Abstract
Asobara japonica (Hymenoptera: Braconidae) is an endoparasitoid wasp that can successfully parasitize a wide range of host species across the Drosophila genus, including the invasive crop pest Drosophila suzukii. Parasitoids are capable of regulating the host metabolism to produce the nutritional metabolites for the survival of their offspring. Here, we intend to investigate the metabolic changes in D. melanogaster hosts after parasitization by A. japonica, using the non-targeted LC-MS (liquid chromatography-mass spectrometry) metabolomics analysis. In total, 3043 metabolites were identified, most of which were not affected by A. japonica parasitization. About 205 metabolites were significantly affected in parasitized hosts in comparison to non-parasitized hosts. The changed metabolites were divided into 10 distinct biochemical groups. Among them, most of the lipid metabolic substances were significantly decreased in parasitized hosts. On the contrary, most of metabolites associated with the metabolism of amino acids and sugars showed a higher abundance of parasitized hosts, and were enriched for a wide range of pathways. In addition, eight neuromodulatory-related substances were upregulated in hosts post A. japonica parasitization. Our results reveal that the metabolites are greatly changed in parasitized hosts, which might help uncover the underlying mechanisms of host manipulation that will advance our understanding of host–parasitoid coevolution.
1. Introduction
Parasitoid wasps are a large group of hymenopteran insects, comprising an estimated number of 150,000 to 600,000 species [1]. They are free-living insects as adults; however, their wasp larvae must parasitize the various life stages of other arthropods, referred to as hosts [2,3]. The female wasps lay eggs in (endoparasitoids) or on (ectoparasitoids) the host body, following the hatched wasp larvae consuming and finally killing their hosts [1,4]. Consequently, parasitoids are ideal biocontrol agents to constrain the destructive pest populations in agriculture ecosystems [5,6,7].
Most of the endoparasitoids belong to koinobionts, whose hosts are allowed to continue growing post parasitism, and thus provide enough nutrients for wasp offspring development and survival [1]. As such, the endoparasitoids have evolved many effective strategies to escape or suppress the host immune response [5,6,8]. Moreover, successful parasitization also requires the regulation of host nutritional metabolism to allow the wasp eggs to hatch and wasp larvae to develop [9,10,11].
Different with insect predators that attain nutrients through external preys, the endoparasitoid species typically rely on the resources derived from a single host to complete their immature development, and usually fails to synthetize some essential nutrients (e.g., lipids) by themselves [9,12,13]. So far, some endoparasitoids have been reported to manipulate the nutrients of their hosts post parasitization. For instance, quantities of free amino acids and proteins were varied in Devorgilla canescens-infected Anagasta kuehniella hosts [14]; the contents of total lipids and proteins were decreased and the contents of glycogen were increased in parasitized Lymantria dispar [15]; and the contents of proteins were decreased and the contents of lipids were increased in Spodoptera littoralis after parasitization by Chelonus inanitus [16]. Recently, results from Leptopilina boulardi, an endoparasitoid wasp of the Drosophila species, showed that parasitization reduced the lipid content in host hemolymph, but lipids were accumulated in host body fat cells [9,17]. Further functional analysis revealed that the lipid nutrients were necessary for the survival of parasitoid offspring [9,17,18]. Collectively, these findings propose that the parasitoids have the ability to regulate the composition of host nutrients to meet the dietary demands of their progeny.
Nutritional metabolites participate in various pathways which are associated with insect development and reproduction. Over the last decade, metabolomics has become one of the most advanced technologies in life science studies, and this technique has also been greatly applied in insects [19], including liquid chromatography-mass spectrometry (LC-MS) [20], gas chromatography-mass spectrometry (GC-MS) [21] and nuclear magnetic resonance (NMR) [22]. Based on high-throughput analysis, the changes of metabolites and the relevant metabolic processes have been discovered in some insect species that were either experiencing different environmental stresses [23,24,25], or associated with plant–insect interactions [26,27,28,29]. However, the metabolomics analyses in parasitoid wasps, especially for dissecting host nutritional manipulation strategies, are largely unexplored.
Asobara japonica (Hymenoptera: Braconidae) is a highly generalized larval-pupal endoparasitoid, that parasitizes a number of host species across the Drosophila genus, thus being considered as a model organism for exploring the mechanisms under host–parasitoid interactions [30,31]. Here we applied liquid chromatography-mass spectrometry (LC-MS)-based metabolomics technology to analyze the metabolic changes in identify main metabolic pathways altered by A. japonica parasitization in D. melanogaster hosts. Our data will therefore contribute to the understanding of the host manipulation strategies and provide a new perspective on host–parasitoid coevolution.
2. Materials and Methods
2.1. Insects
The parasitoid A. japonica was collected from Taizhou (28°50′ N, 120°34′ E), Zhejiang, China, in June 2018 [32]. Since then, this strain was maintained in our lab on D. melanogaster W1118 as a regular host, which was raised on standard cornmeal medium at 25 °C, 50% humidity and 16 L:8 D inside plastic fly bottles. The newly emerged adult wasps were provided with an apple juice agar medium until exposure to hosts. The apple juice agar recipe: 27 g agar, 33 g brown sugar and 330 mL pure apple juice diluted in 1000 mL of water [33].
2.2. Experimental Procedures and Sample Collection
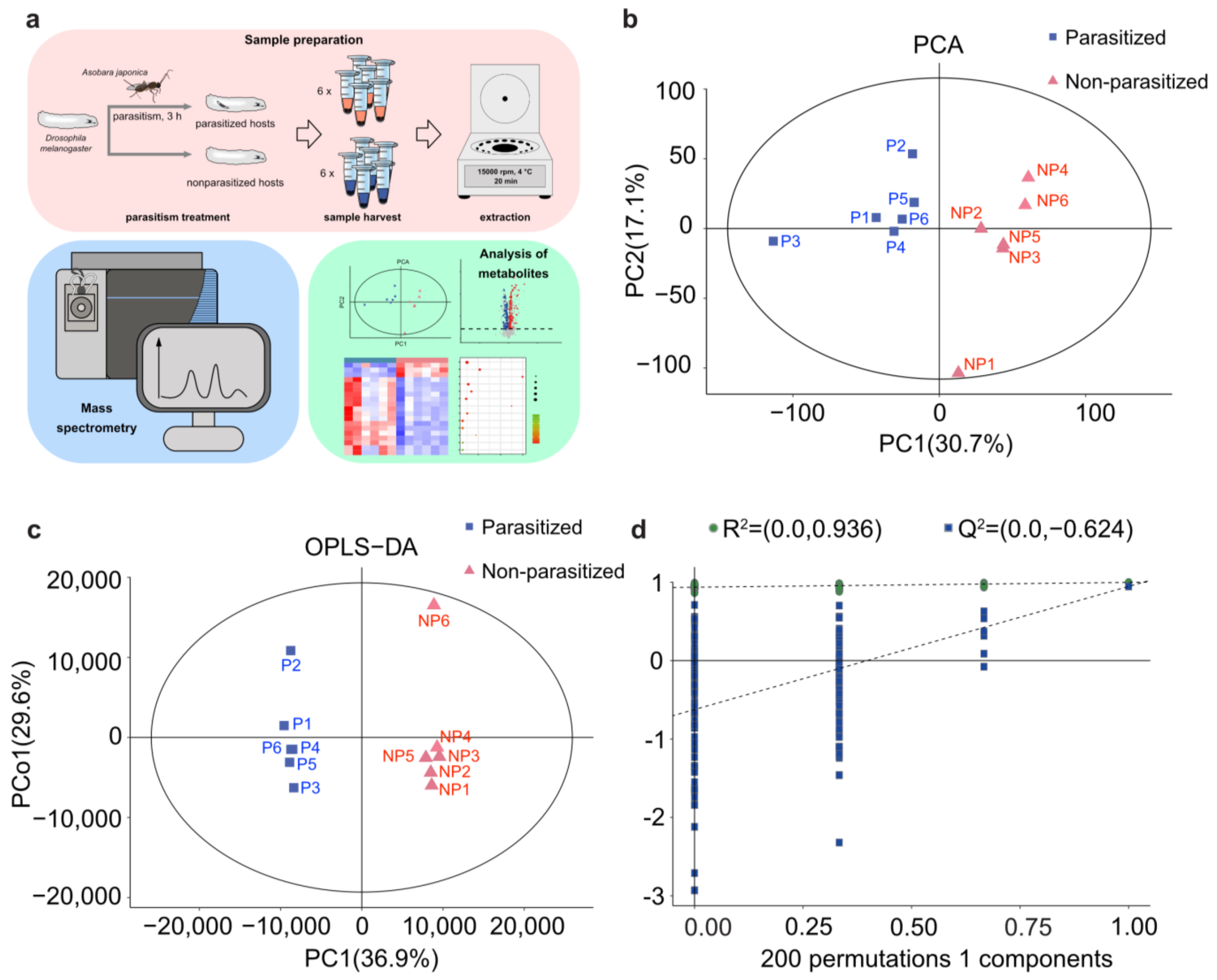
Three-day-old A. japonica females were used to parasitize the 2nd instar Drosophila host larvae for 3 h, to ensure that approximately all the hosts were parasitized. The number of female wasps was based on the parasitoid/host ratio of ~1:10. The Drosophila host larvae were collected after 3 h parasitization, along with the non-parasitized hosts at the same age (control), washed in 75% alcohol, and rinsed with ddH2O 3 times. After air-drying, the Drosophila host larvae were moved to tubes containing 150 μL of 80% methanol for grinding, and then centrifuged at 15,000 rpm for 20 min at 4 °C. The supernatant was extracted into 1.5 mL centrifuge tubes, frozen in liquid nitrogen, and stored at −80 °C for further use. In this study, we collected two different groups, including A. japonica-parasitized and non-parasitized Drosophila host larvae, for metabolomics analysis, and analysis of each group was repeated six times. Approximately 130 host larvae were used in each repetition (Figure 1a).
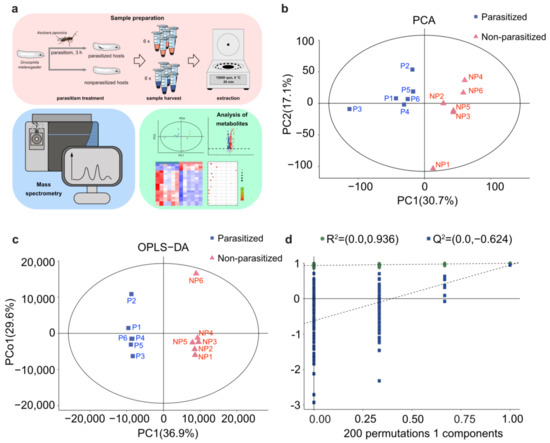
Figure 1.
PCA and OPLS-DA analyses of metabolic differences. (a) Workflow for metabolomics analysis. (b) Principal component (PC) analysis (PCA) score plot of the metabolite profiles of D. melanogaster larvae in the parasitized group and non-parasitized group. Data points are displayed as sample projections on the two primary axes. The oval represents the 95% confidence interval of the model using Hotelling’s T2 statistics. Variances explained by the first two components (PC1 and PC2) are shown in parentheses. (c) Orthogonal partial least-squares discriminant analysis (OPLS-DA) score plot shows a clear separation of metabolic profiles between the parasitized and non-parasitized D. melanogaster larvae. Variances explained by the principal components (PC1 and PCo1) are shown in parentheses. Abbreviations: PC1, predictive component 1; PCo1, predictive component orthogonal 1; NP, non-parasitized host; P, parasitized host. (d) Permutation tests of OPLS-DA models. The permutation test was carried out with 200 random permutations, and the resulting R2 and Q2 values were plotted. Green circle, R2; Blue square, Q2.
2.3. Sample Pretreatment
Samples were pretreated as described previously, with minor modifications [34,35]. Briefly, a total of 50 μL supernatant from each sample was transferred into a 1.5 mL microcentrifuge tube containing 20 μL internal standard buffer (L-2-chlorophenylalanine in methanol, 0.06 mg/mL), and vortexed for 10 s. Then, 200 μL protein precipitant of methanol: acetonitrile at 2:1 ratio (v/v) was added to each sample and vortexed for 1 min, followed by ultrasonicating in an ice-water bath for 10 min, and placed at −20 °C for 30 min. Treated samples were centrifuged at 13,000 rpm at 4 °C for 10 min, and all supernatants were extracted into individual vials for volatilization. Thereafter, 200 μL methanol–water solution at 1:4 ratio (v/v) was added to the dried samples for dissolution. The samples were vortexed for 30 s, ultrasonicated for 3 min, placed at −20 °C for 2 h, and then centrifuged at 13,000 rpm at 4 °C for 10 min. Finally, a supernatant of 150 μL from each sample was filtered through 0.22 μm microfilters and stored at −80 °C for metabolomics analysis (OE Biotech, Shanghai, China).
2.4. LC-MS/MS Analysis
Nontargeted LC-MS analyses were performed using the UPLC-Q-Exactive Orbitrap/MS (Shimadzu Corporation, Kyoto, Japan) platform, which was equipped with a heated electrospray ionization (ESI) source (Thermo Fisher Scientific, Waltham, MA, USA), to analyze the metabolic profiling in both ESI-positive and ESI-negative ion modes. An ACQUITY UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm), with a maintained temperature of 45 °C, was applied in both positive and negative modes. Mobile phases consisted of 0.1% formic acid in water (A) and acetonitrile (B), and the gradient elution conditions were as follows: 0 min, 5% B; 2 min, 5% B; 4 min, 30% B; 8 min, 50% B; 10 min, 80% B; 14 min, 100% B; 15 min, 100% B; 15.1 min, 5% B; and 16 min, 5% B. The flow rate was 0.35 mL/min, and the injection volume was 2 μL.
The mass range was set at m/z 125–1000. The resolution of the full scan was set to 70,000 and 17,500 for HCD MS/MS. When operating the mass spectrometer, the spray voltage was set to 3500 V for positive or 3500 V for negative; sheath gas-flow rate was set to 40 arbitrary units for positive or 35 arbitrary units for negative; auxiliary gas-flow rate was set to 10 arbitrary units for positive or 8 arbitrary units for negative; capillary temperature was set to 320 °C. For MS/MS analysis, the collision energy was set to 10, 20 and 40 eV.
2.5. Data Analysis
The LC-MS raw data were analyzed using the Progenesis QI software V2.3 (Nonlinear, Newcastle upon Tyne, UK). Precursor tolerance was set at 5 ppm, product tolerance was set at 10 ppm, and product ion threshold was set at 5%. Metabolites were identified based on precise mass-to-charge ratio (m/z), secondary fragments, and isotopic distribution. Public databases, such as human metabolome database (HMDB, http://www.hmdb.ca/, accessed on 10 December 2022); Lipidmaps (v2.3, http://www.lipidmaps.org/, accessed on 1 June 2022) and Metlin (https://metlin.scripps.edu/, accessed on 1 June 2022), were used for qualitative analysis.
The LC-MS raw data were imported into R package for multivariate statistical analysis, such as principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA). PCA was carried out to initially visualize the overall distribution among the samples and the stability of the whole analysis process. OPLS-DA was used to distinguish the metabolites that differ between parasitized and non-parasitized control groups. Variable importance in the projection (VIP) was obtained from OPLS-DA to rank the overall contribution of each variable item to group discrimination. The univariate analysis used t-tests, and fold-change (FC) analysis was also used to compare the metabolites between the two groups. FC was calculated as the ratio of the peak area of parasitized and non-parasitized groups. Additionally, the screening criteria for differential metabolites was the VIP values from the OPLS-DA model > 1 and the p-value from the Student’s t-test < 0.05, based on the previous reports [36,37,38]. Here, the default 7-fold cross-validation was applied with 1/7 of the samples being excluded from the mathematical model in each round, to guard against overfitting.
Differential metabolites were mapped onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, to acquire detailed pathway information. The KEGG metabolic pathways were then predicted from the differential metabolites showing a p-value < 0.05. Statistical analyses were performed in GraphPad Prism version 9.0 (GraphPad Software).
3. Results
3.1. PCA and OPLS-DA Analyses of Metabolic Difference in Parasitized Host
To comprehensively analyze the profiles of metabolites, nontargeted LC-MS was performed in A. japonica-parasitized and non-parasitized Drosophila host larvae (Figure 1a). The PCA analysis showed a clear separation between the A. japonica-parasitized and non-parasitized samples, and they were all within the 95% confidence interval of the Hotelling’s T2 ellipse (Figure 1b). The results indicated that no outlier was present, and all samples were suitable for the following analysis. Meanwhile, the OPLS-DA model showed that the parasitized samples were clustered on the left part of the x-axis, and the non-parasitized samples were clustered on the right (Figure 1c), revealing that significant differences existed between the two groups. Seven-fold cross validation and 200 response permutation testing (RPT) was then performed in the OPLS-DA model. The resulting R2 was 0.936 and Q2 was −0.624 (Figure 1d), reflecting that the OPLS-DA did not exceed the fit. The explanatory ability (R2Y(CUM)) and the predictive ability (Q2(CUM)) for the sample were 0.997 and 0.946, respectively, suggesting that the established model conformed to the real situation of the data. These results indicate that the metabolites of D. melanogaster hosts were greatly changed after parasitization by A. japonica.
3.2. Changes in Metabolites Post A. japonica Parasitization
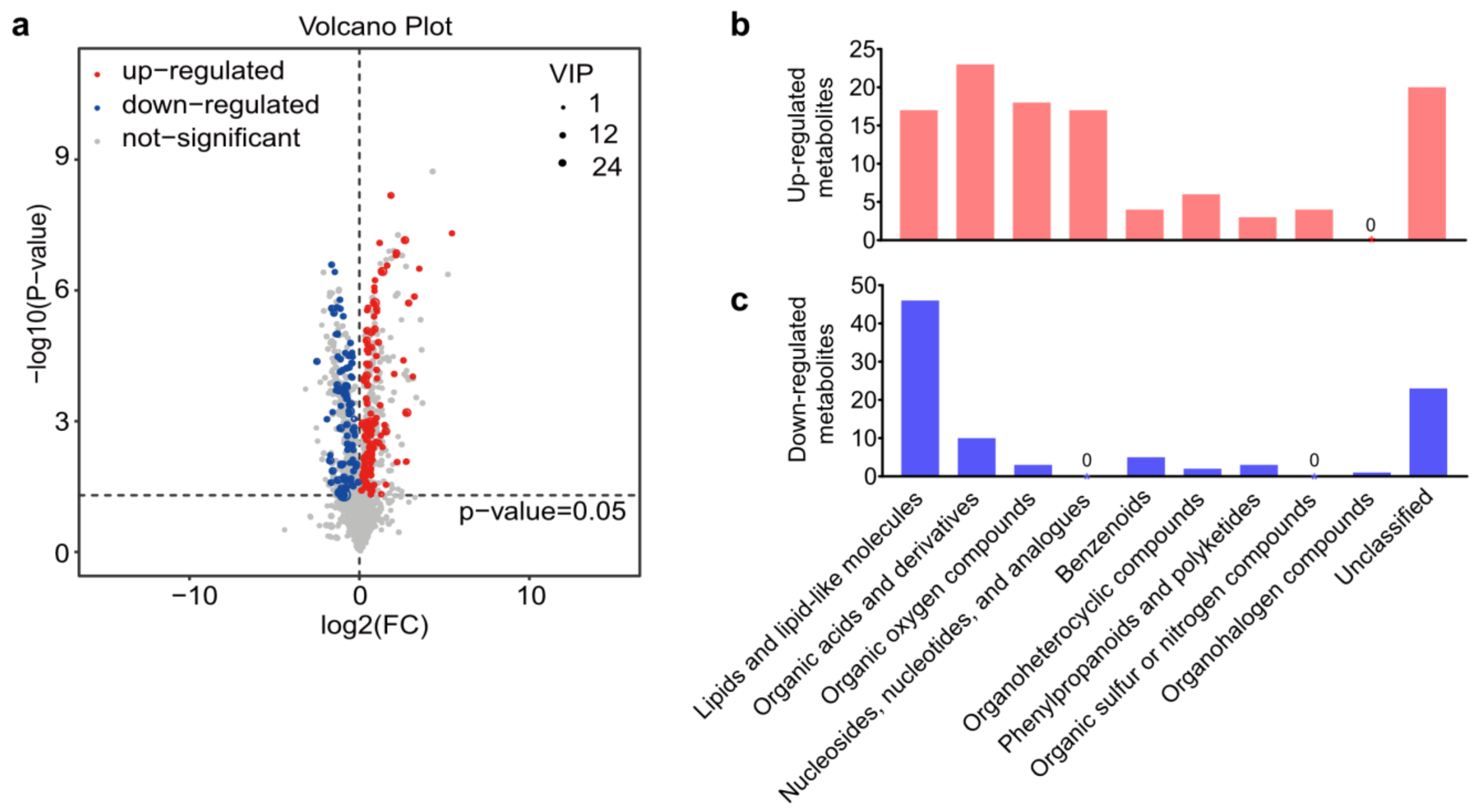
A total of 8563 peaks were detected by LC-MS, including 3339 peaks from the negative ion model and 5224 peaks from the positive ion model, respectively. In addition, 3043 metabolites that consisted of 868 negative ion mode metabolites (ESI−) and 2175 positive ion mode metabolites (ESI+) were identified with annotations. After the exogenous compounds in LC-MS were removed, the differentially abundant metabolites were selected according to the VIP values from the OPLS-DA model (VIP > 1) and the p-value from the Student’s t-test (p < 0.05). Volcano plots showed all the 205 differentially expressed metabolites that identified between A. japonica-parasitized and non-parasitized D. melanogaster hosts (Figure 2a), including 112 upregulated metabolites and 93 downregulated metabolites (Table S1). Those differential metabolites were divided into ten distinct biochemical groups (Figure 2b,c): lipids and lipid-like molecules (63/205, 30.73%); organic acids and derivatives (33/205, 16.10%); organic oxygen compounds (21/205, 10.24%); nucleosides, nucleotides, and analogues (17/205, 8.29%); benzenoids (9/205, 4.39%); organoheterocyclic compounds (8/205, 3.90%); phenylpropanoids and polyketides (6/205, 2.93%); organic sulfur or nitrogen compounds (4/205, 1.95%); organohalogen compounds (1/205, 0.49%) and unclassified metabolites (43/205, 20.98%). We next mainly focused on the top three groups of differential metabolites.
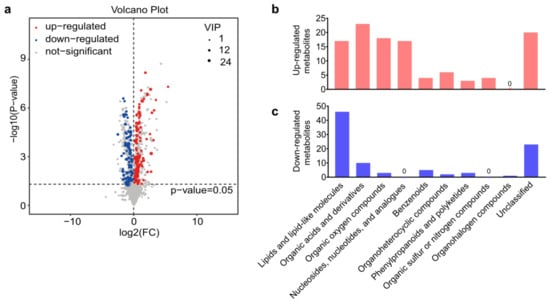
Figure 2.
Differential metabolites between parasitized and non-parasitized hosts. (a) Volcano plot of the 205 differential metabolites. Each point in the volcano diagram represents a metabolite, and the greater the scattered point, the greater the value of variable importance is in the projection (VIP). (b) The classification of upregulated metabolites in parasitized hosts. (c) The classification of downregulated metabolites in parasitized hosts. Abbreviations: FC, fold change; VIP, variable important in projection.
3.3. Differences in Lipids and Lipid-like Molecules Post A. japonica Parasitization
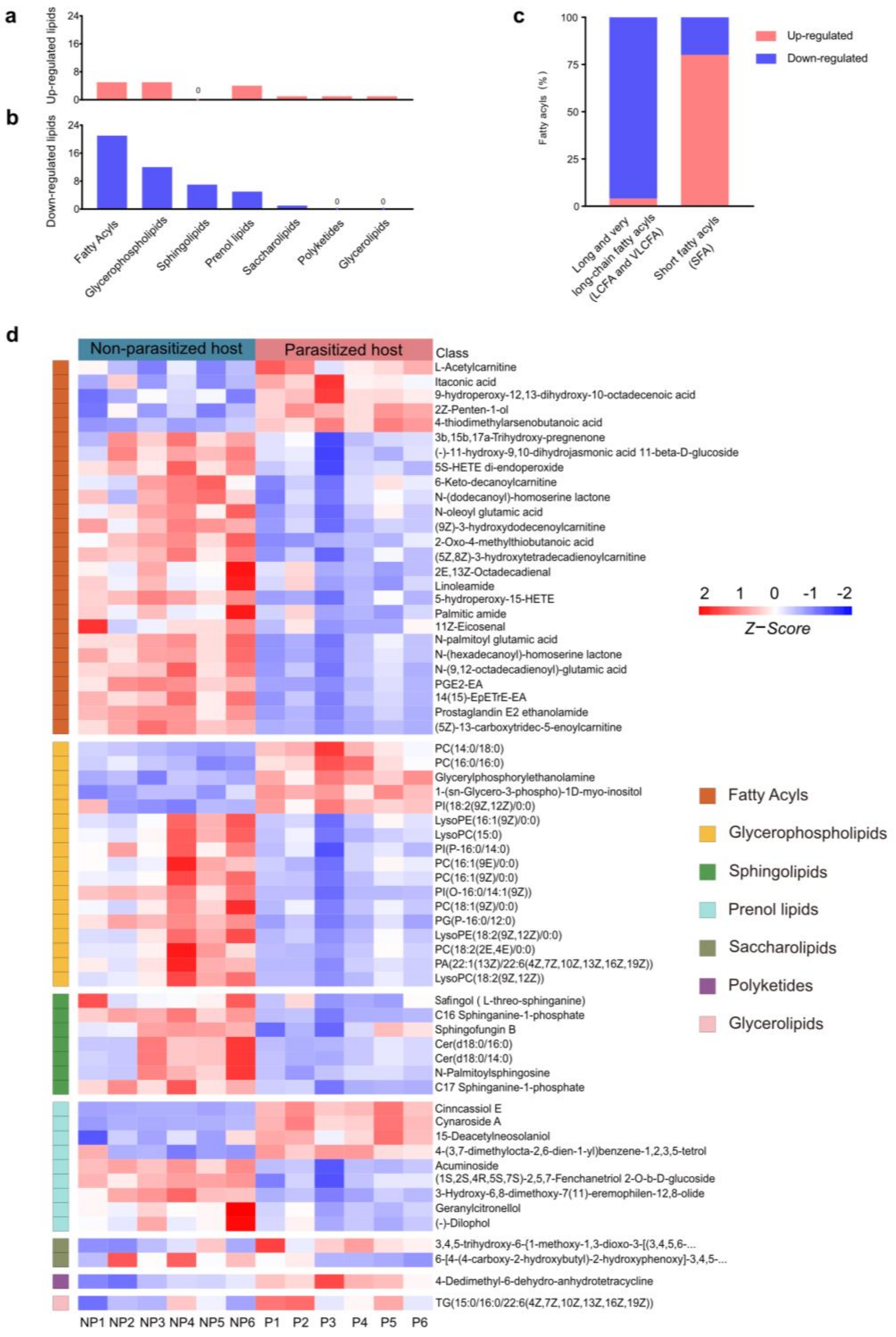
The most altered group of metabolites was the lipids and lipid-like molecules, which can be divided into fatty acyls (FA), glycerophospholipids (GP), sphingolipids (SP), prenol lipids (PR), saccharolipids (SL), polyketides (PK) and glycerolipids (GL), according to different structures and synthetic pathways [39] (Figure 3a,b,d). In this group, a total of 63 differential metabolites were identified, most (46/63, 73.02%) of which were downregulated in parasitized hosts, including FA (21/46, 45.65%), GP (12/46, 26.09%), SP (7/46, 15.22%), PR (5/46, 10.87%) and SL (1/46, 2.17%) (Table S1 and Figure 3a,b). Generally, FA containing more than 13 carbon atoms is referred to as a long-chain fatty acyl (C: 13-21, LCFA) or very long-chain fatty acyl (C > 21, VLCFA), while the FA containing less than 13 carbon atoms is determined as short fatty acyl (SFA) [40]. Among the total of five upregulated FAs and 21 downregulated FAs, LCFAs and VLCFAs accounted for a large proportion (21/26), and almost all were declined in parasitized hosts (20/21, Figure 3c). As for the remaining five differentially expressed SFAs (L-Acetylcarnitine, Itaconic acid, 2Z-Penten-1-ol, 4-thiodimethylarsenobutanoic acid, 2-Oxo-4-methylthiobutanoic acid), there was only one decreased substance (2-oxo-4-methylthiobutanoate) (Figure 3c). Though most of the lipids and lipid-like molecules were decreased, the cluster heatmap showed that parasitization increased some metabolites in six out of the seven groups, except in SP, where the metabolites were all decreased (Figure 3d).
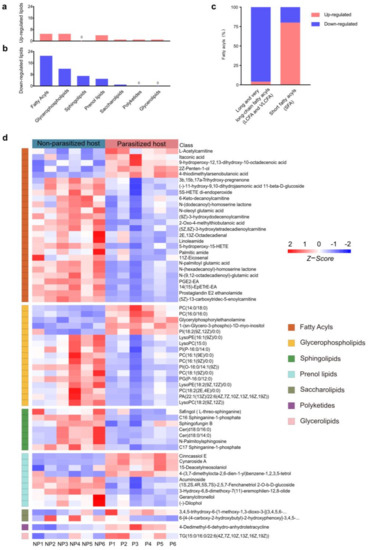
Figure 3.
Differences in lipids and lipid-like molecules post A. japonica parasitization. (a) The classification of upregulated lipids and lipid-like molecules. (b) The classification of downregulated lipids and lipid-like molecules. (c) Distribution of differential long-chain fatty acyls (LCFAs) or very long-chain fatty acyls (VLCFAs) and short fatty acyls (SFAs) in the classification of fatty acyls (FAs). (d) Heatmap for comparison of lipids and lipid-like molecules between parasitized and non-parasitized hosts. Each column represents one NP or P sample, and each row represents one differentially expressed metabolite. The Z-score of each sample is plotted according to the red–blue color scale. The red and blue colors indicate high abundance and low abundance, respectively. Abbreviations: NP, non-parasitized host; P, parasitized host.
3.4. Differences in Organic Acids and Their Derivatives Post A. japonica Parasitization
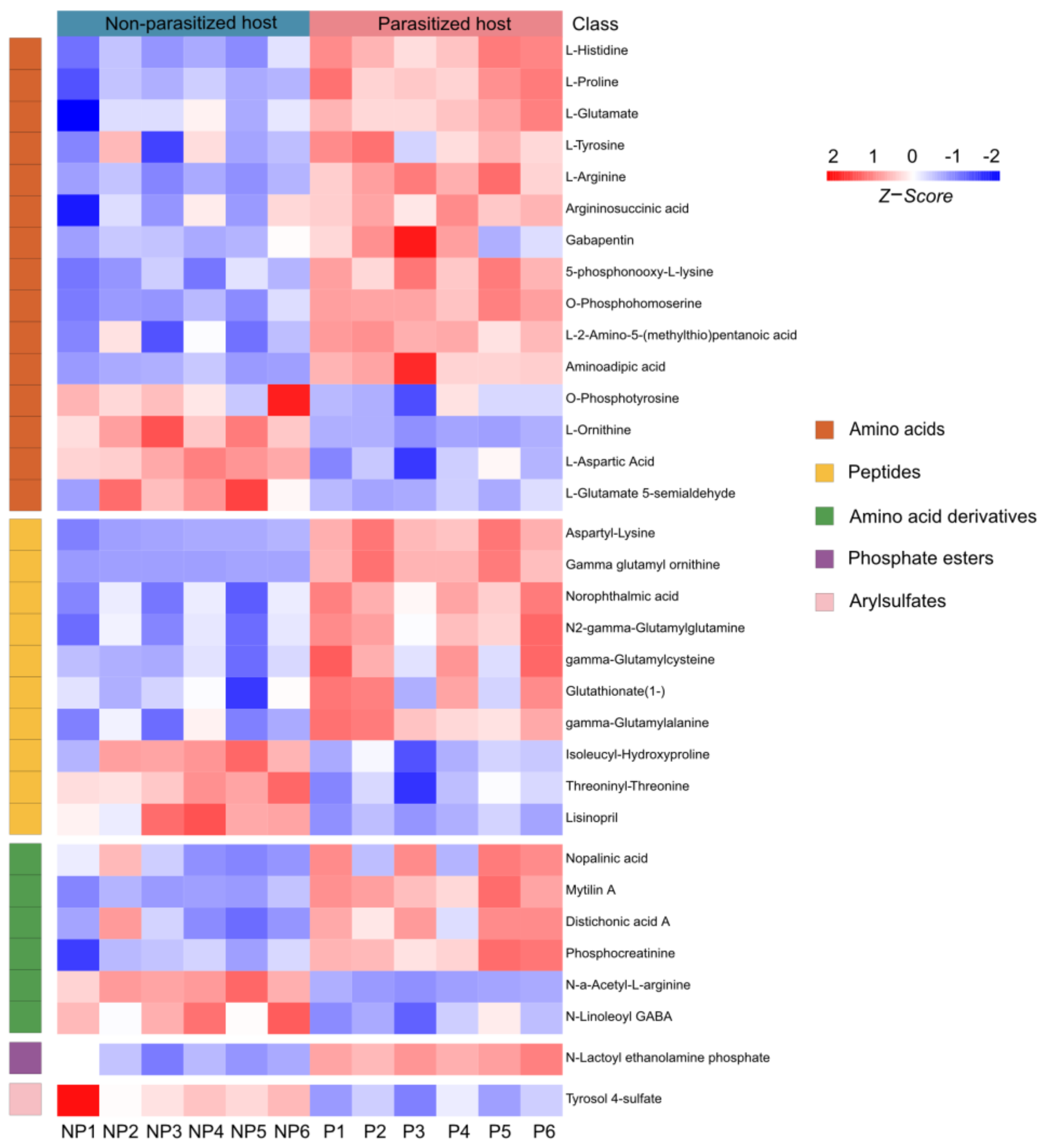
A total of 33 differential metabolites of organic acids and derivatives were annotated, including the classes of amino acids (15/33, 45.45%), peptides (10/33, 30.30%), amino acid derivatives (6/33, 18.18%), phosphate esters (1/33, 3.03%) and arylsulfates (1/33, 3.03%) (Table S1 and Figure 4). Amino acids are necessary in the nutritional and metabolic processes of insects. It has been reported that amino acids from hosts are essential for the hatching of parasitoid eggs [41]. In our study, the majority of amino acids (11/15, 73.33%) were increased, except O-phosphotyrosine, L-ornithine, L-aspartic acid and L-glutamate 5-semialdehyde (Figure 4). Moreover, we also found 10 peptides of which the expression changed post parasitization, with three of them (isoleucyl-hydroxyproline, threoninyl-threonine and lisinopril) being downregulated and seven (aspartyl-lysine, gamma glutamyl ornithine, norophthalmic acid, N2-gamma-glutamylglutamine, gamma-glutamylcysteine, glutathionate(1-), and gamma-glutamylalanine) being upregulated (Figure 4). As for the class of the amino acid derivatives, parasitization significantly increased the contents of four substances (nopalinic acid, mytilin A, distichonic acid A and phosphocreatinine), while decreasing those of the remaining two substances (N-a-acetyl-L-arginine and N-Linoleoyl GABA) in parasitized hosts (Figure 4). Compared with the healthy Drosophila hosts, the metabolite of phosphate ester (N-lactoyl ethanolamine phosphate) was increased, while the arylsulfate (tyrosol 4-sulfate) was decreased after parasitization (Figure 4).
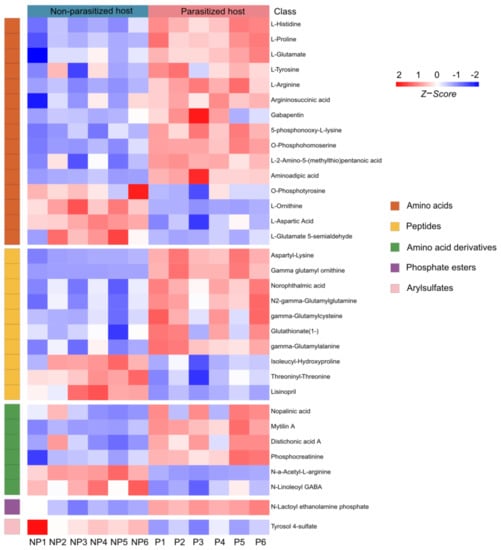
Figure 4.
Differences in organic acids and derivatives post A. japonica parasitization. Heatmap of the group of organic acids and derivatives changing in parasitized and non-parasitized hosts. Each column represents one NP or P sample, and each row represents one differentially expressed metabolite. The Z-score of each sample is plotted according to the red–blue color scale. The red and blue colors indicate high abundance and low abundance, respectively. Abbreviations: NP, non-parasitized host; P, parasitized host.
3.5. Changes in Organic Oxygen Compounds Post A. japonica Parasitization
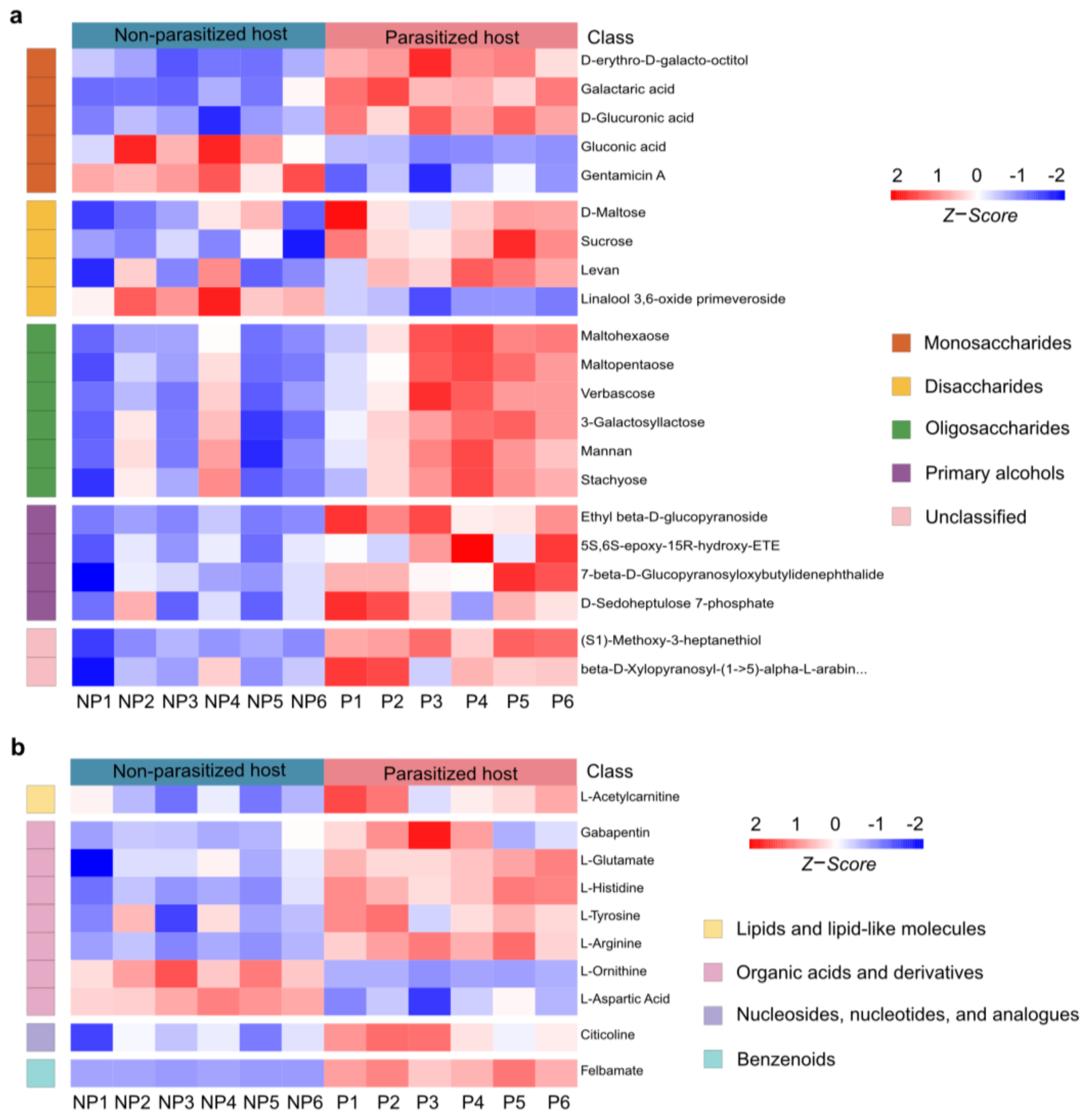
The differential organic oxygen compounds detected in A. japonica-parasitized and non-parasitized hosts have been further divided into monosaccharides, disaccharides, oligosaccharides, primary alcohols and other metabolites (Figure 5a), which came to a total of 21 metabolites. As shown in Figure 5a, parasitization significantly unregulated most (18/21, 85.71%) of the organic oxygen compounds in hosts, while two monosaccharides (gluconic acid and gentamicin A) and one disaccharide (linalool 3,6-oxide primeveroside) were largely downregulated. Interestingly, different types of organic oxygen compounds were found to be involved in the immune responses. For examples, levan, an increased disaccharide, has been used as an anti-irritant, anti-oxidant and anti-inflammatory agent [42]; mannan, an increased oligosaccharide, can function as an immunosuppressant in response to fungus infection [43]; another upregulated oligosaccharide, stachyose, is able to increase both glutathione peroxidase and superoxide dismutase activities, which could remarkably decrease the inflammation response [44]. Taken together, these results suggest that the changed carbohydrate metabolites not only provide the necessary carbon and energy sources for parasitoid offspring, but also might help increase host immunity.
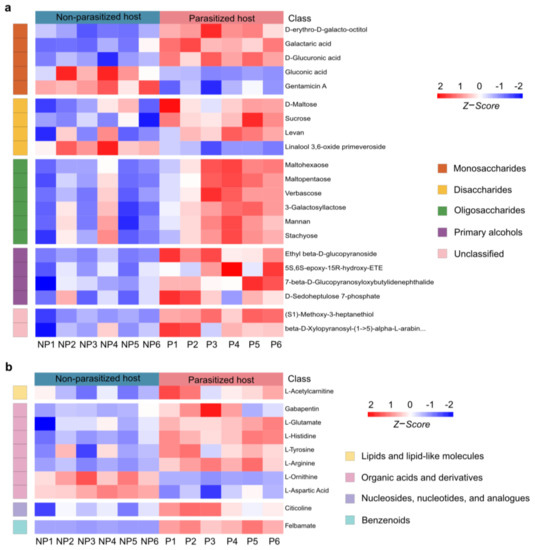
Figure 5.
Changes in organic oxygen compounds and neuromodulators post A. japonica parasitization. (a) Heatmap of the groups of organic oxygen compounds changing in parasitized and non-parasitized hosts. (b) Heatmap for comparison of neuromodulators between parasitized and non-parasitized hosts. Each column represents one NP or P sample, and each row represents one differentially expressed metabolite. The Z-score of each sample is plotted according to the red–blue color scale. The red and blue colors indicate high abundance and low abundance, respectively. Abbreviations: NP, non-parasitized host; P, parasitized host.
3.6. Changes in Neuromodulators Post A. japonica Parasitization
Neuromodulators are an important type of metabolite, which is widely present in insects and mainly controls their numerous behaviors. As a functional active ingredient, neuromodulators have attracted extensive attention in parasitoid–host manipulation [45,46,47,48]. Compared to the non-parasitized hosts, a total of 10 neuromodulators were identified to be changed in A. japonica-parasitized hosts. They were from the groups of lipids and lipid-like molecules (1/10, 10%), organic acids and derivatives (7/10, 70%), nucleosides, nucleotides, and analogues (1/10, 10%) and benzenoids (1/10, 10%) (Figure 5b). Organic acids and derivatives were the most abundant neuromodulators among the four biochemical groups, with five of them (gabapentin, L-glutamate L-histidine L-tyrosine L-arginine) being upregulated and two of them (L-ornithine and L-aspartic Acid) being downregulated. The remaining three differential neuromodulators in the group of lipids and lipid-like molecules (L-acetylcarnitine), nucleosides, nucleotides, and analogues (citicoline), and benzenoids (felbamate) were significantly increased after parasitization. Remarkably, felbamate, a nerve suppressor and antiseizure drugs (ASDs), was the most upregulated neuromodulator upon A. japonica infection. (42.47-fold, Table S1).
3.7. Metabolic Pathway Analysis
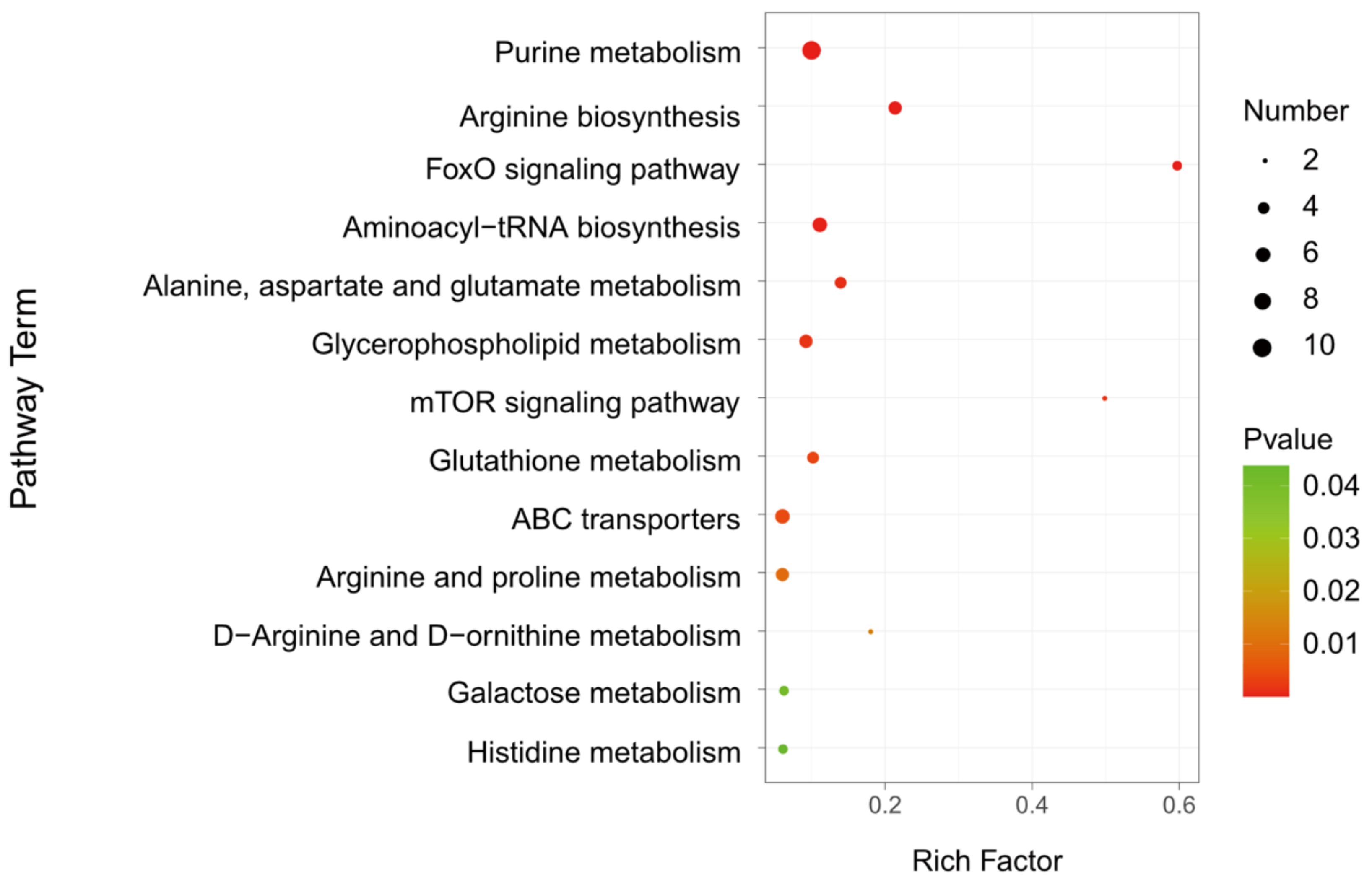
To identify the metabolic pathways altered in parasitized hosts, 205 differential metabolites were further carried out by the KEGG analysis. A total of 66 differential metabolites were identified, which were distributed in 47 metabolic pathways (Table S2). Taking a p-value ≤ 0.05 as the threshold, 13 pathways satisfying the condition were significantly enriched by different metabolites (Figure 6 and Table 1), including purine metabolism (KEGG: dme00230; p = 2.51 × 10−6), arginine biosynthesis (KEGG: dme00220; p = 3 × 10−5), FoxO signaling pathway (KEGG: dme04068; p = 4.44 × 10−5), aminoacyl-tRNA biosynthesis (KEGG: dme00970; p = 0.00019), alanine, aspartate and glutamate metabolism (KEGG: dme00250; p = 0.00110), glycerophospholipid metabolism (KEGG: dme00564; p = 0.00160), mTOR signaling pathway (KEGG: dme04150; p = 0.00165), glutathione metabolism (KEGG: dme00480; p = 0.00349), ABC transporters (KEGG: dme02010; p = 0.00426), arginine and proline metabolism (KEGG: dme00330; p = 0.00937), D-Arginine and D-ornithine metabolism (KEGG: dme00472; p = 0.01398), galactose metabolism (KEGG: dme00052; p = 0.04148) and histidine metabolism (KEGG: dme00340; p = 0.04379). The metabolic pathways, with over five different metabolites between A. japonica-parasitized and non-parasitized hosts, were as follows: purine metabolism, arginine biosynthesis, aminoacyl-tRNA biosynthesis, glycerophospholipid metabolism, ABC transporters, and arginine and proline metabolism. There were 10 differential metabolites in the pathway of purine metabolism, and all of them were upregulated in parasitized hosts (Table 1). In the pathways of aminoacyl-tRNA biosynthesis and ABC transporters, 83.33% (5/6) amino acids were upregulated, except L-aspartic acid, which was also enriched in the pathways of arginine biosynthesis, alanine, aspartate and glutamate metabolism and histidine metabolism. As for the pathway of glycerophospholipid metabolism, more than 60% (4/6) metabolites, including citicoline, glycerylphosphorylethanolamine, PC(16:0/16:0) and phosphocholine, were upregulated, while the contents of lysoPC(15:0) and PC(16:1(9Z)/0:0) in the pathway were downregulated. In addition, all the metabolites in the FoxO signaling pathway, mTOR signaling pathway and galactose metabolism, were increased in parasitized Drosophila hosts. As for the pathways including glutathione metabolism, arginine and proline metabolism and D-Arginine and D-ornithine metabolism, 75% (3/4), 60% (3/5) and 50% (1/2) of the metabolites were increased, respectively.
