Mining for Active Molecules in Probiotic Supernatant by Combining Non-Targeted Metabolomics and Immunoregulation Testing
Abstract
:1. Introduction
2. Results
2.1. Probiotic Strains and Their Respective Supernatant Immunomodulatory Response
2.2. FT-ICR-MS Analysis and Data Processing Prior Statistics
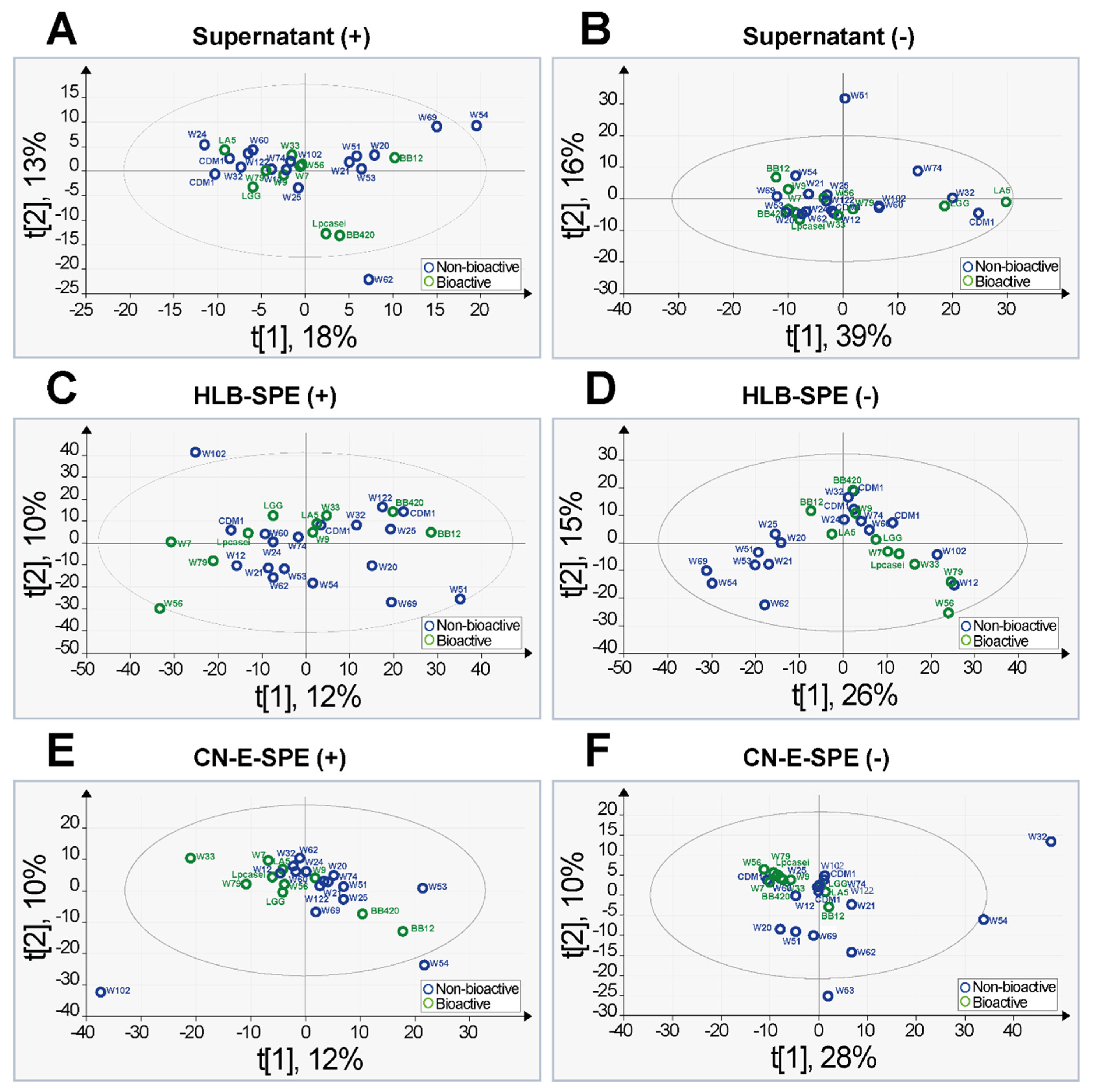
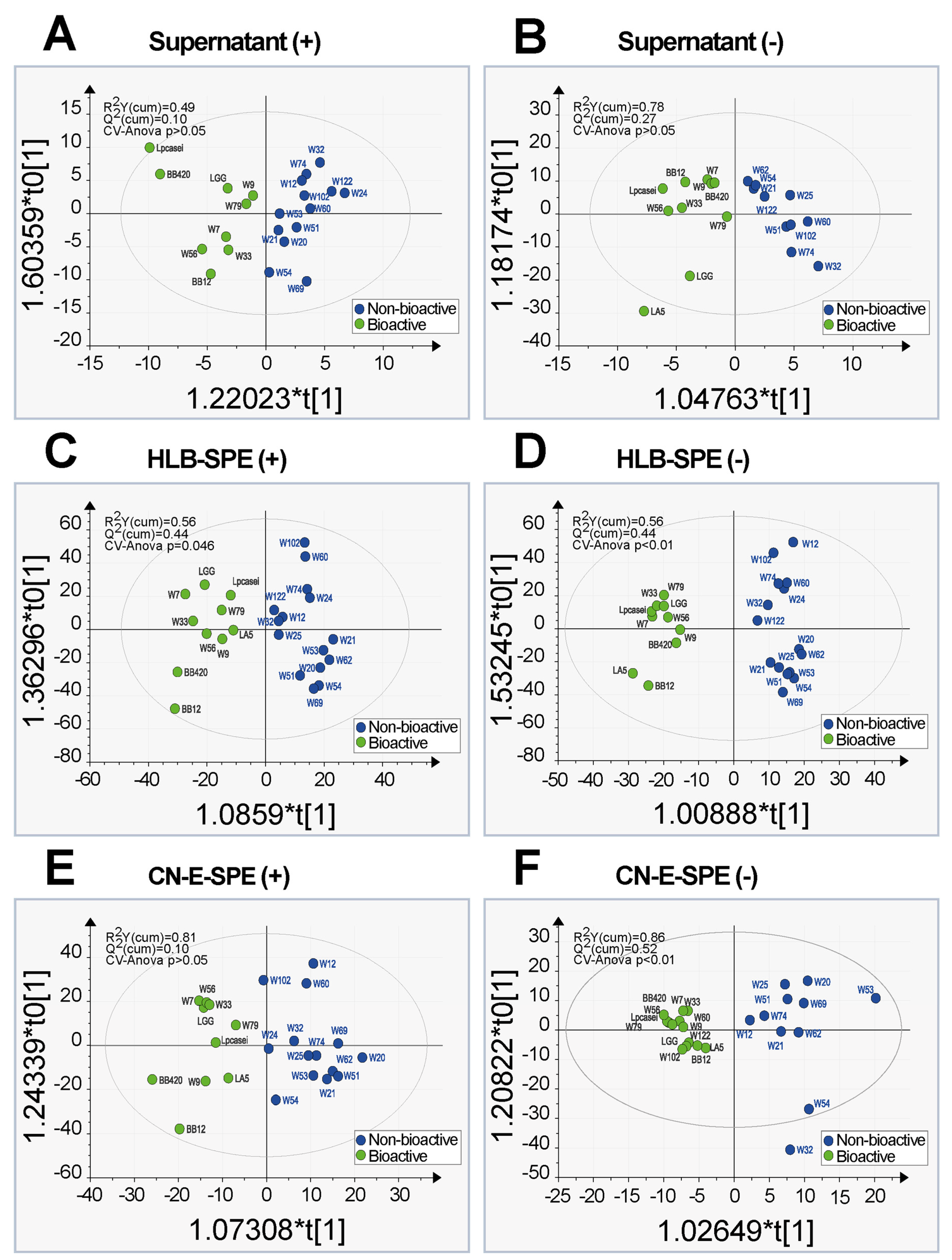
2.3. Principal Component Analysis and Orthogonal Partial Least Square Discriminative Analysis
2.4. Metabolic Pathway Assessment
2.5. Tryptophan Metabolism
3. Discussion
3.1. The Approach to Sample Generation and Untargeted Metabolomics
3.2. Immunomodulatory Supernatants and Bioactive Pathways
3.3. Implications and Limitations of the Study
4. Materials and Methods
4.1. Probiotic Strains Cultivation and Supernatant Collection
4.2. Bioassays for Immunomodulatory Activity in Probiotic Supernatants
4.3. Sample Pre-Treatment Prior to Chemical Analysis
4.4. FT-ICR-MS Chemical Analyses
4.5. Data Processing and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buchen, L. The new germ theory: What can microbiologists who study human bowels learn from those who study the bowels of the earth? Nature 2010, 468, 492–495. [Google Scholar] [CrossRef]
- Strachan, D.P. Hay fever, hygiene, and household size. BMJ 1989, 299, 1259–1260. [Google Scholar] [CrossRef] [Green Version]
- Metchnikoff, E.; Mitchell, P.C. The Prolongation of Life; Optimistic Studies; G.P. Putnam’s Sons: London, UK, 1908. [Google Scholar]
- Podolsky, S.H. Metchnikoff and the microbiome. Lancet 2012, 380, 1810–1811. [Google Scholar] [CrossRef]
- Raghuwanshi, S.; Misra, S.; Sharma, R.; Bisen, P.S. Probiotics: Nutritional therapeutic tool. J. Probiotics Health 2018, 6, 194. [Google Scholar] [CrossRef]
- Holgate, S.T.; Lack, G. Improving the management of atopic disease. Arch. Dis. Child. 2005, 90, 826–831. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Salminen, S.; Arvilommi, H.; Kero, P.; Koskinen, P.; Isolauri, E. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 2001, 357, 1076–1079. [Google Scholar] [CrossRef]
- Isolauri, E.; Arvola, T.; Sütas, Y.; Moilanen, E.; Salminen, S. Probiotics in the management of atopic eczema. Clin. Exp. Allergy 2000, 30, 1604–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeşilova, Y.; Çalka, Ö.; Akdeniz, N.; Berktaş, M. Effect of probiotics on the treatment of children with atopic dermatitis. Ann. Dermatol. 2012, 24, 189–193. [Google Scholar] [CrossRef] [Green Version]
- Niers, L.; Martín, R.; Rijkers, G.; Sengers, F.; Timmerman, H.; Van Uden, N.; Smidt, H.; Kimpen, J.; Hoekstra, M. The effects of selected probiotic strains on the development of eczema (the PandA Study). Allergy 2009, 64, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Osborn, D.A.; Sinn, J.K.H. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst. Rev. 2007, 4, CD006475. [Google Scholar] [CrossRef] [PubMed]
- Michail, S. The role of probiotics in allergic diseases. Allergy Asthma Clin. Immunol. 2009, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. Food and Drug Administration. Guidance for Clinical Investigators, Sponsors, and IRBs: Investigational New Drug Applications (INDs)—Determining Whether Human Research Studies Can Be Conducted without an IND; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2013; p. 10.
- Katan, M.B. Why the European Food Safety Authority was right to reject health claims for probiotics. Benef. Microbes 2012, 3, 85–89. [Google Scholar] [CrossRef]
- Yan, F.; Polk, D.B. Probiotics and immune health. Curr. Opin. Gastroenterol. 2011, 27, 496–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frei, R.; Lauener, R.P.; Crameri, R.; O’Mahony, L. Microbiota and dietary interactions: An update to the hygiene hypothesis? Allergy 2012, 67, 451–461. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elina, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Prince, E.K.; Pohnert, G. Searching for signals in the noise: Metabolomics in chemical ecology. Anal. Bioanal. Chem. 2010, 396, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Bazanella, M.; Maier, T.V.; Clavel, T.; Lagkouvardos, I.; Lucio, M.; Maldonado-Gòmez, M.X.; Autran, C.; Walter, J.; Bode, L.; Schmitt-Kopplin, P.; et al. Randomized controlled trial on the impact of early-life intervention with bifidobacteria on the healthy infant fecal microbiota and metabolome. Am. J. Clin. Nutr. 2017, 106, 1274–1286. [Google Scholar] [CrossRef] [Green Version]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [Green Version]
- Antón, J.; Lucio, M.; Pena, A.; Cifuentes, A.; Brito-Echeverria, J.; Moritz, F.; Tziotis, D.; López, C.; Urdiain, M.; Schmitt-Kopplin, P.; et al. High metabolomic microdiversity within co-occurring isolates of the extremely halophilic bacterium salinibacter ruber. PLoS ONE 2013, 8, e64701. [Google Scholar]
- Kremb, S.; Mueller, C.; Schmitt-Kopplin, P.; Voolstra, C.R. Bioactive potential of marine macroalgae from the Central Red Sea (Saudi Arabia) assessed by high throughput imaging-based phenotypic profiling. Mar. Drugs 2017, 15, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Rourke, A.; Kremb, S.; Bader, T.M.; Helfer, M.; Schmitt-Kopplin, P.; Gerwick, W.H.; Brack-Werner, R.; Voolstra, C.R. Alkaloids from the sponge Stylissa carteri present prospective scaffolds for the inhibition of Human immunodeficiency Virus 1 (HIV-1). Mar. Drugs 2016, 14, 28. [Google Scholar] [CrossRef] [Green Version]
- Mueller, C.; Kremb, S.; Gonsior, M.; Brack-Werner, R.; Voolstra, C.R.; Schmitt-Kopplin, P. Advanced identification of global bioactivity hotspots via screening of the metabolic fingerprint of entire ecosystems. Sci. Rep. 2020, 10, 1319. [Google Scholar] [CrossRef]
- Zhernov, Y.V.; Kremb, S.; Helfer, M.; Schindler, M.; Harir, M.; Mueller, C.; Hertkorn, N.; Avvakumova, N.P.; Konstantinov, A.I.; Brack-Werner, R.; et al. Supramolecular combinations of humic polyanions as potent microbicides with polymodal anti-HIV-activities. New J. Chem. 2016, 41, 212–224. [Google Scholar] [CrossRef] [Green Version]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular Microbial Metabolomics: The State of the Art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Dörries, K.; Lalk, M. Metabolic footprint analysis uncovers strain specific overflow metabolism and d-isoleucine production of staphylococcus aureus COL and HG001. PLoS ONE 2013, 8, e81500. [Google Scholar] [CrossRef] [Green Version]
- Behrends, V.; Ebbels, T.M.D.; Williams, H.D.; Bundy, J.G. Time-resolved metabolic footprinting for nonlinear modeling of bacterial substrate utilization. Appl. Environ. Microbiol. 2009, 75, 2453–2463. [Google Scholar] [CrossRef] [Green Version]
- Mapelli, V.; Olsson, L.; Nielsen, J. Metabolic footprinting in microbiology: Methods and applications in functional genomics and biotechnology. Trends Biotechnol. 2008, 26, 490–947. [Google Scholar] [CrossRef]
- Kepert, I.; Fonseca, J.; Müller, C.; Milger, K.; Hochwind, K.; Kostric, M.; Fedoseeva, M.; Ohnmacht, C.; Dehmel, S.; Nathan, P.; et al. d-Tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 2017, 139, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Krauss-Etschmann, S.; Hartmann, A.; Schmitt-Kopplin, P.; Schloter, M. Methods and Compositions for Treating Inflammatory Diseases. U.S. Patent 10,857,128, 8 December 2020. [Google Scholar]
- Chubukov, V.; Sauer, U. Environmental Dependence of Stationary-Phase Metabolism in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 2901–2909. [Google Scholar] [CrossRef] [Green Version]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. BioSyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Hair, J.F.; Hult, G.T.M.; Ringle, C.M.; Sarstedt, M. A Primer on Partial Least Squares Structural Equation Modeling (PLS-SEM), 2nd ed.; SAGE Publications Inc.: Los Angeles, CA, USA, 2017; Chapter 6. [Google Scholar]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayma, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Harir, M.; Uhl, J.; Kanawati, B.; Lucio, M.; Smirnov, K.S.; Koch, B.P.; Schmitt-Kopplin, P.; Hertkorn, N. How representative are dissolved organic matter (DOM) extracts? A comprehensive study of sorbent selectivity for DOM isolation. Water Res. 2017, 116, 316–323. [Google Scholar] [CrossRef]
- Jha, B.; Kavita, K.; Westphal, J.; Hartmann, A.; Philippe Schmitt-Kopplin, P. Quorum Sensing Inhibition by Asparagopsis taxiformis, a Marine Macro Alga: Separation of the Compound that Interrupts Bacterial Communication. Mar. Drugs 2013, 11, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Buszewski, B.; Szultka, M. Past, Present, and Future of Solid Phase Extraction: A Review. Crit. Rev. Anal. Chem. 2012, 42, 198–213. [Google Scholar] [CrossRef]
- Gostner, J.M.; Becker, K.; Kofler, H.; Strasser, B.; Fuchs, D. Tryptophan Metabolism in Allergic Disorders. Int. Arch. Allergy Immunol. 2016, 169, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Pallotta, M.; Orabona, C.; Volpi, C.; Vacca, C.; Belladonna, M.L.; Bianchi, R.; Servillo, G.; Brunacci, C.; Calvitti, M.; Bicciato, S.; et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011, 12, 870–878. [Google Scholar] [CrossRef] [Green Version]
- Aldajani, W.A.; Salazar, F.; Sewell, H.F.; Knox, A.; Ghaemmaghami, A.M. Expression and regulation of immune-modulatory enzyme indoleamine 2,3-dioxygenase (IDO) by human airway epithelial cells and its effect on T cell activation. Oncotarget 2016, 7, 57606–57617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Leek, A.P.; Yanishevsky, Y.; Kozyrskyj, A.L. The Kynurenine Pathway as a Novel Link between Allergy and the Gut Microbiome. Front. Immunol. 2017, 8, 1374. [Google Scholar] [CrossRef] [Green Version]
- Mellor, A.L.; Munn, D.H. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004, 4, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Von Bubnoff, D.; Bieber, T. The indoleamine 2,3-dioxygenase (IDO) pathway controls allergy. Allergy 2012, 67, 718–725. [Google Scholar] [CrossRef]
- Von Bubnoff, D.; Wilms, H.; Scheler, M.; Brenk, M.; Koch, S.; Biber, T. Human myeloid dendritic cells are refractory to tryptophan metabolites. Hum. Immunol. 2011, 72, 791–797. [Google Scholar] [CrossRef]
- Belladonna, M.L.; Grohmann, U.; Guidetti, P.; Volpi, C.; Bianchi, R.; Fioretti, M.C.; Schwarcz, R.; Fallarino, F.; Puccetti, P. Kynurenine Pathway Enzymes in Dendritic Cells Initiate Tolerogenesis in the Absence of Functional IDO. J. Immunol. 2006, 177, 130–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manni, G.; Mondanelli, G.; Scalisi, G.; Pallotta, M.T.; Nardi, D.; Padiglioni, E.; Romani, R.; Talesa, V.N.; Puccetti, P.; Fallarino, F.; et al. Pharmacologic Induction of Endotoxin Tolerance in Dendritic Cells by l-Kynurenine. Front. Immunol. 2020, 11, 292. [Google Scholar] [CrossRef]
- Murray, P.J. Amino acid auxotrophy as a system of immunological control nodes. Nat. Immunol. 2016, 17, 132–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondanelli, G.; Bianchi, R.; Pallotta, M.T.; Orabona, C.; Albini, E.; Iacono, A.; Belladonna, M.L.; Vacca, C.; Fallarino, F.; Macchiarulo, A.; et al. A Relay Pathway between Arginine and Tryptophan Metabolism Confers Immunosuppressive Properties on Dendritic Cells. Immunity 2017, 46, 233–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agus, A.; Planchais, J.; Soko, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neavin, D.R.; Liu, D.; Ray, B.; Weinshilboum, R.M. The Role of the Aryl Hydrocarbon Receptor (AHR) in Immune and Inflammatory Diseases. Int. J. Mol. Sci. 2018, 19, 3851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.T.; Kimura, A.; Nakahama, T.; Chinen, I.; Masuda, K.; Nohara, K.; Fujii-Kuriyama, Y.; Kishimoto, T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc. Natl. Acad. Sci. USA 2010, 107, 19961–19966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasabe, J.; Masataka Suzuki, M. Emerging Role of d-Amino Acid Metabolism in the Innate Defense. Front. Microbiol. 2018, 9, 933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliashkevich, A.; Alvarez, L.; Cava, F. New Insights into the Mechanisms and Biological Roles of d-Amino Acids in Complex Eco-Systems. Front. Microbiol. 2018, 9, 683. [Google Scholar] [CrossRef] [Green Version]
- Radvok, A.D.; Moe, L.A. Bacterial synthesis of d-amino acids. Appl. Microbiol. Biotechnol. 2014, 98, 5363–5374. [Google Scholar]
- Notarangelo, F.M.; Wang, C.-D.; Horning, K.J.; Schwarcz, R. Role of d-amino acid oxidase in the production of kynurenine pathway metabolites from d-tryptophan in mice. J. Neurochem. 2016, 136, 804–814. [Google Scholar] [CrossRef] [Green Version]
- Sridharan, G.V.; Choi, K.; Klemashevich, C.; Wu, C.; Prabakaran, D.; Pan, L.B.; Steinmeyer, S.; Mueller, C.; Yousofshahi, M.; Alaniz, R.C.; et al. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat. Commun. 2014, 5, 5492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, A.; Sun, B.; Fu, Z.; Yu, D. Roles of aminoacyl-tRNA synthetases in immune regulation and immune diseases. Cell Death Dis. 2019, 10, 901. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B.; Brown, M.; Davey, H.M.; Dunn, W.B.; Spasic, I.; Oliver, S.G. Metabolic footprinting and Systems Biology: The medium is the message. Nat. Rev. Microbiol. 2005, 3, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Suokko, A.; Palva, A.; Varmanen, P. New convenient defined media for [(35)S] methionine labelling and proteomic analyses of probiotic lactobacilli. Lett. Appl. Microbiol. 2006, 42, 202–209. [Google Scholar] [CrossRef]
- