1067. [Google Scholar] [CrossRef]
- Houseley, J.M.; Garcia-Casado, Z.; Pascual, M.; Paricio, N.; O’Dell, K.M.C.; Monckton, D.G.; Artero, R.D. Noncanonical RNAs From Transcripts of the Drosophila Muscleblind Gene. J. Hered. 2006, 97, 253–260. [Google Scholar] [CrossRef]
- Irion, U. Drosophila Muscleblind Codes for Proteins with One and Two Tandem Zinc Finger Motifs. PLoS ONE 2012, 7, e34248. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Patop, I.L.; Krishnamoorthy, A.; Bartok, O.; Maya, R.; Lerner, N.; Ashwall-Fluss, R.; Konakondla, J.V.V.; Beatus, T.; Kadener, S. CircMbl Functions in Cis and in Trans to Regulate Gene Expression and Physiology in a Tissue-Specific Fashion. Cell Rep. 2022, 39, 110740. [Google Scholar] [CrossRef]
- Xiong, X.-P.; Liang, W.; Liu, W.; Xu, S.; Li, J.-L.; Tito, A.; Situ, J.; Martinez, D.; Wu, C.; Perera, R.J.; et al. The Circular RNA Edis Regulates Neurodevelopment and Innate Immunity. PLoS Genet. 2022, 18, e1010429. [Google Scholar] [CrossRef]
- Liu, W.; Liang, W.; Xiong, X.-P.; Li, J.-L.; Zhou, R. A Circular RNA Edis-Relish-Castor Axis Regulates Neuronal Development in Drosophila. PLoS Genet. 2022, 18, e1010433. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-Wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.; Medina, P.; Cooper, D.A.; Escobedo, S.E.; Rounds, J.; Brennan, K.J.; Vincent, C.; Miura, P.; Doerge, R.; Weake, V.M. Transcriptome Profiling of Aging Drosophila Photoreceptors Reveals Gene Expression Trends That Correlate with Visual Senescence. BMC Genom. 2017, 18, 894. [Google Scholar] [CrossRef] [PubMed]
- Stegeman, R.; Hall, H.; Escobedo, S.E.; Chang, H.C.; Weake, V.M. Proper Splicing Contributes to Visual Function in the Aging Drosophila Eye. Aging Cell 2018, 17, e12817. [Google Scholar] [CrossRef]
- Weigelt, C.M.; Sehgal, R.; Tain, L.S.; Cheng, J.; Eßer, J.; Pahl, A.; Dieterich, C.; Grönke, S.; Partridge, L. An Insulin-Sensitive Circular RNA That Regulates Lifespan in Drosophila. Mol. Cell 2020, 79, 268–279.e5. [Google Scholar] [CrossRef]
- Gao, L.; Chang, S.; Xia, W.; Wang, X.; Zhang, C.; Cheng, L.; Liu, X.; Chen, L.; Shi, Q.; Huang, J.; et al. Circular RNAs from BOULE Play Conserved Roles in Protection against Stress-Induced Fertility Decline. Sci. Adv. 2020, 6, eabb7426. [Google Scholar] [CrossRef] [PubMed]
- Montigny, A.; Tavormina, P.; Duboe, C.; San Clémente, H.; Aguilar, M.; Valenti, P.; Lauressergues, D.; Combier, J.-P.; Plaza, S. Drosophila Primary MicroRNA-8 Encodes a MicroRNA-Encoded Peptide Acting in Parallel of MiR-8. Genome Biol. 2021, 22, 118. [Google Scholar] [CrossRef]
- Dozier, C.; Montigny, A.; Viladrich, M.; Culerrier, R.; Combier, J.-P.; Besson, A.; Plaza, S. Small ORFs as New Regulators of Pri-MiRNAs and MiRNAs Expression in Human and Drosophila. IJMS 2022, 23, 5764. [Google Scholar] [CrossRef] [PubMed]
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).