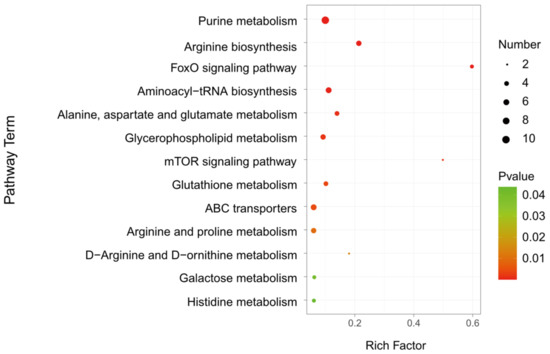
Figure 6.
KEGG analysis of the differential metabolites in D. melanogaster hosts post A. japonica parasitization. KEGG analysis of differential metabolites in parasitized and non-parasitized hosts. The larger the rich factor, the greater the enrichment degree; the color from green to red indicates that the p-value decreasing. The larger the point, the more metabolites enriched in the pathway.

Table 1.
Significantly enriched metabolic pathways, identified in different metabolites.
4. Discussion
It is well known that parasitoids need to change host metabolism to ensure the development and survival of their offspring; however, the underlying mechanisms are largely unknown [5,9]. Metabolomics analysis is extremely helpful to explore the molecular bases of host responses to parasitoid infection. Here, we compared metabolic profiling between A. japonica-parasitized and non-parasitized D. melanogaster hosts, and identified 3043 metabolites in the two groups of the host larvae. Among them, a total of 205 differential metabolites, consisting of 112 upregulated and 93 downregulated metabolites, were obtained, which belonged to 13 different metabolic pathways.
Lipids are widely distributed in insect individuals and are known to play important roles in growth and survival. Parasitoid wasps have evolved to modify host lipid metabolism and consume host fat as necessary nutrition, because they are incapable of synthesizing lipids [3,12,49,50]. Therefore, to some extent, lipid profiles are probably the best-characterized metabolic factor, of which the expression is associated with parasitoid infection [51,52]. In this study, A. japonica parasitism resulted in remarkable changes in host lipid metabolism. We found 63 lipids and lipid-like molecules changing their expression post parasitization, and more than 70% of these changed substances were downregulated, especially FAs, GPs and SPs. The decreased content of FAs may result in decreased activity of GP and SP metabolism, because FAs are building blocks of membrane lipids (e.g., GP and SP), as well as other nonpolar lipids with important storage functions [25]. Interestingly, in Lysiphlebia japonica-parasitized Aphis gossypii hosts, it has been reported that an increased abundance of FAs and GPs were detected [53,54]. Their findings were opposite to our results, indicating that different lipid manipulating strategies were taken in response to parasitizaion by different parasitoid species. SP, one of the necessary cellular lipid components, participates in multiple physiological activities, such as intercellular communication, cell proliferation and differentiation [55,56]. In 2017, Gao et al. have found that SPs were less abundant in the parasitized hosts, the cotton-melon aphid [54], which was similar to our results. In those differential FA lipids, we further found that almost all LCFAs and VLCFAs were declined in parasitized hosts, and almost all SFAs were upregulated, indicating that parasitoids require a high amount of SFAs, but not LCFAs and VLCFAs, from their hosts, which possibly benefits their development and reproduction.
Amino acids are one of the most important compositions of organic acids and derivatives, which are found to be crucial in egg-hatching [41]. Infection by parasitoid wasps usually leads to noticeable alterations in the levels of amino acids within their hosts. Our results confirmed the findings that 11 amino acids were increased in parasitized hosts, including L-histidine, L-proline, L-glutamate, L-tyrosine, L-arginine, argininosuccinic acid, gabapentin, 5-phosphonooxy-L-lysine, O-phosphohomoserine, L-2-Amino-5-(methylthio)pentanoic acid and aminoadipic acid. In addition, we also found levels of four amino acids were decreased, including O-phosphotyrosine, L-ornithine, L-aspartic acid and L-glutamate 5-semialdehyde. It has been reported that L-histidine and L-glutamate are momentous in the transition of insect development and metamorphosis [57]. L-proline is an endogenous neutral amino acid, which is essential for physiological and behavioral actions in the nervous system of vertebrates [58,59]. It has also been reported that L-arginine, argininosuccinic acid and aminoadipic acid can help insects synthesize immune effectors to participate in the immune priming response [60]. As such, our results suggested that parasitoids might need high amounts of L-histidine, L-glutamate and L-proline, to trigger their metamorphosis and neuronal development, along with high amounts of amino acids such as L-arginine, argininosuccinic acid and aminoadipic acid, to enhance their immunity. Moreover, previous studies have found that the content of protein amino acids (e.g., L-glutamate and L-histidine) were increased in hosts due to Dastarcus helophoroides or Aphidius ervi parasitization [61,62], which was consistent with our findings. Previous studies also showed that a reduction of tyrosine concentration was observed in Cotesia congregata-parasitized Manduca sexta host larvae and Toxoneuron nigriceps-parasitized Heliothis virescens host larvae [41,63]. However, the content of tyrosine was identified to be higher in host larvae post A. japonica parasitization. Thus, it is urgently necessary to identify the key elements that are in charge of the expression of amino acids within parasitized hosts, and to investigate the important functions of the amino acids in different parasitoid–host interaction.
Carbohydrates are the primary energy sources for longevity and fecundity of parasitoids. The principal carbohydrate sources available to parasitoid adults are nectar and honeydew, which contain abundant sucrose and other prevalent sugars [64,65,66]. Moreover, the host sugars are also essential for parasitoids to complete the developmental stages of their juveniles. For example, as the main storage sugar in insects, trehalose is used to generate glucose for glycolysis [67], and previous studies have shown that parasitism largely alters the level of trehalose and glucose in hosts [15,68,69,70]. In our study, sugar was also the most important part of the organic oxygen compounds in the changed metabolites. However, both the contents of trehalose and glucose were not significantly changed in A. japonica-parasitized host larvae at the first 3 h infection (Table S1). Nevertheless, we found that the majority of other sugars were upregulated in parasitized host larvae, which included three monosaccharides (D-erythro-D-galacto-octitol, galactaric acid and D-glucuronic acid), three disaccharides (D-maltose, sucrose and levan) and six oligosaccharides (maltohexaose, maltopentaose, verbascose, 3-galactosyllactose, mannan and stachyose). Previous reports have shown that the sugar nutrients can promote longevity, fecundity and population sizes of the insects [71,72]. In addition, three saccharides (levan, mannan and stachyose) were found to be involved in the immune responses [42,43,44]. Based on these data, we propose that the increased sugar metabolites are possibly beneficial to parasitoid embryonic and postembryonic development, and/or manipulate host immunity.
Parasites have great abilities to change host behaviors by manipulating neuromodulatory systems [45,46]. The behavioral responses are fascinating and varied, including suppression of host feeding behavior by increasing the octopamine level [47], and a transient paralysis of the host by blocking both acetylcholine and GABA, that mediate synaptic transmissions [48]. In A. japonica-parasitized host larvae, we also found changes in 10 neuromodulators, which belonged to four groups of metabolites (e.g., lipids and lipid-like molecules, organic acids and derivatives, nucleosides, nucleotides, and analogues, and benzenoids). The 10 neuromodulators were L-acetylcarnitine, gabapentin, L-glutamate, L-histidine, L-tyrosine, L-arginine, L-ornithine, L-aspartic acid, citicoline and felbamate. It has been reported that parasitism by A. japonica could cause a transient paralysis of the hosts [73]. Thus, it is possible that these differential neuromodulators are responsible to the behavioral regulation post A. japonica parasitization.
Endoparasitoids and ectoparasitoids are two kinds of parasitic insects with completely different egg-laying strategies. Ectoparasitoids lay their eggs on host integument; meanwhile, endoparasitoids exploit hosts by injecting egg(s) into their bodies. A. japonica is a typical larval-pupal endoparasitoid, and can successfully parasitize a large number of species across the Drosophila genus, including D. suzukii, an invasive pest insect [30,74,75]. To manipulate the host metabolism, parasitoids usually inject several parasitic factors along with the oviposition, which include venom, polydnaviruses and virus-like particles [5,8,33,76]. So far, the mechanisms of these parasitic factors regulating host metabolism are largely unclear. Since venom is the only reported parasitic factor of A. japonica, it is thus necessary to identify the venom composition in future studies, which will help dissect the underlying mechanisms.
5. Conclusions
Our findings provide a useful resource for understanding how parasitoids regulate the metabolism status of infected hosts. These results will broaden our knowledge on the nutritional requirements for the development and survival of parasitoids, and contribute to develop advanced pest-management strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo13030336/s1, Table S1: Different metabolites between A. japonica-parasitized and nonparasitized hosts; Table S2: KEGG classification of metabolites in A. japonica-parasitized or non-parasitized hosts.
Author Contributions
Conceptualization, J.C.; methodology, investigation, software, and resources, S.L., J.Z. and Y.S.; validation, data curation and formal analysis, S.L., T.F., W.S., Y.L. and X.G.; writing—original draft preparation, S.L.; writing—review and editing, J.C. and J.H.; visualization, J.C. and J.H.; supervision, J.C.; project administration, J.H. and X.C.; funding acquisition, J.C. and J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant number 32202375) and the China Postdoctoral Science Foundation (grant number 2022M710127) to J.C., and the National Natural Science Foundation of China (grant number 32172467) to J.H.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The metabolomics data have been deposited in the MetaboLights with accession number MTBLS6193. All data presented in this study are available in the article and supplementary material.
