Enhancing the Production of the Phenolic Extracts of Asparagus Using an Advanced Green Process
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Sample Treatment
2.1.1. Plant Material
2.1.2. Freeze-Drying and Grinding
2.2. Reagents
2.3. Phenolic Compound Extraction
2.3.1. Conventional Solid–Liquid Extraction (SLE)
2.3.2. Pressurized Liquid Extraction (PLE)
2.4. HPLC–ESI-TOF-MS Analysis
3. Results
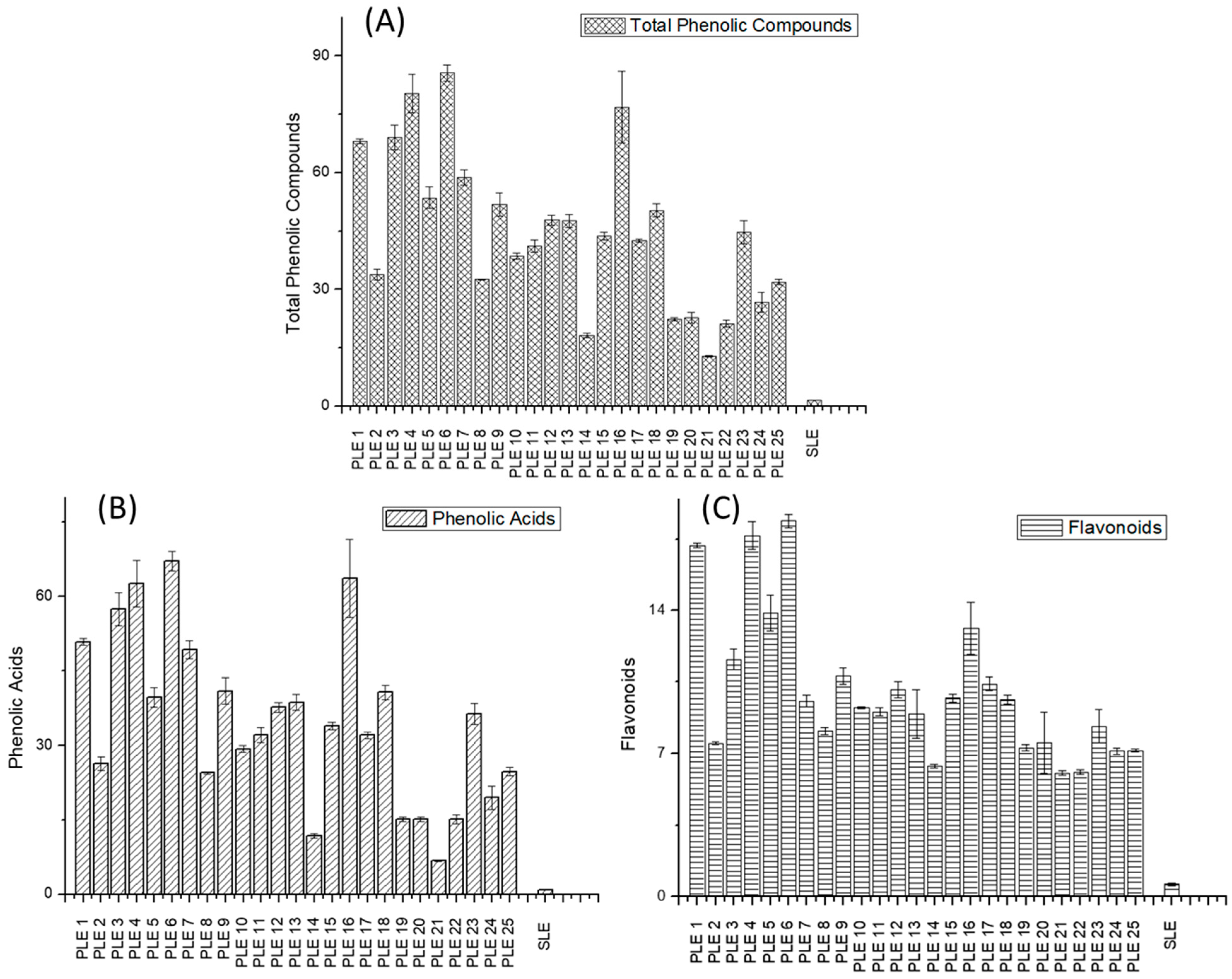
3.1. Characterization and Quantification of Phenolic Compounds in PLE and SLE Extracts by HPLC–ESI-TOF-MS
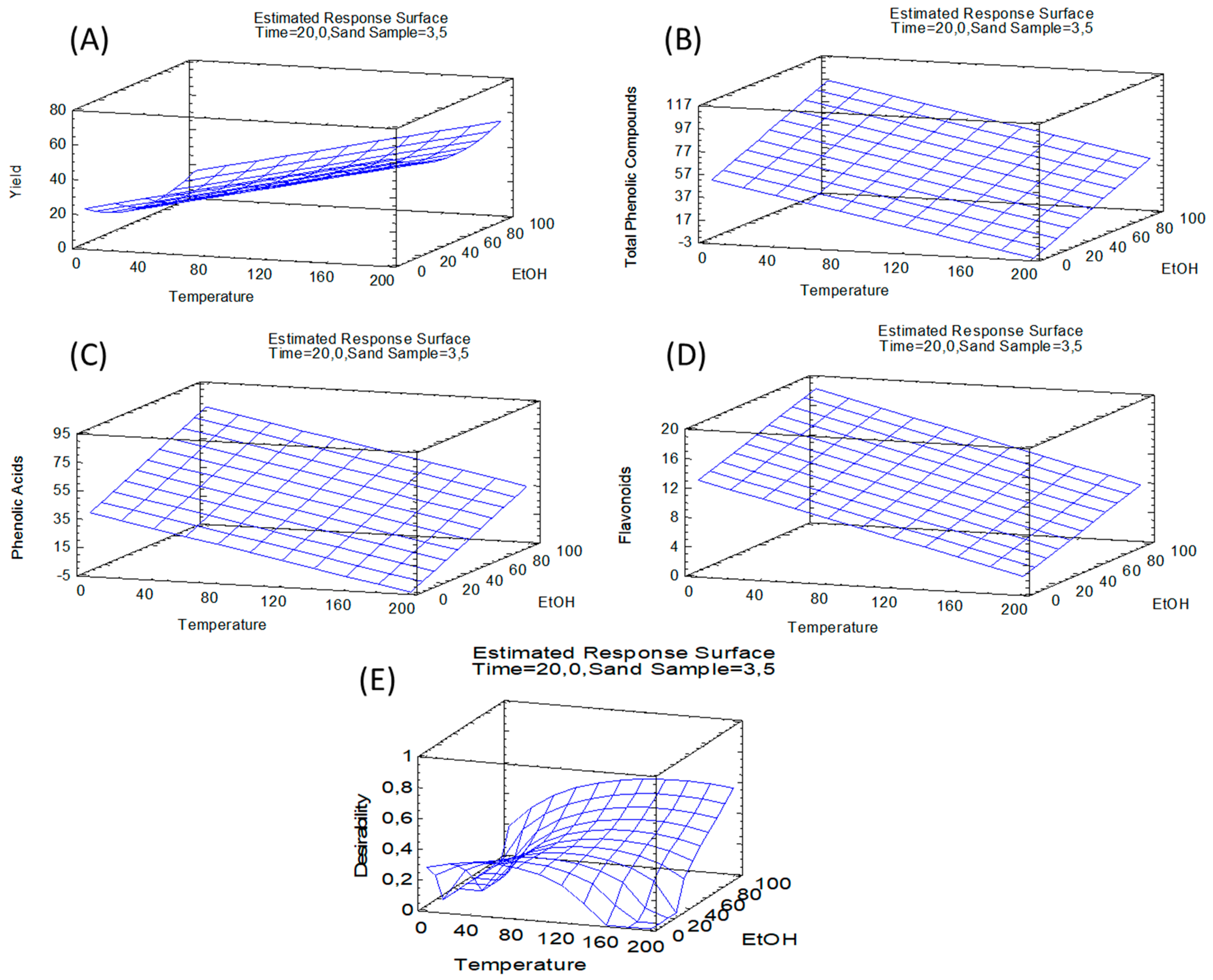
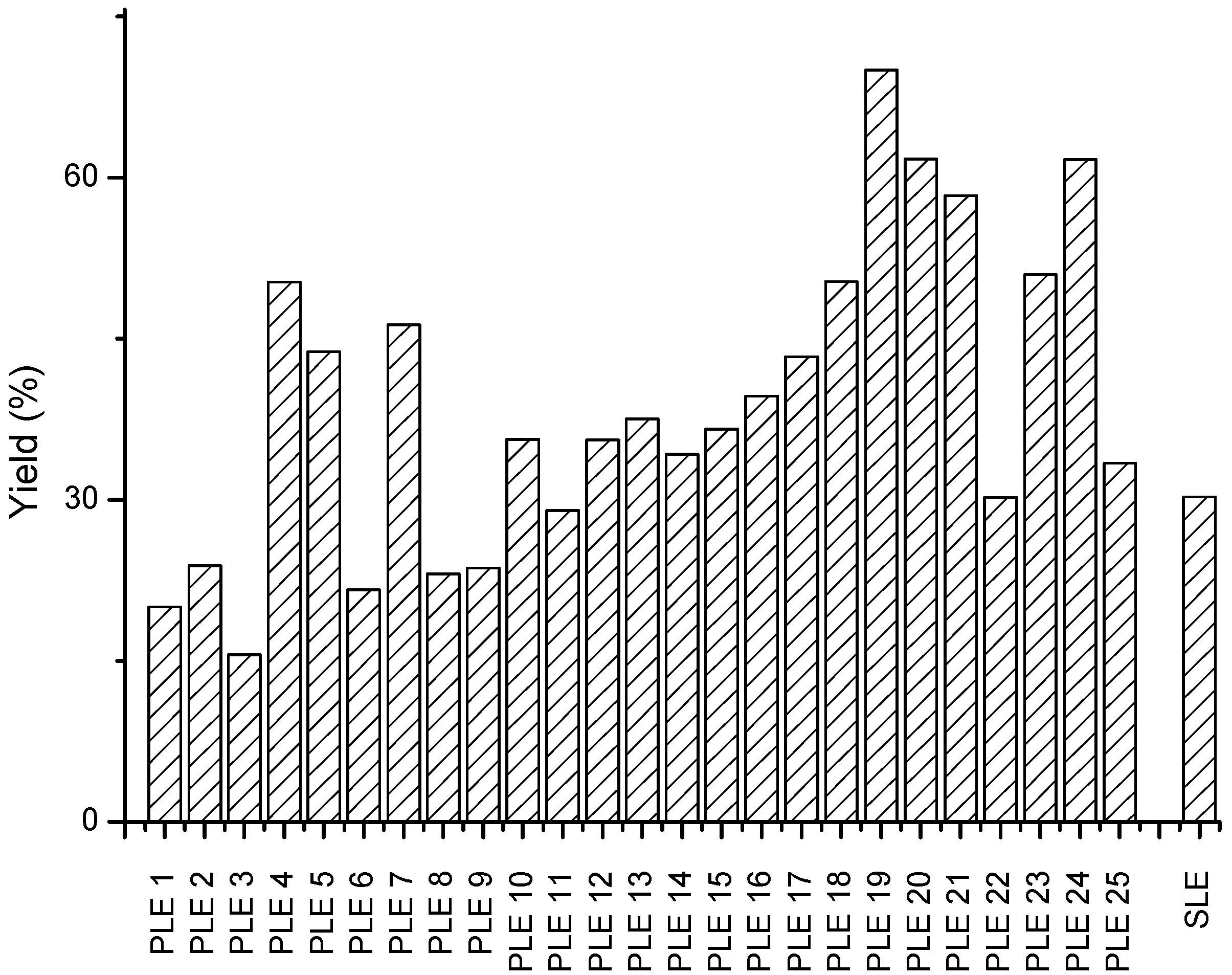
3.2. PLE Optimization of Green Asparagus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez, R.; Jaramillo, S.; Rodríguez, G.; Espejo, J.A.; Guillén, R.; Fernández-Bolaños, J.; Heredia, A.; Jiménez, A. Antioxidant activity of ethanolic extracts from several asparagus cultivars. J. Agric. Food Chem. 2005, 53, 5212–5217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Birch, J.; Pei, J.; Mohamed Ahmed, I.A.; Yang, H.; Dias, G.; Abd El-Aty, A.M.; Bekhit, A.E.-D. Identification of Six Phytochemical Compounds from Asparagus officinalis L. Root Cultivars from New Zealand and China Using UAE-SPE-UPLC-MS/MS: Effects of Extracts on H2O2-Induced Oxidative Stress. Nutrients 2019, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.G.; Bae, J.H.; Namieśnik, J.; Barasxh, D.; Nemirovski, A.; Katrich, E.; Gorinstein, S. Detection of Bioactive Compounds in Organically and Conventionally Grown Asparagus Spears. Food Anal. Methods 2018, 11, 309–318. [Google Scholar] [CrossRef]
- Chiriac, E.R.; Chiţescu, C.L.; Geană, E.I.; Gird, C.E.; Socoteanu, R.P.; Boscencu, R. Advanced analytical approaches for the analysis of polyphenols in plants matrices—A review. Separations 2021, 8, 65. [Google Scholar] [CrossRef]
- Villalva, M.; Santoyo, S.; Salas-Pérez, L.; Siles-Sánchez, M.d.l.N.; Rodríguez García-Risco, M.; Fornari, T.; Reglero, G.; Jaime, L. Sustainable Extraction Techniques for Obtaining Antioxidant and Anti-Inflammatory Compounds from the Lamiaceae and Asteraceae Species. Foods 2021, 10, 2067. [Google Scholar] [CrossRef]
- Kwiatkowski, M.; Kravchuk, O.; Skouroumounis, G.K.; Taylor, D.K. Microwave-assisted and conventional phenolic and colour extraction from grape skins of commercial white and red cultivars at veraison and harvest. J. Clean. Prod. 2020, 275, 122671. [Google Scholar] [CrossRef]
- Fuentes, J.A.M.; López-Salas, L.; Borrás-Linares, I.; Navarro-Alarcón, M.; Segura-Carretero, A.; Lozano-Sánchez, J. Development of an Innovative Pressurized Liquid Extraction Procedure by Response Surface Methodology to Recover Bioactive Compounds from Carao Tree Seeds. Foods 2021, 10, 398. [Google Scholar] [CrossRef]
- Woloszyn, N.; Krabbe, R.D.; Fischer, B.; Bernardi, J.L.; Duarte, P.F.; Puton, B.M.S.; Cansian, R.L.; Paroul, N.; Junges, A. Use of pressurized liquid extraction technique to obtain extracts with biological and antioxidant activity from Mentha pulegium, Equisetum giganteum and Sida cordifolia. Chem. Pap. 2022, 76, 5775–5788. [Google Scholar] [CrossRef]
- Ho, T.C.; Kiddane, A.T.; Khan, F.; Cho, Y.J.; Park, J.S.; Lee, H.J.; Kim, G.D.; Kim, Y.M.; Chun, B.S. Pressurized liquid extraction of phenolics from Pseuderanthemum palatiferum (Nees) Radlk. leaves: Optimization, characterization, and biofunctional properties. J. Ind. Eng. Chem. 2022, 108, 418–428. [Google Scholar] [CrossRef]
- Dobroslavić, E.; Garofulić, I.E.; Šeparović, J.; Zorić, Z.; Pedisić, S.; Dragović-Uzelac, V. Pressurized Liquid Extraction as a Novel Technique for the Isolation of Laurus nobilis L. Leaf Polyphenols. Molecules 2022, 27, 5099. [Google Scholar] [CrossRef]
- Katsinas, N.; Bento da Silva, A.; Enríquez-de-Salamanca, A.; Fernández, N.; Bronze, M.R.; Rodríguez-Rojo, S. Pressurized Liquid Extraction Optimization from Supercritical Defatted Olive Pomace: A Green and Selective Phenolic Extraction Process. ACS Sustain. Chem. Eng. 2021, 9, 5590–5602. [Google Scholar] [CrossRef]
- Barrales, F.M.; Silveira, P.; Menezes Barbosa, P.P.M.; Roggia Ruviaro, A.; Paulino, B.N.; Pastores, G.M.; Alves Macedo, G.; Martinez, J. Recovery of phenolic compounds from citrus by-products using pressurized liquids—An application to orange peel. Food Bioprod. Process. 2018, 112, 9–21. [Google Scholar] [CrossRef]
- Regulation, European Commission, (EC). No. 547/2000 (2000). Supplementing the Annex to Regulation (EC) No 2400/96 on the entry of certain names in the ‘Register of protected designations of origin and protected geographical indications’ provided for in Council Regulation (EEC) No 2081/92 on the protection of geographical indications and designations of origin for agricultural products and foodstuffs. Off. J. Eur. Union 2000, L67/8, 1–4. Available online: https://op.europa.eu/en/publication-detail/-/publication/a8716468-0703-48fe-9cfc-dd09a1d44c26/language-en/format-PDF/source-search (accessed on 1 May 2022).
- Yu, Q.; Li, J.; Fan, L. Effect of Drying Methods on the Microstructure, Bioactivity Substances, and Antityrosinase Activity of Asparagus Stems. J. Agric. Food Chem. 2019, 67, 1537–1545. [Google Scholar] [CrossRef]
- Bataglion, G.A.; Da Silva, F.M.A.; Eberlin, M.N.; Koolen, H.H.F. Determination of the phenolic composition from Brazilian tropical fruits by UHPLC-MS/MS. Food Chem. 2015, 180, 280–287. [Google Scholar] [CrossRef]
- Park, M. Sucrose delays senescence and preserves functional compounds in Asparagus officinalis L. Biochem. Biophys. Res. Commun. 2016, 480, 241–247. [Google Scholar] [CrossRef]
- Motoki, S.; Kitazawa, H.; Maeda, T.; Suzuki, T.; Chiji, H.; Nishihara, E.; Shinohara, Y. Effects of various asparagus production methods on rutin and protodioscin contents in spears and cladophylls. Biosci. Biotechnol. Biochem. 2012, 76, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Sánchez, C.; Lozano-Sánchez, J.; Rodríguez-Pérez, C.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Comprehensive, untargeted, and qualitative RP-HPLC-ESI-QTOF/MS2 metabolite profiling of green asparagus (Asparagus officinalis). J. Food Compos. Anal. 2016, 46, 78–87. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, C.; Pedregosa, F.; Borrás-Linares, I.; Lozano-Sánchez, J.; Segura-Carretero, A. Identification of bioactive compounds of Asparagus officinalis L.: Permutation test allows differentiation among “triguero” and hybrid green varieties. Molecules 2021, 26, 1640. [Google Scholar] [CrossRef]
- Solana, M.; Boschiero, I.; Dall’Acqua, S.; Bertucco, A. A comparison between supercritical fluid and pressurized liquid extraction methods for obtaining phenolic compounds from Asparagus officinalis L. J. Supercrit. Fluids 2015, 100, 201–208. [Google Scholar] [CrossRef]
- Chen, X.H.; Ma, L.H.; Dong, Y.W.; Song, H.; Pu, Y.; Zhou, Q.Y. Evaluation of the differences in phenolic compounds and antioxidant activities of five green asparagus (Asparagus officinalis L.) cultivars. Qual. Assur. Saf. Crop. Foods 2017, 9, 479–487. [Google Scholar] [CrossRef]
- Liyana-Pathirana, C.; Shahidi, F. Optimization of extraction of phenolic compounds from wheat using response surface methodology. Food Chem. 2005, 93, 47–56. [Google Scholar] [CrossRef]
- Figueroa, J.G.; Borrás-Linares, I.; Lozano-Sánchez, J.; Quirantes-Piné, R.; Segura-Carretero, A. Optimization of drying process and pressurized liquid extraction for recovery of bioactive compounds from avocado peel by-product. Electrophoresis 2018, 39, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Rudke, A.R.; Mazzutti, S.; Andrade, K.S.; Vitali, L.; Ferreira, S.R.S. Optimization of green PLE method applied for the recovery of antioxidant compounds from buriti (Mauritia flexuosa L.) shell. Food Chem. 2019, 298, 125061. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.V.F.; Portugal, L.A.; dos Anjos, J.P.; de Jesus, O.N.; de Oliveira, E.J.; David, J.P.; Davida, J.M. Accelerated solvent extraction of phenolic compounds exploiting a Box-Behnken design and quantification of five flavonoids by HPLC-DAD in Passiflora species. Microchem. J. 2017, 132, 28–35. [Google Scholar] [CrossRef]
- Leyva-Jiménez, F.J.; Lozano-Sánchez, J.; Borrás-Linares, I.; Arráez-Román, D.; Segura-Carretero, A. Comparative study of conventional and pressurized liquid extraction for recovering bioactive compounds from Lippia citriodora leaves. Food Res. Int. 2018, 109, 213–222. [Google Scholar] [CrossRef]
- Viganó, J.; Brumer, I.Z.; Braga, P.A.C.; Silva, J.K.; Júnior, M.R.M.; Reyes, F.G.R.; Martínez, J. Pressurized liquids extraction as an alternative process to readily obtain bioactive compounds from passion fruit rinds. Food Bioprod. Process. 2016, 100, 382–390. [Google Scholar] [CrossRef]
- Herrero, M.; Castro-Puyana, M.; Mendiola, J.A.; Ibañez, E. Compressed Fluids for the Extraction of Bioactive Compounds. Trends Anal. Chem. 2013, 43, 67–83. [Google Scholar] [CrossRef]

| Peak | RT 1 (min) | m/z (Exp) | m/z (Theor) | Error (ppm) | mSigma | Molecular Formula | Proposed Compound | PLE and SLE Extracts |
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 191.0571 | 191.0561 | −4.3 | 7.9 | C7H12O6 | Quinic acid | PLE *, SLE |
| 2 | 9.2 | 353.0883 | 353.0878 | −1.4 | 22.1 | C16H18O9 | Chlorogenic acid | PLE *, SLE |
| 3 | 10.5 | 337.0915 | 337.0929 | 6.9 | 1.5 | C16H18O8 | Coumaroylquinic acid | PLE 1,2,3,4,5,6,7,8,9, 10,11,12,13,14,15,16,17, 18,19,21,22,23,24,25 |
| 4 | 11 | 367.1039 | 367.1035 | 1.1 | 21 | C17H20O9 | Feruloylquinic acid | PLE *, SLE |
| 5 | 11.2 | 311.0007 | 311.0028 | 1.4 | 6.8 | C18H30O4 | UK 1 | PLE * |
