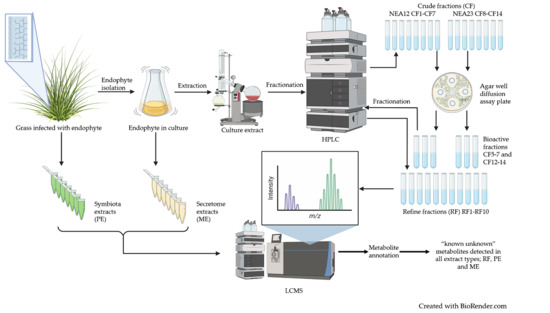
A Metabolomic Study of Epichloë Endophytes for Screening Antifungal Metabolites
Abstract
1. Introduction
2. Results
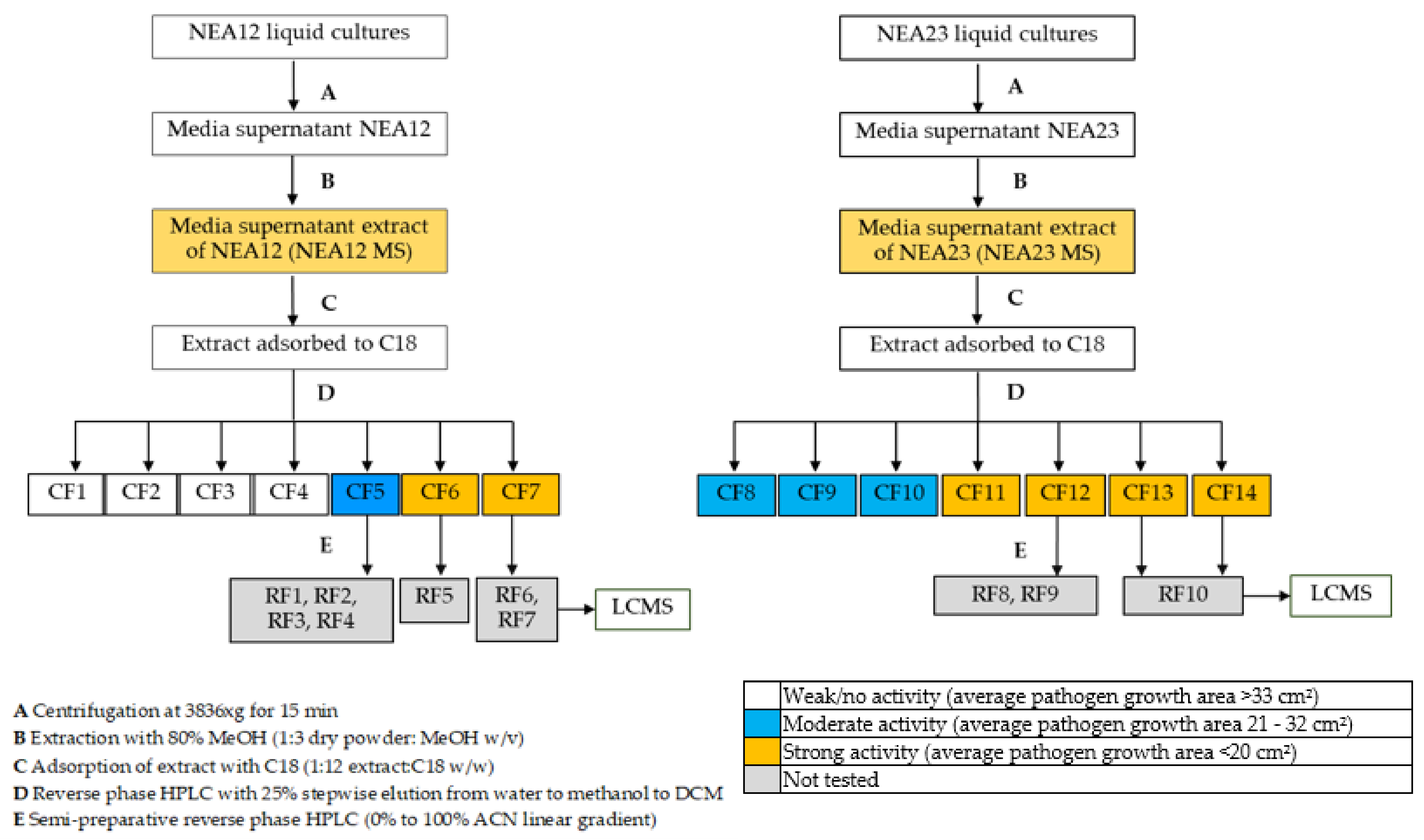
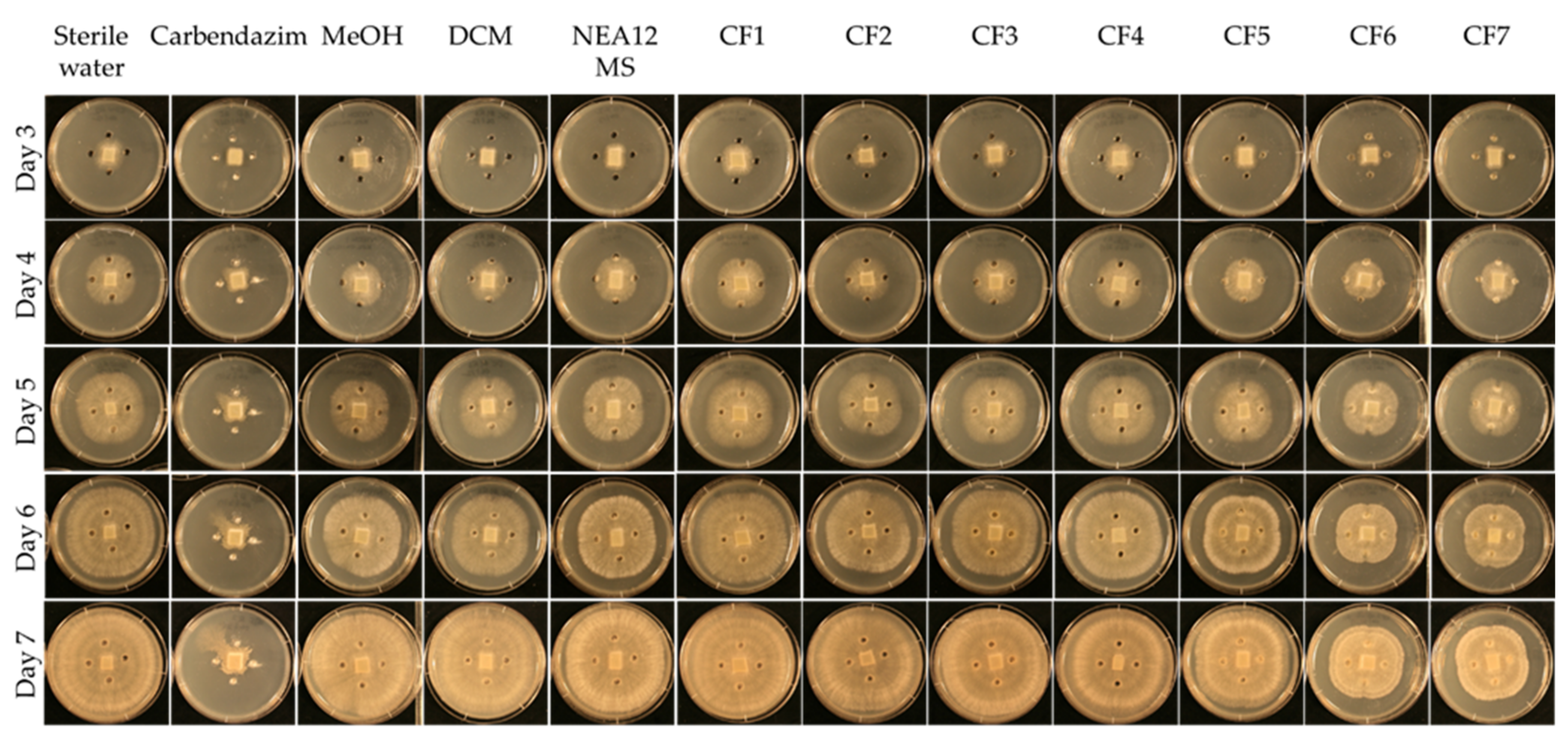
2.1. Extraction of Antifungal Metabolites
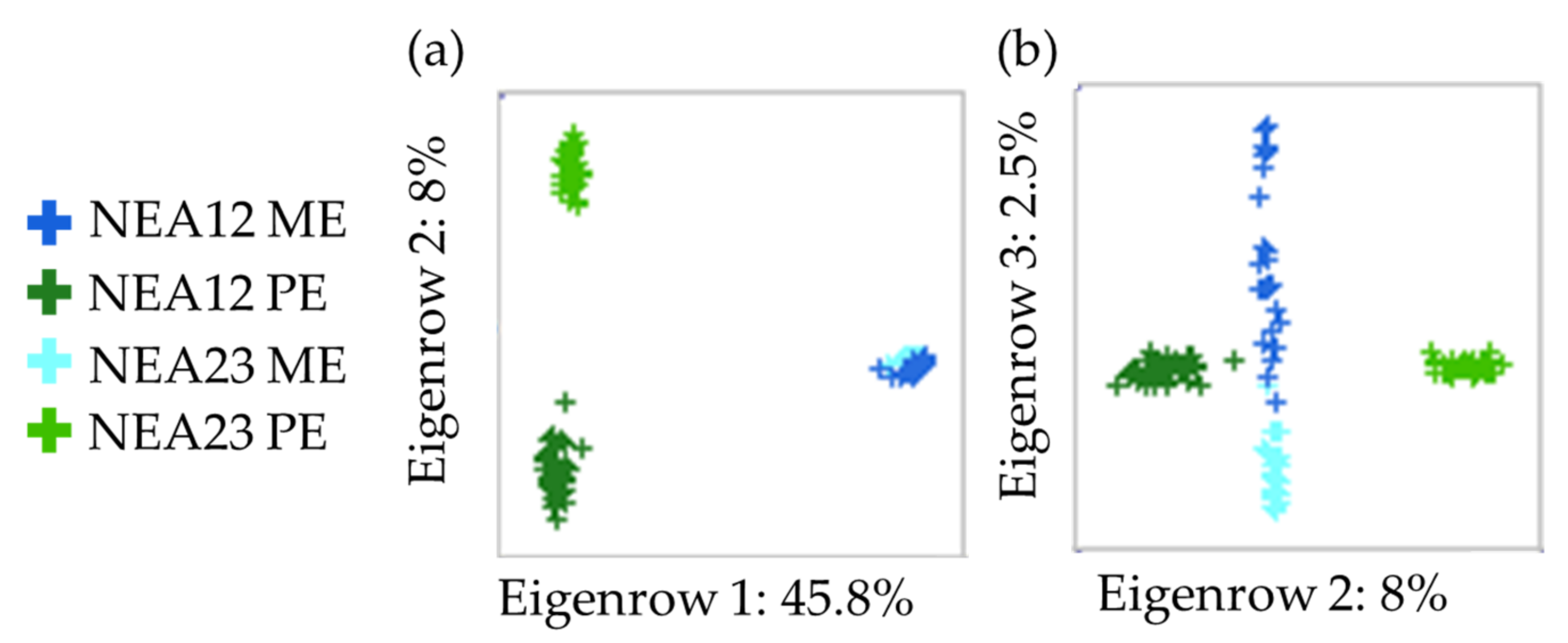
2.2. Metabolite Profiling
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Fungal Cultures
4.2.1. Endophyte Cultures
4.2.2. Pathogen Cultures
4.3. Chemicals
4.4. Extraction and Fractionation of In Vitro Culture
4.4.1. Crude Extracts
4.4.2. Solid Phase Fractionation of Crude Extracts
4.4.3. Agar Well Diffusion Assay for Crude Fractions
4.4.4. Fractionation Using Semi Preparative High-Performance Liquid Chromatography
4.5. Media Supernatant Extraction of Endophyte Cultures
4.5.1. Preparation of Liquid Cultures
4.5.2. Extraction
4.6. Plant Material Extraction
4.7. LCMS Data Acquisition and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilkins, P. Breeding perennial ryegrass for agriculture. Euphytica 1991, 52, 201–214. [Google Scholar] [CrossRef]
- Waller, R.A.; Sale, P.W.G. Persistence and productivity of perennial ryegrass in sheep pastures in south-western Victoria: A review. Aus. J. Experi. Agricul. 2001, 41, 117–144. [Google Scholar] [CrossRef]
- Vargas, J.M. Management of Turfgrass Diseases; CRC Press: New York, NY, USA, 2018. [Google Scholar]
- Dairy Australia. Forage Value Index. Available online: https://www.dairyaustralia.com.au/feed-and-nutrition/growing-feed-for-the-herd/growing-pastures/forage-value-index#.YXXbevlBxPZ (accessed on 25 October 2021).