Acknowledgments
We thank Qichao Zhang and Lan Pang for their technical support during the course of this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pennacchio, F.; Strand, M.R. Evolution of developmental strategies in parasitic Hymenoptera. Annu. Rev. Èntomol. 2006, 51, 233–258. [Google Scholar] [CrossRef]
- Fei, M.; Gols, R.; Harvey, J.A. The Biology and Ecology of Parasitoid Wasps of Predatory Arthropods. Annu. Rev. Èntomol. 2022, 68, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Beckage, N.E.; Gelman, D.B. Wasp parasitoid disruption of host development: Implications for new biologically based strategies for insect control. Annu. Rev. Èntomol. 2004, 49, 299–330. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.A.; Strand, M.R. The developmental strategies of endoparasitoid wasps vary with host feeding ecology. Ecology 2002, 83, 2439–2451. [Google Scholar] [CrossRef]
- Huang, J.; Chen, J.; Fang, G.; Pang, L.; Zhou, S.; Zhou, Y.; Pan, Z.; Zhang, Q.; Sheng, Y.; Lu, Y.; et al. Two novel venom proteins underlie divergent parasitic strategies between a generalist and a specialist parasite. Nat. Commun. 2021, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- A Schlenke, T.; Morales, J.; Govind, S.; Clark, A. Contrasting Infection Strategies in Generalist and Specialist Wasp Parasitoids of Drosophila melanogaster. PLoS Pathog. 2007, 3, 1486–1501. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, S.; Wang, Y.; Shi, M.; Chen, X.; Huang, J. Biocontrol characteristics of the fruit fly pupal parasitoid Trichopria drosophilae (Hymenoptera: Diapriidae) emerging from different hosts. Sci. Rep. 2018, 8, 13323. [Google Scholar] [CrossRef]
- Asgari, S.; Rivers, D.B. Venom Proteins from Endoparasitoid Wasps and Their Role in Host-Parasite Interactions. Annu. Rev. Èntomol. 2011, 56, 313–335. [Google Scholar] [CrossRef]
- Zhou, S.; Lu, Y.; Chen, J.; Pan, Z.; Pang, L.; Wang, Y.; Zhang, Q.; Strand, M.R.; Chen, X.-X.; Huang, J. Parasite reliance on its host gut microbiota for nutrition and survival. ISME J. 2022, 16, 2574–2586. [Google Scholar] [CrossRef]
- Toledo, D.A.M.; D’Avila, H.; Melo, R.C.N. Host Lipid Bodies as Platforms for Intracellular Survival of Protozoan Parasites. Front. Immunol. 2016, 7, 174. [Google Scholar] [CrossRef]
- Pennacchio, F.; Caccia, S.; Digilio, M.C. Host regulation and nutritional exploitation by parasitic wasps. Curr. Opin. Insect Sci. 2014, 6, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Visser, B.; Ellers, J. Lack of lipogenesis in parasitoids: A review of physiological mechanisms and evolutionary implications. J. Insect Physiol. 2008, 54, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Visser, B.; Le Lann, C.; Blanken, F.J.D.; Harvey, J.A.; van Alphen, J.J.M.; Ellers, J. Loss of lipid synthesis as an evolutionary consequence of a parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 8677–8682. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.C.; Ganesalingam, V.K. Changes in the Composition of Host Haemolymph after Attack by an Insect Parasitoid. Nature 1970, 227, 191–192. [Google Scholar] [CrossRef] [PubMed]
- Bischof, C.; Ortel, J. The effects of parasitism by Glyptapanteles liparidis (Braconidae: Hymenoptera) on the hemolymph and total body composition of gypsy moth larvae (Lymantria dispar, Lymantriidae: Lepidoptera). Parasitol. Res. 1996, 82, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Kaeslin, M.; Pfister-Wilhelm, R.; Lanzrein, B. Influence of the parasitoid Chelonus inanitus and its polydnavirus on host nutritional physiology and implications for parasitoid development. J. Insect Physiol. 2005, 51, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Paredes, J.C.; Herren, J.K.; Schüpfer, F.; Lemaitre, B. The Role of Lipid Competition for Endosymbiont-Mediated Protection against Parasitoid Wasps in Drosophila. Mbio 2016, 7, e01006-16. [Google Scholar] [CrossRef]
- Palm, W.; Sampaio, J.L.; Brankatschk, M.; Carvalho, M.; Mahmoud, A.; Shevchenko, A.; Eaton, S. Lipoproteins in Drosophila melanogaster—Assembly, function, and influence on tissue lipid composition. PLoS Genet. 2012, 8, e1002828. [Google Scholar] [CrossRef]
- Snart, C.J.; Hardy, I.C.; Barrett, D.A. Entometabolomics: Applications of modern analytical techniques to insect studies. Èntomol. Exp. Appl. 2015, 155, 1–17. [Google Scholar] [CrossRef]
- Zhou, Y.; Qin, D.Q.; Zhang, P.W.; Chen, X.T.; Liu, B.J.; Cheng, D.M.; Zhang, Z.X. Integrated LC–MS and GC–MS-based untargeted metabolomics studies of the effect of azadirachtin on Bactrocera dorsalis larvae. Sci. Rep. 2020, 10, 2306. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, H.; Meng, Q.; Wang, M.; Zhou, G.; Li, X.; Wang, H.; Miao, L.; Qin, Q.; Zhang, J. Metabolic insights into the cold survival strategy and overwintering of the common cutworm, Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae). J. Insect Physiol. 2017, 100, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hao, F.; Hu, J.; Zhang, W.; Wan, L.; Zhu, L.; Tang, H.; He, G. Revealing Different Systems Responses to Brown Planthopper Infestation for Pest Susceptible and Resistant Rice Plants with the Combined Metabonomic and Gene-Expression Analysis. J. Proteome Res. 2010, 9, 6774–6785. [Google Scholar] [CrossRef] [PubMed]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Wilinski, D.; Winzeler, J.; Duren, W.; Persons, J.L.; Holme, K.J.; Mosquera, J.; Khabiri, M.; Kinchen, J.M.; Freddolino, P.L.; Karnovsky, A.; et al. Rapid metabolic shifts occur during the transition between hunger and satiety in Drosophila melanogaster. Nat. Commun. 2019, 10, 4052. [Google Scholar] [CrossRef]
- Guan, X.L.; Cestra, G.; Shui, G.; Kuhrs, A.; Schittenhelm, R.B.; Hafen, E.; van der Goot, F.G.; Robinett, C.C.; Gatti, M.; Gonzalez-Gaitan, M.; et al. Biochemical Membrane Lipidomics during Drosophila Development. Dev. Cell 2013, 24, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Hunt, E.J.; Pritchard, J.; Bennett, M.J.; Zhu, X.; Barrett, D.A.; Allen, T.; Bale, J.S.; Newbury, H.J. The Arabidopsis thaliana/Myzus persicae model system demonstrates that a single gene can influence the interaction between a plant and a sap-feeding insect. Mol. Ecol. 2006, 15, 4203–4213. [Google Scholar] [CrossRef]
- Maag, D.; Erb, M.; Glauser, G. Metabolomics in plant-herbivore interactions: Challenges and applications. Èntomol. Exp. Appl. 2015, 157, 18–29. [Google Scholar] [CrossRef]
- Kang, K.; Yue, L.; Xia, X.; Liu, K.; Zhang, W. Comparative metabolomics analysis of different resistant rice varieties in response to the brown planthopper Nilaparvata lugens Hemiptera: Delphacidae. Metabolomics 2019, 15, 62. [Google Scholar] [CrossRef]
- Bertrand, C.; Gonzalez-Coloma, A.; Prigent-Combaret, C. Plant Metabolomics to the Benefit of Crop Protection and Growth Stimulation. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2021; Volume 98, pp. 107–132. [Google Scholar]
- Ideo, S.; Watada, M.; Mitsui, H.; Kimura, M.T. Host range of Asobara japonica (Hymenoptera: Braconidae), a larval parasitoid of drosophilid flies. Èntomol. Sci. 2008, 11, 1–6. [Google Scholar] [CrossRef]
- Poyet, M.; Havard, S.; Prevost, G.; Chabrerie, O.; Doury, G.; Gibert, P.; Eslin, P. Resistance of Drosophila suzukii to the larval parasitoids Leptopilina heterotoma and Asobara japonica is related to haemocyte load. Physiol. Entomol. 2013, 38, 45–53. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Pan, Z.; Zhu, J.; Wang, Z.; Shi, M.; Chen, X.; Huang, J. The complete mitochondrial genome of Asobara japonica (Hymenoptera: Braconidae). Mitochondrial DNA Part B 2020, 5, 1279–1281. [Google Scholar] [CrossRef]
- Chen, J.; Fang, G.; Pang, L.; Sheng, Y.; Zhang, Q.; Zhou, Y.; Zhou, S.; Lu, Y.; Liu, Z.; Zhang, Y.; et al. Neofunctionalization of an ancient domain allows parasites to avoid intraspecific competition by manipulating host behaviour. Nat. Commun. 2021, 12, 5489. [Google Scholar] [CrossRef]
- Feng, X.; Gao, G.; Yu, C.; Zhu, A.; Chen, J.; Chen, K.; Wang, X.; Abubakar, A.S.; Chen, P. Transcriptome and metabolome analysis reveals anthocyanin biosynthesis pathway associated with ramie (Boehmeria nivea (L.) Gaud.) leaf color formation. BMC Genom. 2021, 22, 684. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Pan, H.; Shao, H.; Qian, C.; Han, J.; Li, Y.; Lou, Y. UPLC/MS-based untargeted metabolomics reveals the changes in muscle metabolism of electron beam irradiated Solenocera melantho during refrigerated storage. Food Chem. 2022, 367, 130713. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, K.; Ji, W.; Xu, H.; Liu, Y.; Wang, S.; Wang, Z.; Gao, F.; Lin, Z.; Ji, T. Metabolomic analysis of honey bees (Apis mellifera) response to carbendazim based on UPLC-MS. Pestic. Biochem. Physiol. 2021, 179, 104975. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, Y.; Qin, D.; Chen, J.; Zhang, Z. Metabolic Changes in Larvae of Predator Chrysopa sinica Fed on Azadirachtin-Treated Plutella xylostella Larvae. Metabolites 2022, 12, 158. [Google Scholar] [CrossRef]