| 6 | 11.5 | 355.101 | 355.1035 | 5.9 | 42 | C16H20O9 | Feruloyl hexose | PLE 1,2,3,4,5,6,7,8,9, 10,11,12,13,14,15,16,17, 18,19,23,24,25, SLE |
| 7 | 11.8 | 311.0263 | 311.0256 | −1.7 | 63.5 | C9H12O12 | UK 2 | PLE * |
| 8 | 12.3 | 311.0005 | 311.0031 | −1.4 | 16.5 | C18H30O4 | UK 3 | PLE * |
| 9 | 12.6 | 771.2017 | 771.1989 | −2.2 | 28.9 | C33H40O21 | Quercetin glucosyl rutinoside | PLE *, SLE |
| 10 | 13.2 | 609.1562 | 609.1555 | −0.2 | 25.6 | C27C31O16 | Rutin | PLE *, SLE |
| 11 | 13.9 | 463.0883 | 463.0882 | 0.4 | 10.3 | C21H20O12 | Quercetin-glucoside | PLE *, SLE |
| 12 | 14.1 | 593.1525 | 593.1512 | −0.6 | 22.1 | C27H30O15 | Kaempferol-rutinoside | PLE *, SLE |
| 13 | 14.2 | 623.1619 | 623.1618 | 0.7 | 19.4 | C28H32O16 | Isorhamnetin-rutinoside | PLE *, SLE |
| 14 | 16 | 523.3837 | 523.2760 | 5.3 | 32.9 | C24H44O12 | UK 4 | PLE * |
| 15 | 21.1 | 329.2326 | 329.2333 | 3.7 | 12.5 | C18H34O5 | Trihydroxy-octadecaenoic acid | PLE * |
| 16 | 21.3 | 383.1128 | 383.1136 | 4.2 | 1.1 | C21H20O7 | Dicoumaroylglycerol | PLE *, SLE |
| 17 | 21.6 | 413.1239 | 413.1242 | 4.2 | 22.5 | C22H22O8 | Coumaroylferuloyl glycerol | PLE *, SLE |
| 18 | 21.9 | 443.1325 | 443.1348 | 5.9 | 7.9 | C23H24O9 | Diferuloyl glycerol | PLE *, SLE |
| 19 | 41.1 | 277.2161 | 277.2173 | 3.8 | 62.4 | C18H30O2 | Linolenic acid | PLE * |
| 20 | 43.7 | 279.2536 | 279.2540 | 4 | 28.2 | C18H32O2 | Linoleic acid | PLE * |
| Y1 | |||||
|---|---|---|---|---|---|
| Variable | Sum of squares | d.f. | Mean Square | F-Ratio | p-Value |
| X1: Temperature | 1748.27 | 1 | 1748.27 | 3793.99 | 0.0103 |
| X2: % EtOH | 49.8912 | 1 | 49.8912 | 108.27 | 0.061 |
| X3: Extraction time | 265.038 | 1 | 265.038 | 575.17 | 0.0265 |
| X4: S–S | 20.6204 | 1 | 20.6204 | 44.75 | 0.0945 |
| X2X2 | 140.293 | 1 | 140.293 | 304.45 | 0.0364 |
| X2X3 | 46.6476 | 1 | 46.6476 | 101.23 | 0.0631 |
| X2X4 | 487.792 | 1 | 487.792 | 1058.58 | 0.0196 |
| X3X3 | 44.17 | 1 | 44.17 | 95.85 | 0.0648 |
| X3X4 | 321.16 | 1 | 321.16 | 696.96 | 0.0241 |
| X4X4 | 236.344 | 1 | 236.344 | 512.9 | 0.0281 |
| Lack-of-fit | 1416.09 | 13 | 108.93 | 236.39 | 0.0502 |
| Pure error | 0.4608 | 1 | 0.4608 | ||
| Total (corr.) | 5048.77 | 24 | |||
| R2 | 0.719427 | ||||
| Y2 | |||||
| Variable | Sum of squares | d.f. | Mean Square | F-Ratio | p-Value |
| X1: Temperature | 3725.07 | 1 | 3725.07 | 275.52 | 0.0383 |
| X2: % EtOH | 2980.15 | 1 | 2980.15 | 220.43 | 0.0428 |