- Cogan, N.O.I.; Smith, K.F.; Yamada, T.; Francki, M.G.; Vecchies, A.C.; Jones, E.S.; Spangenberg, G.C.; Forster, J.W. QTL analysis and comparative genomics of herbage quality traits in perennial ryegrass (Lolium perenne L.). Theore. App. Gen. 2005, 110, 364–380. [Google Scholar] [CrossRef][Green Version]
- Pearson, A.; Cogan, N.O.; Baillie, R.C.; Hand, M.L.; Bandaranayake, C.K.; Erb, S.; Wang, J.; Kearney, G.A.; Gendall, A.R.; Smith, K.F. Identification of QTLs for morphological traits influencing waterlogging tolerance in perennial ryegrass (Lolium perenne L.). Theore. App. Gen. 2011, 122, 609–622. [Google Scholar] [CrossRef]
- Yamada, T.; Jones, E.; Cogan, N.; Vecchies, A.; Nomura, T.; Hisano, H.; Shimamoto, Y.; Smith, K.; Hayward, M.; Forster, J. QTL analysis of morphological, developmental, and winter hardiness-associated traits in perennial ryegrass. Crop Sci. 2004, 44, 925–935. [Google Scholar]
- Leuchtmann, A.; Bacon, C.W.; Schardl, C.L.; White, J.F., Jr.; Tadych, M. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 2014, 106, 202–215. [Google Scholar] [CrossRef]
- Schardl, C.L. The Epichloë, symbionts of the grass subfamily Poöideae. Ann. Miss. Bot. Gard. 2010, 97, 646–665. [Google Scholar] [CrossRef]
- Yue, Q.; Miller, C.J.; White, J.F.; Richardson, M.D. Isolation and characterization of fungal inhibitors from Epichloë festucae. J. Agri. Food Chem. 2000, 48, 4687–4692. [Google Scholar] [CrossRef]
- Schardl, C.L.; Balestrini, R.; Florea, S.; Zhang, D.; Scott, B. Epichloë endophytes: Clavicipitaceous symbionts of grasses. In Plant Relationships; Springer: Berlin/Heidelberg, Germany, 2009; pp. 275–306. [Google Scholar]
- Vázquez-de-Aldana, B.R.; Romo, M.; García-Ciudad, A.; Petisco, C.; García-Criado, B. Infection with the fungal endophyte Epichloë festucae may alter the allelopathic potential of red fescue. Ann. App. Biol. 2011, 159, 281–290. [Google Scholar] [CrossRef]
- Karpyn Esqueda, M.; Yen, A.L.; Rochfort, S.; Guthridge, K.M.; Powell, K.S.; Edwards, J.; Spangenberg, G.C. A review of perennial ryegrass endophytes and their potential use in the management of African black beetle in perennial grazing systems in Australia. Front Plant Sci. 2017, 8, 3. [Google Scholar] [CrossRef]
- Xia, C.; Li, N.; Zhang, Y.; Li, C.; Zhang, X.; Nan, Z. Role of Epichloë endophytes in defense responses of cool-season grasses to pathogens: A review. Plant Dis. 2018, 102, 2061–2073. [Google Scholar] [CrossRef]
- Singh, L.P.; Gill, S.S.; Tuteja, N. Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal. Behav. 2011, 6, 175–191. [Google Scholar] [CrossRef]
- Ruppert, K.G.; Matthew, C.; McKenzie, C.M.; Popay, A.J. Impact of Epichloë endophytes on adult Argentine stem weevil damage to perennial ryegrass seedlings. Entomol. Exp. Appl. 2017, 163, 328–337. [Google Scholar] [CrossRef]
- Reddy, P.; Hemsworth, J.; Guthridge, K.M.; Vinh, A.; Vassiliadis, S.; Ezernieks, V.; Spangenberg, G.C.; Rochfort, S.J. Ergot alkaloid mycotoxins: Physiological effects, metabolism and distribution of the residual toxin in mice. Sci. Rep. 2020, 10, 9714. [Google Scholar] [CrossRef]
- Siegel, M.R.; Latch, G.C. Expression of antifungal activity in agar culture by isolates of grass endophytes. Mycologia 1991, 83, 529–537. [Google Scholar] [CrossRef]
- Gallagher, R.; Campbell, A.; Hawkes, A.; Holland, P.; McGaveston, D.; Pansier, E.; Harvey, I. Ryegrass staggers: The presence of lolitrem neurotoxins in perennial ryegrass seed. N. Z. Vet. J. 1982, 30, 183–184. [Google Scholar] [CrossRef]
- Tor-Agbidye, J. Correlation of endophyte toxins (ergovaline and lolitrem B) with clinical disease: Fescue foot and perennial ryegrass staggers. Vet. Hum. Tox. 1993, 43, 140–146. [Google Scholar]
- Leuchtmann, A.; Schmidt, D.; Bush, L.P. Different levels of protective alkaloids in grasses with stroma-forming and seed-transmitted Epichloë/Neotyphodium endophytes. J. Chem. Ecol. 2000, 26, 1025–1036. [Google Scholar] [CrossRef]
- Guerre, P. Ergot alkaloids produced by endophytic fungi of the genus Epichloë. Toxins 2015, 7, 773–790. [Google Scholar] [CrossRef]
- Young, C.A.; Felitti, S.; Shields, K.; Spangenberg, G.; Johnson, R.D.; Bryan, G.T.; Saikia, S.; Scott, B. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fung. Genet. Biol. 2006, 43, 679–693. [Google Scholar] [CrossRef]
- Young, C.A.; Bryant, M.K.; Christensen, M.J.; Tapper, B.A.; Bryan, G.T.; Scott, B. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genom. 2005, 274, 13–29. [Google Scholar] [CrossRef]
- Saikia, S.; Takemoto, D.; Tapper, B.A.; Lane, G.A.; Fraser, K.; Scott, B. Functional analysis of an indole-diterpene gene cluster for lolitrem B biosynthesis in the grass endosymbiont Epichloë festucae. FEBS Lett. 2012, 586, 2563–2569. [Google Scholar] [CrossRef]
- Ludlow, E.J.; Vassiliadis, S.; Ekanayake, P.N.; Hettiarachchige, I.K.; Reddy, P.; Sawbridge, T.I.; Rochfort, S.J.; Spangenberg, G.C.; Guthridge, K.M. Analysis of the indole diterpene gene cluster for biosynthesis of the epoxy-janthitrems in Epichloë endophytes. Microorganisms 2019, 7, 560. [Google Scholar] [CrossRef]
- Fleetwood, D.J.; Scott, B.; Lane, G.A.; Tanaka, A.; Johnson, R.D. A complex ergovaline gene cluster in Epichloë endophytes of grasses. App. Environ. Microb. 2007, 73, 2571–2579. [Google Scholar] [CrossRef]
- Tanaka, A.; Tapper, B.A.; Popay, A.; Parker, E.J.; Scott, B. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 2005, 57, 1036–1050. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef]
- Young, C.A.; Tapper, B.A.; May, K.; Moon, C.D.; Schardl, C.L.; Scott, B. Indole-diterpene biosynthetic capability of Epichloë endophytes as predicted by ltm gene analysis. App. Enviro. Microb. 2009, 75, 2200–2211. [Google Scholar] [CrossRef]
- Reddy, P.; Guthridge, K.; Vassiliadis, S.; Hemsworth, J.; Hettiarachchige, I.; Spangenberg, G.; Rochfort, S. Tremorgenic mycotoxins: Structure diversity and biological activity. Toxins 2019, 11, 302. [Google Scholar] [CrossRef]
- Gallagher, R.T.; Hawkes, A.D.; Steyn, P.S.; Vleggaar, R. Tremorgenic neurotoxins from perennial ryegrass causing ryegrass staggers disorder of livestock: Structure elucidation of lolitrem B. J. Chem. Soc. Chem. Comm. 1984, 614–616. [Google Scholar] [CrossRef]
- Paterson, J.; Forcherio, C.; Larson, B.; Samford, M.; Kerley, M. The effects of fescue toxicosis on beef cattle productivity. J. Anim. Sci. 1995, 73, 889–898. [Google Scholar] [CrossRef]
- Rowan, D.D. Lolitrems, peramine and paxilline: Mycotoxins of the ryegrass/endophyte interaction. Agric. Ecosyst. Environ. 1993, 44, 103–122. [Google Scholar] [CrossRef]
- Koulman, A.; Lane, G.A.; Christensen, M.J.; Fraser, K.; Tapper, B.A. Peramine and other fungal alkaloids are exuded in the guttation fluid of endophyte-infected grasses. Phytochemistry 2007, 68, 355–360. [Google Scholar] [CrossRef]
- Christensen, M. Antifungal activity in grasses infected with Acremonium and Epichloë endophytes. Australas. Pl. Pathol. 1996, 25, 186–191. [Google Scholar] [CrossRef]
- Holzmann-Wirth, A.; Dapprich, P.; Eierdanz, S.; Heerz, D.; Paul, V. Anti-fungal substances extracted from Neotyphodium endophytes. In Proceedings of the 3rd International Conference on Harmful and Beneficial Microorganisms in Grassland, Pasture and Turf, Paderborn, Germany, 26–29 September 2000; pp. 65–69. [Google Scholar]
- Koshino, H.; Yoshihara, T.; Okuno, M.; Sakamura, S.; Tajimi, A.; Shimanuki, T. Gamahonolides A, B, and gamahorin, novel antifungal compounds from stromata of Epichloë typhina on Phleum pratense. Biosci. Biotechnol. Biochem. 1992, 56, 1096–1099. [Google Scholar] [CrossRef]
- Steinebrunner, F.; Schiestl, F.P.; Leuchtmann, A. Ecological role of volatiles produced by Epichloë: Differences in antifungal toxicity. FEMS Microbiol. Ecol. 2008, 64, 307–316. [Google Scholar] [CrossRef]
- Mousa, W.K.; Raizada, M.N. The diversity of anti-microbial secondary metabolites produced by fungal endophytes: An interdisciplinary perspective. Front. Microbiol. 2013, 4, 65. [Google Scholar] [CrossRef]
- Song, Q.Y.; Nan, Z.B.; Gao, K.; Song, H.; Tian, P.; Zhang, X.X.; Li, C.J.; Xu, W.B.; Li, X.Z. Antifungal, phytotoxic, and cytotoxic activities of metabolites from Epichloë bromicola, a fungus obtained from Elymus tangutorum grass. J. Agric. Food Chem. 2015, 63, 8787–8792. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, X.; Li, C.; Christensen, M.J.; Nan, Z. Antifungal activity and phytochemical investigation of the asexual endophyte of Epichloë sp. from Festuca sinensis. Sci. China Life Sci. 2015, 58, 821–826. [Google Scholar] [CrossRef]
- Niones, J.T.; Takemoto, D. Viba, a homologue of a transcription factor for fungal heterokaryon incompatibility, is involved in antifungal compound production in the plant-symbiotic fungus Epichloë festucae. Eukaryotic Cell 2015, 14, 13–24. [Google Scholar] [CrossRef]
- Tian, Z.; Wang, R.; Ambrose, K.V.; Clarke, B.B.; Belanger, F.C. The Epichloë festucae antifungal protein has activity against the plant pathogen Sclerotinia homoeocarpa, the causal agent of dollar spot disease. Sci. Rep. 2017, 7, 5643. [Google Scholar] [CrossRef]
- Fernando, K.; Reddy, P.; Hettiarachchige, I.K.; Spangenberg, G.C.; Rochfort, S.J.; Guthridge, K.M. Novel antifungal activity of Lolium-associated Epichloë endophytes. Microorganisms 2020, 8, 955. [Google Scholar] [CrossRef]
- Tian, P.; Nan, Z.; Li, C.; Spangenberg, G. Effect of the endophyte Neotyphodium lolii on susceptibility and host physiological response of perennial ryegrass to fungal pathogens. Eur. J. Plant Pathol. 2008, 122, 593–602. [Google Scholar] [CrossRef]
- Clarke, B.B.; White, J.F.; Hurley, R.H.; Torres, M.S.; Sun, S.; Huff, D.R. Endophyte-mediated suppression of dollar spot disease in fine fescues. Plant Dis. 2006, 90, 994–998. [Google Scholar] [CrossRef]
- Li, F.; Guo, Y.e.; Christensen, M.J.; Gao, P.; Li, Y.; Duan, T. An arbuscular mycorrhizal fungus and Epichloë festucae var. lolii reduce Bipolaris sorokiniana disease incidence and improve perennial ryegrass growth. Mycorrhiza 2018, 28, 159–169. [Google Scholar] [CrossRef]
- Wiewióra, B.; Żurek, G.; Żurek, M. Endophyte-mediated disease resistance in wild populations of perennial ryegrass (Lolium perenne). Fungal Ecol. 2015, 15, 1–8. [Google Scholar] [CrossRef]
- Welty, R.; Barker, R.; Azevedo, M. Response of field-grown tall fescue infected by Acremonium coenophialum to Puccinia graminis subsp. Graminicola. Plant Dis. 1993, 77, 574–575. [Google Scholar] [CrossRef]
- Pańka, D.; Jeske, M.; Troczyński, M. Effect of Neotyphodium uncinatum endophyte on meadow fescue yielding, health status and ergovaline production in host-plants. J. Plant Prot. Res. 2011, 51, 362–370. [Google Scholar] [CrossRef]
- Clarke, R.; Eagling, D. Effects of pathogens on perennial pasture grasses. N. Z. J. Agric. Res. 1994, 37, 319–327. [Google Scholar] [CrossRef][Green Version]
- Gwinn, K.; Gavin, A. Relationship between endophyte infestation level of tall fescue seed lots and Rhizoctonia zeae seedling disease. Plant Dis. 1992, 76, 911–914. [Google Scholar] [CrossRef]
- Burpee, L.L.; Bouton, J.H. Effect of eradication of the endophyte Acremonium coenophialum on epidemics of Rhizoctonia blight in tall fescue. Plant Dis. 1993, 77, 157–159. [Google Scholar] [CrossRef]
- Bonos, S.A.; Wilson, M.M.; Meyer, W.A.; Reed Funk, C. Suppression of red thread in fine fescues through endophyte-mediated resistance. Appl. Turfgrass Sci. 2005, 2, 1–7. [Google Scholar] [CrossRef]
- Vassiliadis, S.; Elkins, A.C.; Reddy, P.; Guthridge, K.M.; Spangenberg, G.C.; Rochfort, S.J. A simple LC–MS method for the quantitation of alkaloids in endophyte-infected perennial ryegrass. Toxins 2019, 11, 649. [Google Scholar] [CrossRef]
- Fernando, K.; Reddy, P.; Vassiliadis, S.; Spangenberg, G.C.; Rochfort, S.J.; Guthridge, K.M. The known antimammalian and insecticidal alkaloids are not responsible for the antifungal activity of Epichloë endophytes. Plants 2021, 10, 2486. [Google Scholar] [CrossRef]
- Malviya, N.; Malviya, S. Bioassay guided fractionation-an emerging technique influence the isolation, identification and characterization of lead phytomolecules. Int. J. Hosp. Pharm. 2017, 2. [Google Scholar] [CrossRef][Green Version]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Stromstedt, A.A.; Felth, J.; Bohlin, L. Bioassays in natural product research–strategies and methods in the search for anti-inflammatory and antimicrobial activity. Phytochem. Anal. 2014, 25, 13–28. [Google Scholar] [CrossRef]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef]
- Christensen, M.J.; Latch, G.C.M.; Tapper, B.A. Variation within isolates of Acremonium endophytes from perennial ryegrasses. Mycol. Res. 1991, 95, 918–923. [Google Scholar] [CrossRef]
- Hiroyuki, K.; Satoshi, T.; Shun-ichi, T.; Yoshihara, T.; Sakamura, S.; Shimanuki, T.; Sato, T.; Tajimi, A. New fungitoxic sesquiterpenoids, chokols A-G, from stromata of Epichloë typhina and the absolute configuration of Chokol E. Agric. Biol. Chem. 1989, 53, 789–796. [Google Scholar] [CrossRef]
- Koshino, H.; Togiya, S.; Yoshihara, T.; Sakamura, S.; Shimanuki, T.; Sato, T.; Tajimi, A. Four fungitoxic C-18 hydroxy unsaturated fatty acids from stromata of Epichloë typhina. Tetrahedron Lett. 1987, 28, 73–76. [Google Scholar] [CrossRef]
- Yoshihara, T.; Togiya, S.; Koshino, H.; Sakamura, S.; Shimanuki, T.; Sato, T.; Tajimi, A. Three fungitoxic cyclopentanoid sesquiterpenes from stromata of Epichloë typhina. Tetrahedron Lett. 1985, 26, 5551–5554. [Google Scholar] [CrossRef]
- Niones, J.T.; Takemoto, D. An isolate of Epichloë festucae, an endophytic fungus of temperate grasses, has growth inhibitory activity against selected grass pathogens. J. Gen. Plant Pathol. 2014, 80, 337–347. [Google Scholar] [CrossRef]
- Purev, E.; Kondo, T.; Takemoto, D.; Niones, J.T.; Ojika, M. Identification of ε-poly-l-lysine as an antimicrobial product from an Epichloë endophyte and isolation of fungal ε-PL synthetase gene. Molecules 2020, 25, 1032. [Google Scholar] [CrossRef]
- Saravana Kumar, P.; Yuvaraj, P.; Gabrial Paulraj, M.; Ignacimuthu, S.; Abdullah Al-Dhabi, N. Bio-prospecting of soil Streptomyces and its bioassay-guided isolation of microbial derived auxin with antifungal properties. J. Mycol. Med. 2018, 28, 462–468. [Google Scholar] [CrossRef]
- Nothias, L.-F.L.; Nothias-Esposito, M.L.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef]
- Weller, M.G. A unifying review of bioassay-guided fractionation, effect-directed analysis and related techniques. Sensors 2012, 12, 9181–9209. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Da Silva Lima, G.; da Rocha, A.M.; dos Santos, G.F.; D’Silva, A.F.; Marriel, I.E.; Takahashi, J.A. Metabolic response of Aspergillus sydowii to OSMAC modulation produces acetylcholinesterase inhibitors. Phytochem. Lett. 2018, 24, 39–45. [Google Scholar] [CrossRef]
- Hewage, R.T.; Aree, T.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. One strain-many compounds (OSMAC) method for production of polyketides, azaphilones, and an isochromanone using the endophytic fungus Dothideomycete sp. Phytochemistry 2014, 108, 87–94. [Google Scholar] [CrossRef]
- Bayram, Ö.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.-J.; Keller, N.P.; Yu, J.-H. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Nielsen, K.F.; Holm, G.; Uttrup, L.; Nielsen, P. Mould growth on building materials under low water activities. Influence of humidity and temperature on fungal growth and secondary metabolism. Internat. Biodeter. Biodegrad. 2004, 54, 325–336. [Google Scholar] [CrossRef]
- Cao, M.; Fraser, K.; Jones, C.; Stewart, A.; Lyons, T.; Faville, M.; Barrett, B. Untargeted Metabotyping Lolium perenne Reveals Population-Level Variation in Plant Flavonoids and Alkaloids. Front. Plant Sci. 2017, 8, 133. [Google Scholar] [CrossRef]
- Rasmussen, S.; Parsons, A.J.; Fraser, K.; Xue, H.; Newman, J.A. Metabolic profiles of Lolium perenne are differentially affected by nitrogen supply, carbohydrate content, and fungal endophyte infection. Plant Physiol. 2008, 146, 1440–1453. [Google Scholar] [CrossRef]
- Rasmussen, S.; Parsons, A.J.; Newman, J.A. Metabolomics analysis of the Lolium perenne–Neotyphodium lolii symbiosis: More than just alkaloids? Phytochem. Rev. 2009, 8, 535–550. [Google Scholar] [CrossRef]
- Rasmussen, S.; Parsons, A.J.; Popay, A.; Xue, H.; Newman, J.A. Plant-endophyte-herbivore interactions: More than just alkaloids? Plant Signal. Behav. 2008, 3, 974–977. [Google Scholar] [CrossRef]
- Venugopalan, A.; Srivastava, S. Endophytes as in vitro production platforms of high value plant secondary metabolites. Biotechnol. Adv. 2015, 33, 873–887. [Google Scholar] [CrossRef]
- Ekanayake, P.N.; Kaur, J.; Tian, P.; Rochfort, S.J.; Guthridge, K.M.; Sawbridge, T.I.; Spangenberg, G.C.; Forster, J.W. Genomic and metabolic characterisation of alkaloid biosynthesis by asexual Epichloë fungal endophytes of tall fescue pasture grasses. Genome 2017, 60, 496–509. [Google Scholar] [CrossRef]
- Ekanayake, P.; Rabinovich, M.; Guthridge, K.; Spangenberg, G.; Forster, J. Phylogenomics of fescue grass-derived fungal endophytes based on selected nuclear genes and the mitochondrial gene complement. BMC Evol. Biol. 2013, 13, 270. [Google Scholar] [CrossRef]
- Ekanayake, P.N.; Hand, M.L.; Spangenberg, G.C.; Forster, J.W.; Guthridge, K.M. Genetic diversity and host specificity of fungal endophyte taxa in Fescue pasture grasses. Crop Sci. 2012, 52, 2243. [Google Scholar] [CrossRef]
- Kaur, J.; Ekanayake, P.N.; Tian, P.; van Zijll de Jong, E.; Dobrowolski, M.P.; Rochfort, S.J.; Mann, R.C.; Smith, K.F.; Forster, J.W.; Guthridge, K.M.; et al. Discovery and characterisation of novel asexual Epichloë endophytes from perennial ryegrass (Lolium perenne L.). Crop Past. Sci. 2015, 66, 1058. [Google Scholar] [CrossRef]
- Hettiarachchige, I.K.; Ekanayake, P.N.; Mann, R.C.; Guthridge, K.M.; Sawbridge, T.I.; Spangenberg, G.C.; Forster, J.W. Phylogenomics of asexual Epichloë fungal endophytes forming associations with perennial ryegrass. BMC Evol. Biol. 2015, 15, 72. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Fenollosa, E.; Salguero-Gómez, R.; Munné-Bosch, S. What is the minimal optimal sample size for plant ecophysiological studies? Plant Physiol. 2018, 178, 953–955. [Google Scholar] [CrossRef]
- Guthridge, K.; Dupal, M.; Kölliker, R.; Jones, E.; Smith, K.; Forster, J. AFLP analysis of genetic diversity within and between populations of perennial ryegrass (Lolium perenne L.). Euphytica 2001, 122, 191–201. [Google Scholar] [CrossRef]
- De Oliveira, G.P.; de Almeida Martins, B.; Lima, M.T.N.S.; Takahashi, J.A. Modulation of Fungal Metabolome by Biotic Stress. In Advancing Frontiers in Mycology & Mycotechnology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 599–626. [Google Scholar]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef]
- Wishart, D.S. Computational strategies for metabolite identification in metabolomics. Bioanalysis 2009, 1, 1579–1596. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]



| Endophyte Strain | Source | DF 1 | Seq SS 2 | Contribution | Adj SS 3 | Adj MS 4 | F-Value | p-Value |
|---|---|---|---|---|---|---|---|---|
| Factor | 11 | 7054.1 | 96.63% | 7054.1 | 641.282 | 122.33 | <0.001 | |
| NEA12 | Error | 47 | 246.4 | 3.37% | 246.4 | 5.242 | ||
| Total | 58 | 7300.5 | 100.00% | |||||
| Factor | 11 | 7948.0 | 97.47% | 7948.0 | 722.550 | 168.00 | <0.001 | |
| NEA23 | Error | 48 | 206.4 | 2.53% | 206.4 | 4.301 | ||
| Total | 59 | 8154.5 | 100.00% |
| Metabolite No. | Mass | m/z | RT | Presence/Absence in Refined Fractions | Presence in Planta (Number of Symbiota) 1 | Q-Value 2 | Directed Effect Size (NEA23 PE: NEA12 PE) | ||
|---|---|---|---|---|---|---|---|---|---|
| NEA23 RFs | NEA12 RFs | NEA23 PE | NEA12 PE | ||||||
| M1 | 242.0412 | 243.0485 | 2.91 | + | − | 9 | 23 | 0.189 | NA |
| M2 | 255.2558 | 256.2631 | 11.19 | + | − | 30 | 47 | 0.004 | 1.59 |
| M3 | 364.1280 | 365.1353 | 9.14 | + | + | 30 | 48 | 0.891 | NA |
| M4 | 365.3625 | 366.3697 | 12.95 | + | − | 30 | 48 | <0.001 | 1.47 |
| M5 | 365.3652 | 366.3725 | 12.84 | + | − | 30 | 48 | <0.001 | 1:3.92 |
| M6 | 365.3682 | 366.3755 | 12.95 | + | − | 30 | 48 | <0.001 | 1.57 |
| M7 | 367.4173 | 368.4245 | 13.50 | + | − | 30 | 48 | <0.001 | 1:1.68 |
| M8 | 407.3153 | 408.3226 | 8.36 | − | + | 0 | 9 | NA | NA |
| M9 | 426.2376 | 427.2449 | 10.80 | + | − | 25 | 40 | 0.191 | NA |
| M10 | 426.2375 | 427.2448 | 10.90 | + | − | 30 | 45 | 0.749 | NA |
| M11 | 452.3370 | 453.3442 | 8.61 | − | + | 3 | 4 | 0.316 | NA |
| M12 | 452.3344 | 453.3417 | 8.70 | − | + | 3 | 3 | 0.661 | NA |
| M13 | 452.3341 | 453.3414 | 8.72 | − | + | 0 | 2 | NA | NA |
| M14 | 457.3394 | 458.3467 | 9.92 | + | − | 29 | 48 | 0.919 | NA |
| M15 | 462.2965 | 463.3038 | 9.83 | + | − | 30 | 48 | 0.792 | NA |
| M16 | 466.3496 | 467.3569 | 9.25 | − | + | 30 | 47 | 0.248 | NA |
| M17 | 483.3763 | 484.3835 | 9.24 | + | + | 4 | 47 | 0.363 | NA |
| M18 | 496.3602 | 497.3674 | 8.71 | + | + | 15 | 39 | 0.004 | 1:7.06 |
| M19 | 527.4024 | 528.4096 | 9.24 | + | + | 20 | 43 | <0.001 | 2.49 |
| M20 | 532.3574 | 533.3647 | 9.24 | + | + | 8 | 48 | 0.772 | NA |
| M21 | 546.3005 | 547.3077 | 11.85 | + | − | 17 | 9 | 0.024 | 1:2.39 |
| M22 | 571.4285 | 572.4358 | 9.24 | + | + | 5 | 24 | 0.945 | NA |
| M23 | 576.3837 | 577.3909 | 9.23 | + | + | 3 | 38 | <0.001 | 3.93 |
| M24 | 589.4180 | 590.4253 | 9.89 | + | − | 30 | 40 | <0.001 | 5.12 |
| M25 | 590.4236 | 591.4308 | 11.79 | + | − | 30 | 48 | <0.001 | 1:2.20 |
| M26 | 594.3594 | 595.3666 | 9.82 | + | − | 30 | 48 | <0.001 | 1.31 |
| M27 | 594.3744 | 595.3817 | 9.90 | + | − | 30 | 48 | <0.001 | 1.82 |
| M28 | 607.4645 | 608.4718 | 11.83 | + | − | 3 | 10 | 0.234 | NA |
| M29 | 615.4546 | 616.4618 | 9.24 | + | + | 3 | 17 | <0.001 | 11.86 |
| M30 | 620.4100 | 621.4173 | 9.24 | + | + | 7 | 29 | 0.218 | NA |
| M31 | 634.4644 | 635.4717 | 11.85 | + | − | 4 | 2 | NA | NA |
| M32 | 651.4907 | 652.4980 | 11.85 | + | − | 1 | 12 | NA | NA |
| M33 | 662.4203 | 663.4276 | 9.80 | + | − | 30 | 48 | <0.001 | 1.71 |
| M34 | 662.4171 | 663.4243 | 14.30 | + | + | 30 | 48 | <0.001 | 1:4.53 |
| M35 | 662.4447 | 663.4520 | 14.38 | + | + | 30 | 48 | <0.001 | 1.25 |
| M36 | 663.4473 | 664.4546 | 14.33 | + | + | 30 | 48 | <0.001 | 1:1.71 |
| M37 | 678.3797 | 679.3869 | 11.94 | + | − | 30 | 48 | 0.235 | NA |
| M38 | 678.4871 | 679.4944 | 11.95 | + | − | 27 | 48 | 0.453 | NA |
| M39 | 684.4323 | 685.4395 | 14.23 | − | + | 29 | 48 | <0.001 | 1:9.06 |
| M40 | 684.4297 | 685.4369 | 14.53 | − | + | 30 | 48 | <0.001 | 1.58 |
| M41 | 684.4313 | 685.4385 | 14.58 | − | + | 30 | 48 | <0.001 | 1.64 |
| M42 | 685.4159 | 686.4232 | 14.29 | − | + | 30 | 48 | <0.001 | 1:3.27 |
| M43 | 695.5170 | 696.5243 | 11.87 | + | − | 1 | 9 | NA | NA |
| M44 | 695.5172 | 696.5245 | 11.96 | + | − | 5 | 32 | 0.086 | NA |
| M45 | 721.4687 | 722.4759 | 11.97 | + | − | 24 | 37 | 0.174 | NA |
| M46 | 762.5085 | 763.5158 | 14.59 | − | + | 30 | 48 | 0.177 | NA |
| M47 | 762.5091 | 763.5164 | 14.68 | − | + | 30 | 48 | <0.001 | 1.74 |
| M48 | 762.5095 | 763.5168 | 14.74 | − | + | 30 | 47 | <0.001 | 2.04 |
| M49 | 762.5071 | 763.5144 | 14.38 | + | + | 30 | 48 | <0.001 | 1.23 |
| M50 | 762.5090 | 763.5163 | 14.54 | − | + | 30 | 48 | <0.001 | 1.54 |
| M51 | 763.4952 | 764.5024 | 14.64 | − | + | 30 | 48 | <0.001 | 1.80 |
| M52 | 763.5110 | 764.5182 | 14.42 | + | + | 30 | 48 | <0.001 | 2.48 |
| M53 | 763.5109 | 764.5182 | 14.33 | + | + | 30 | 48 | <0.001 | 1:1.90 |
| M54 | 787.5037 | 788.5109 | 12.10 | + | − | 26 | 39 | <0.001 | 1:1.19 |
| M55 | 788.5186 | 789.5259 | 11.90 | + | − | 30 | 43 | 0.919 | NA |
| M56 | 788.5210 | 789.5283 | 11.94 | + | − | 29 | 40 | 0.419 | NA |
| M57 | 802.5324 | 803.5397 | 12.41 | + | − | 30 | 48 | 0.607 | NA |
| M58 | 802.5319 | 803.5392 | 12.46 | + | − | 30 | 48 | <0.001 | 1.24 |
| M59 | 802.5335 | 803.5408 | 12.26 | + | − | 30 | 48 | <0.001 | 1:1.34 |
| M60 | 802.5342 | 803.5415 | 12.33 | + | − | 30 | 48 | 0.960 | NA |
| M61 | 858.5970 | 859.6043 | 12.59 | + | − | 3 | 6 | <0.001 | 1.18 |
| Crude MS Extract Strain | Crude Fraction | Water (%) | Methanol (%) | DCM (%) | Dry Weight/(g) |
|---|---|---|---|---|---|
| NEA12 MS | CF1 | 100 | 0 | - | 40.69 |
| CF2 | 80 | 20 | - | 10.17 | |
| CF3 | 50 | 50 | - | 4.36 | |
| CF4 | 20 | 80 | - | 22.01 | |
| CF5 | 0 | 100 | 0 | 14.23 | |
| CF6 | - | 50 | 50 | 7.38 | |
| CF7 | - | 0 | 100 | 10.41 | |
| NEA23 MS | CF8 | 100 | 0 | - | 32.87 |
| CF9 | 80 | 20 | - | 8.13 | |
| CF10 | 50 | 50 | - | 7.83 | |
| CF11 | 20 | 80 | - | 7.26 | |
| CF12 | 0 | 100 | 0 | 3.29 | |
| CF13 | - | 50 | 50 | 2.30 | |
| CF14 | - | 0 | 100 | 3.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernando, K.; Reddy, P.; Guthridge, K.M.; Spangenberg, G.C.; Rochfort, S.J. A Metabolomic Study of Epichloë Endophytes for Screening Antifungal Metabolites. Metabolites 2022, 12, 37. https://doi.org/10.3390/metabo12010037
Fernando K, Reddy P, Guthridge KM, Spangenberg GC, Rochfort SJ. A Metabolomic Study of Epichloë Endophytes for Screening Antifungal Metabolites. Metabolites. 2022; 12(1):37. https://doi.org/10.3390/metabo12010037
Chicago/Turabian StyleFernando, Krishni, Priyanka Reddy, Kathryn M. Guthridge, German C. Spangenberg, and Simone J. Rochfort. 2022. "A Metabolomic Study of Epichloë Endophytes for Screening Antifungal Metabolites" Metabolites 12, no. 1: 37. https://doi.org/10.3390/metabo12010037
APA StyleFernando, K., Reddy, P., Guthridge, K. M., Spangenberg, G. C., & Rochfort, S. J. (2022). A Metabolomic Study of Epichloë Endophytes for Screening Antifungal Metabolites. Metabolites, 12(1), 37. https://doi.org/10.3390/metabo12010037