- Tao, S.; Wang, J.; Liu, M.; Sun, F.; Li, B.; Ye, C. Haemolymph metabolomic differences in silkworms (Bombyx mori L.) under mulberry leaf and two artificial diet rearing methods. Arch. Insect Biochem. 2022, 109, e21851. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Jia, Z.; Moulson, C.L.; Pei, Z.; Miner, J.H.; Watkins, P.A. Fatty acid transport protein 4 is the principal very long chain fatty acyl-CoA synthetase in skin fibroblasts. J. Biol. Chem. 2007, 282, 20573–20583. [Google Scholar] [CrossRef]
- Consoli, F.L.; Vinson, S. Host regulation and the embryonic development of the endoparasitoid Toxoneuron nigriceps (Hymenoptera: Braconidae). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 137, 463–473. [Google Scholar] [CrossRef]
- Öner, E.T.; Hernández, L.; Combie, J. Review of Levan polysaccharide: From a century of past experiences to future prospects. Biotechnol. Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.D.; Shibata, N.; Podzorski, R.P.; Herron, M.J. Candida mannan: Chemistry, suppression of cell-mediated immunity, and possible mechanisms of action. Clin. Microbiol. Rev. 1991, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, D.; Gao, A.; Yang, X. Stachyose increases absorption and hepatoprotective effect of tea polyphenols in high fructose-fed mice. Mol. Nutr. Food Res. 2016, 60, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Adamo, S.A. Parasites: Evolution’s neurobiologists. J. Exp. Biol. 2013, 216, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Haspel, G.; Gefen, E.; Ar, A.; Glusman, J.G.; Libersat, F. Parasitoid wasp affects metabolism of cockroach host to favor food preservation for its offspring. J. Comp. Physiol. A 2005, 191, 529–534. [Google Scholar] [CrossRef]
- Adamo, S.; Linn, C.; Beckage, N. Correlation between changes in host behaviour and octopamine levels in the tobacco hornworm Manduca sexta parasitized by the gregarious braconid parasitoid wasp Cotesia congregata. J. Exp. Biol. 1997, 200, 117–127. [Google Scholar] [CrossRef]
- Libersat, F.; Delago, A.; Gal, R. Manipulation of Host Behavior by Parasitic Insects and Insect Parasites. Annu. Rev. Èntomol. 2009, 54, 189–207. [Google Scholar] [CrossRef]
- Engelstädter, J.; Hurst, G.D. The Ecology and Evolution of Microbes that Manipulate Host Reproduction. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 127–149. [Google Scholar] [CrossRef]
- Boots, M.; Sasaki, A. The evolutionary dynamics of local infection and global reproduction in host-parasite interactions. Ecol. Lett. 2000, 3, 181–185. [Google Scholar] [CrossRef]
- Pernas, L.; Bean, C.; Boothroyd, J.C.; Scorrano, L. Mitochondria Restrict Growth of the Intracellular Parasite Toxoplasma gondii by Limiting Its Uptake of Fatty Acids. Cell Metab. 2018, 27, 886–897.e4. [Google Scholar] [CrossRef]
- Charron, A.J.; Sibley, L.D. Host cells: Mobilizable lipid resources for the intracellular parasite Toxoplasma gondii. J. Cell Sci. 2002, 115, 3049–3059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Luo, J.-Y.; Lv, L.-M.; Wang, C.-Y.; Li, C.-H.; Zhu, X.-Z.; Cui, J.-J. Effects of Lysiphlebia japonica (Ashmead) on cotton-melon aphid Aphis gossypii Glover lipid synthesis. Insect Mol. Biol. 2015, 24, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, S.; Luo, J.; Lü, L.; Zhang, L.; Cui, J. Lipidomics and RNA-Seq study of lipid regulation in Aphis gossypii parasitized by Lysiphlebia japonica. Sci. Rep. 2017, 7, 1364. [Google Scholar]
- Zuo, W.; Li, C.; Luan, Y.; Zhang, H.; Tong, X.; Han, M.; Gao, R.; Hu, H.; Song, J.; Dai, F.; et al. Genome-wide identification and analysis of elongase of very long chain fatty acid genes in the silkworm, Bombyx mori. Genome 2018, 61, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.C.; Lee, K.C.; Cho, O.R.; Jung, I.Y.; Cho, S.Y.; Kim, S.Y. Sphingolipids from Bombycis Corpus 101A and Their Neurotrophic Effects. J. Nat. Prod. 2003, 66, 466–469. [Google Scholar] [CrossRef]
- Li, M.-J.; Jiang, G.-F.; Wang, W. Metabolite Changes in Orange Dead Leaf Butterfly Kallima inachus during Ontogeny and Diapause. Metabolites 2022, 12, 804. [Google Scholar] [CrossRef]
- Gage, S.L.; Kramer, C.; Calle, S.; Carroll, M.; Heien, M.; De Grandi-Hoffman, G. Nosema ceranae parasitism impacts olfactory learning and memory and neurochemistry in honey bees (Apis mellifera). J. Exp. Biol. 2018, 221, jeb161489. [Google Scholar] [CrossRef]
- Schulz, D.; Morschel, J.; Schuster, S.; Eulenburg, V.; Gomeza, J. Inactivation of the Mouse L-Proline Transporter PROT Alters Glutamatergic Synapse Biochemistry and Perturbs Behaviors Required to Respond to Environmental Changes. Front. Mol. Neurosci. 2018, 11, 279. [Google Scholar] [CrossRef]
- Wu, G.; Liu, J.; Li, M.; Xiao, Y.; Yi, Y. Prior infection of Galleria mellonella with sublethal dose of Bt elicits immune priming responses but incurs metabolic changes. J. Insect Physiol. 2022, 139, 104401. [Google Scholar] [CrossRef]
- Li, X.-J.; Dong, G.-P.; Fang, J.-M.; Liu, H.-J.; Guo, W.-L. Parasitic behaviour of Dastarcus helophoroides (Fairmaire) (Coleoptera: Bothrideridae) induced changes in free animo acid pools in hemolymph of host Monochamus alternatus Hope (Coleoptera: Cerambycidae). Orient. Insects 2018, 52, 329–341. [Google Scholar] [CrossRef]
- Rahbé, Y.; Digilio, M.; Febvay, G.; Guillaud, J.; Fanti, P.; Pennacchio, F. Metabolic and symbiotic interactions in amino acid pools of the pea aphid, Acyrthosiphon pisum, parasitized by the braconid Aphidius ervi. J. Insect Physiol. 2002, 48, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, T.L.; Starkey, S.R.; Beckage, N.E. Tyrosine and catecholamine levels in the hemolymph of tobacco hornworm larvae, Manduca sexta, parasitized by the braconid wasp, Cotesia congregata, and in the developing parasitoids. Arch. Insect Biochem. 1998, 38, 193–201. [Google Scholar] [CrossRef]
- Idris, A.B.; Grafius, E. Wildflowers as Nectar Sources for Diadegma insulare (Hymenoptera: Ichneumonidae), a Parasitoid of Diamondback Moth (Lepidoptera: Yponomeutidae). Environ. Èntomol. 1995, 24, 1726–1735. [Google Scholar] [CrossRef]
- Wäckers, F.L. A comparison of flowering herbs with respect to their nectar accessibility for the parasitoid Pimpla turionellae. Exp. Appl. Entomol. 1996, 7, 177–182. [Google Scholar]
- Baker, H.G. Floral Nectar Sugar Constituents in Relation to Pollinator Type. In Handbook of Experimental Pollination Biology; Springer: New York, NY, USA, 1983; pp. 117–141. [Google Scholar]
- Nation, J.L. Insect Physiology and Biochemistry; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Mrinalini; Siebert, A.L.; Wright, J.; Martinson, E.; Wheeler, D.; Werren, J.H. Parasitoid venom induces metabolic cascades in fly hosts. Metabolomics 2015, 11, 350–366. [Google Scholar] [CrossRef]
- Thompson, S. Effect of the insect parasite Hyposoter exiguae (Viereck) on the carbohydrate metabolism of its host, Trichoplusia ni (Hübner). J. Insect Physiol. 1986, 32, 287–293. [Google Scholar] [CrossRef]
- Dahlman, D. Trehalose and glucose levels in hemolymph of diet-reared, tobacco leaf-reared and parasitized tobacco hornworm larvae. Comp. Biochem. Physiol. Part A Physiol. 1975, 50, 165–167. [Google Scholar] [CrossRef]
- Mattson, W.J. Herbivory in Relation to Plant Nitrogen Content. Annu. Rev. Ecol. Syst. 1980, 11, 119–161. [Google Scholar] [CrossRef]
- Blanco, C.A.; Portilla, M.; Abel, C.A.; Winters, H.; Ford, R.; Streett, D.; Cohen, A. Soybeanflour and wheat germ proportions in artificial diet and their effect on the growth rates of the tobacco budworm, Heliothis virescens. J. Insect Sci. 2009, 9, 1536–2442. [Google Scholar] [CrossRef]
- Mabiala-Moundoungou, A.; Doury, G.; Eslin, P.; Cherqui, A.; Prévost, G. Deadly venom of Asobara japonica parasitoid needs ovarian antidote to regulate host physiology. J. Insect Physiol. 2010, 56, 35–41. [Google Scholar] [CrossRef]
- Wang, X.; Biondi, A.; Nance, A.H.; Zappalà, L.; Hoelmer, K.A.; Daane, K.M. Assessment of Asobara japonica as a potential biological control agent for the spotted wing drosophila, Drosophila suzukii. Èntomol. Gen. 2021, 41, 1–12. [Google Scholar] [CrossRef]
- Lee, J.C.; Bruck, D.J.; Curry, H.; Edwards, D.; Haviland, D.R.; Van Steenwyk, R.A.; Yorgey, B.M. The susceptibility of small fruits and cherries to the spotted-wing drosophila, Drosophila suzukii. Pest Manag. Sci. 2011, 67, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.R.; Pech, L.L. Immunological compatibility in parasitoid-host relationships. Annu. Rev. Entomol. 1995, 40, 31–56. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).