| X3: Extraction time | 0.356199 | 1 | 0.356199 | 0.03 | 0.8976 |
| X4: S–S | 79.0719 | 1 | 79.0719 | 5.85 | 0.2496 |
| X1X1 | 11.8109 | 1 | 11.8109 | 0.87 | 0.5215 |
| X1X2 | 87.9975 | 1 | 87.9975 | 6.51 | 0.2378 |
| X1X3 | 20.6351 | 1 | 20.6351 | 1.53 | 0.4332 |
| X1X4 | 145.53 | 1 | 145.53 | 10.76 | 0.1883 |
| X2X2 | 3.14735 × 10−5 | 1 | 3.14735 × 10−5 | 0 | 0.999 |
| X2X3 | 120.993 | 1 | 120.993 | 8.95 | 0.2054 |
| X2X4 | 851.39 | 1 | 851.39 | 62.97 | 0.0798 |
| X3X3 | 19.0035 | 1 | 19.0035 | 1.41 | 0.4461 |
| X3X4 | 1.05488 | 1 | 1.05488 | 0.08 | 0.8266 |
| X4X4 | 11.0446 | 1 | 11.0446 | 0.82 | 0.5321 |
| Lack-of-fit | 750.45 | 9 | 83.3833 | 6.17 | 0.2992 |
| Pure error | 13.52 | 1 | 13.52 | ||
| Total (corr.) | 9538.24 | 24 | |||
| R2 | 0.919905 | ||||
| Y3 | |||||
| Variable | Sum of squares | d.f. | Mean Square | F-Ratio | p-Value |
| X1: Temperature | 2446.91 | 1 | 2446.91 | 221.54 | 0.0427 |
| X2: % EtOH | 2294.93 | 1 | 2294.93 | 207.78 | 0.0441 |
| X3: Extraction time | 1.01332 | 1 | 1.01332 | 0.09 | 0.8128 |
| X4: S–S | 40.7444 | 1 | 40.7444 | 3.69 | 0.3056 |
| X1X1 | 1.21651 | 1 | 1.21651 | 0.11 | 0.796 |
| X1X2 | 30.1739 | 1 | 30.1739 | 2.73 | 0.3464 |
| X1X3 | 11.1687 | 1 | 11.1687 | 1.01 | 0.4982 |
| X1X4 | 146.599 | 1 | 146.599 | 13.27 | 0.1705 |
| X2X2 | 1.0669 | 1 | 1.0669 | 0.1 | 0.8082 |
| X2X3 | 105.217 | 1 | 105.217 | 9.53 | 0.1995 |
| X2X4 | 517.419 | 1 | 517.419 | 46.85 | 0.0924 |
| X3X3 | 5.14163 | 1 | 5.14163 | 0.47 | 0.6188 |
| X3X4 | 4.38077 | 1 | 4.38077 | 0.4 | 0.6422 |
| X4X4 | 8.91063 | 1 | 8.91063 | 0.81 | 0.5341 |
| Lack-of-fit | 603.555 | 9 | 67.0617 | 6.07 | 0.3014 |
| Pure error | 11.045 | 1 | 11.045 | ||
| Total (corr.) | 6793.25 | 24 | |||
| R2 | 0.909528 | ||||
| Y4 | |||||
| Variable | Sum of squares | d.f. | Mean Square | F-Ratio | p-Value |
| X1: Temperature | 135.195 | 1 | 135.195 | 1081.56 | 0.0194 |
| X2: % EtOH | 44.5893 | 1 | 44.5893 | 356.71 | 0.0337 |
| X3: Extraction time | 2.4977 | 1 | 2.4977 | 19.98 | 0.1401 |
| X4: S–S | 6.54616 | 1 | 6.54616 | 52.37 | 0.0874 |
| X1X1 | 5.70086 | 1 | 5.70086 | 45.61 | 0.0936 |
| X1X2 | 14.8228 | 1 | 14.8228 | 118.58 | 0.0583 |
| X1X3 | 1.53315 | 1 | 1.53315 | 12.27 | 0.1771 |
| X1X4 | 0.00324709 | 1 | 0.00324709 | 0.03 | 0.8983 |
| X2X2 | 1.11277 | 1 | 1.11277 | 8.9 | 0.2059 |
| X2X3 | 0.49643 | 1 | 0.49643 | 3.97 | 0.2961 |
| X2X4 | 41.479 | 1 | 41.479 | 331.83 | 0.0349 |
| X3X3 | 4.74534 | 1 | 4.74534 | 37.96 | 0.1024 |
| X3X4 | 1.15483 | 1 | 1.15483 | 9.24 | 0.2023 |
| X4X4 | 0.125791 | 1 | 0.125791 | 1.01 | 0.499 |
| Lack-of-fit | 32.3611 | 9 | 3.59568 | 28.77 | 0.1418 |
| Pure error | 0.125 | 1 | 0.125 | ||
| Total (corr.) | 299.83 | 24 | |||
| R2 | 0.891652 | ||||
| Y1 | Y2 | Y3 | Y4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run | Predicted | Exp. | CV | Predicted | Exp. | CV | Predicted | Exp. | CV | Predicted | Exp. | CV |
| PLE 1 | 17.80 | 20.03 | 8.3 | 64.8 | 68 ± 0.6 | 3.4 | 51.0 | 50.8 ± 0.7 | 0.3 | 13.8 | 17.2 ± 0.1 | 10.3 |
| PLE 2 | 34.55 | 23.86 | 25.9 | 46.4 | 34 ± 1 | 17.3 | 35.5 | 26 ± 1 | 16.3 | 9.5 | 7.49 ± 0.06 | 12.5 |
| PLE 3 | 26.08 | 15.6 | 35.6 | 70.2 | 69 ± 3 | 1.1 | 56.1 | 57 ± 3 | 1.7 | 12.3 | 11.6 ± 0.5 | 4.8 |
| PLE 4 | 42.87 | 50.25 | 11.2 | 74.1 | 80 ± 5 | 5.6 | 59.6 | 63 ± 5 | 3.5 | 16.1 | 17.7 ± 0.7 | 5.7 |
| PLE 5 | 40.51 | 43.8 | 5.5 | 42.5 | 54 ± 3 | 16.2 | 32.0 | 40 ± 2 | 15.2 | 12.3 | 13.9 ± 0.9 | 8.0 |
| PLE 6 | 25.97 | 21.61 | 13.0 | 72.4 | 86 ± 2 | 11.8 | 57.5 | 67 ± 2 | 11.0 | 16.6 | 18.4 ± 0.3 | 5.1 |
| PLE 7 | 39.05 | 46.29 | 12.0 | 44.2 | 59 ± 2 | 20.4 | 34.0 | 49 ± 2 | 17.0 | 11.7 | 9.6 ± 0.3 | 10.8 |
| PLE 8 | 17.65 | 23.11 | 19.0 | 44.7 | 32.6 ± 0.1 | 17.0 | 33.4 | 24.5 ± 0.2 | 16.6 | 10.0 | 8.1 ± 0.2 | 11.4 |
| PLE 9 | 24.62 | 23.64 | 2.9 | 71.9 | 52 ± 3 | 23.0 | 58.2 | 41 ± 3 | 19.5 | 11.8 | 10.8 ± 0.4 | 5.4 |
| PLE 10 | 32.27 | 35.61 | 7.0 | 44.9 | 38.5 ± 0.7 | 10.8 | 34.9 | 29.3 ± 0.7 | 12.4 | 9.9 | 9.24 ± 0.05 | 3.5 |
| PLE 11 | 26.54 | 29 | 6.3 | 43.7 | 41.2 ± 1.6 | 4.1 | 33.4 | 32 ± 2 | 2.7 | 10.3 | 9.0 ± 0.2 | 7.0 |
| PLE 12 | 45.03 | 35.58 | 16.6 | 43.3 | 48 ± 1 | 7.0 | 33.9 | 38 ± 1 | 7.5 | 9.1 | 10.1 ± 0.4 | 6.1 |
| PLE 13 | 37.99 | 37.52 | 0.9 | 46.2 | 48 ± 2 | 2.2 | 36.5 | 39 ± 2 | 4.2 | 9.5 | 9 ± 1 | 11.1 |
| PLE 14 | 43.04 | 34.24 | 16.1 | 24.4 | 18.2 ± 0.6 | 15.8 | 17.0 | 11.8 ± 0.5 | 20.1 | 7.4 | 6.4 ± 0.1 | 7.9 |
| PLE 15 | 32.27 | 36.57 | 8.8 | 44.9 | 43.7 ± 0.9 | 1.9 | 34.9 | 34.0 ± 0.7 | 1.9 | 9.9 | 9.7 ± 0.2 | 2.0 |
| PLE 16 | 38.52 | 39.66 | 2.1 | 65.5 | 77 ± 9 | 11.3 | 52.8 | 64 ± 8 | 13.2 | 12.4 | 13 ± 1 | 8.6 |
| PLE 17 | 41.61 | 43.29 | 2.8 | 46.6 | 42.5 ± 0.4 | 6.5 | 36.0 | 32.1 ± 0.7 | 8.1 | 10.7 | 10.4 ± 0.3 | 2.9 |
| PLE 18 | 45.48 | 50.32 | 7.1 | 45.6 | 50 ± 2 | 6.9 | 35.8 | 41 ± 1 | 9.0 | 11.3 | 9.6 ± 0.2 | 8.6 |
| PLE 19 | 54.06 | 70.01 | 18.2 | 19.6 | 22.4 ± 0.3 | 9.3 | 13.7 | 15.1 ± 0.5 | 6.8 | 4.2 | 7.3 ± 0.2 | 23.9 |
| PLE 20 | 58.56 | 61.72 | 3.7 | 17.4 | 23 ± 1 | 13.4 | 12.3 | 15.1 ± 0.5 | 10.1 | 6.4 | 8 ± 1 | 18.4 |
| PLE 21 | 60.02 | 58.34 | 2.0 | 15.7 | 12.9 ± 0.2 | 14.1 | 10.3 | 6.8 ± 0.2 | 22.3 | 7.0 | 6.0 ± 0.1 | 7.6 |
| PLE 22 | 37.16 | 30.22 | 14.6 | 17.9 | 21 ± 1 | 11.8 | 11.7 | 15.1 ± 0.9 | 18.2 | 4.7 | 6.1 ± 0.1 | 11.7 |
| PLE 23 | 62.38 | 50.93 | 14.3 | 47.3 | 45 ± 3 | 4.0 | 37.9 | 36 ± 2 | 2.9 | 10.8 | 8.3 ± 0.8 | 15.5 |
| PLE 24 | 45.59 | 61.69 | 21.2 | 43.4 | 27 ± 3 | 28.0 | 34.4 | 20 ± 2 | 33.1 | 7.0 | 7.1 ± 0.2 | 1.8 |
| PLE 25 | 46.73 | 33.38 | 23.6 | 25.1 | 31.9 ± 0.7 | 11.5 | 18.8 | 24.8 ± 0.8 | 13.0 | 6.0 | 7.15 ± 0.07 | 8.7 |
| Factors | Temperature X1 (°C) | EtOH X2 (%) | Time X3 (min) | Sand—Sample Ratio X4 (w/w) | Theoretical Optimum |
|---|---|---|---|---|---|
| Variable Response | |||||
| Yield | 184.1 | 94.5 | 34.7 | 5.7 | 89.5% |
| TPC | 35.9 | 94.5 | 29.0 | 5.2 | 87.30 mg/g |
| Phenolic acids | 35.9 | 94.1 | 31.5 | 5.7 | 71.02 mg/g |
| Flavonoids | 36.1 | 94.5 | 5.8 | 5.7 | 21.00 mg/g |
| Multiple response | 67.6 | 92.9 | 34.8 | 5.7 | Yield = 66.4% TPC = 78.94 mg/g |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Salas, L.; Borrás-Linares, I.; Quirantes-Piné, R.; Emanuelli, T.; Segura-Carretero, A.; Lozano-Sánchez, J. Enhancing the Production of the Phenolic Extracts of Asparagus Using an Advanced Green Process. Metabolites 2022, 12, 951. https://doi.org/10.3390/metabo12100951
López-Salas L, Borrás-Linares I, Quirantes-Piné R, Emanuelli T, Segura-Carretero A, Lozano-Sánchez J. Enhancing the Production of the Phenolic Extracts of Asparagus Using an Advanced Green Process. Metabolites. 2022; 12(10):951. https://doi.org/10.3390/metabo12100951
Chicago/Turabian StyleLópez-Salas, Lucía, Isabel Borrás-Linares, Rosa Quirantes-Piné, Tatiana Emanuelli, Antonio Segura-Carretero, and Jesús Lozano-Sánchez. 2022. "Enhancing the Production of the Phenolic Extracts of Asparagus Using an Advanced Green Process" Metabolites 12, no. 10: 951. https://doi.org/10.3390/metabo12100951
APA StyleLópez-Salas, L., Borrás-Linares, I., Quirantes-Piné, R., Emanuelli, T., Segura-Carretero, A., & Lozano-Sánchez, J. (2022). Enhancing the Production of the Phenolic Extracts of Asparagus Using an Advanced Green Process. Metabolites, 12(10), 951. https://doi.org/10.3390/metabo12100951










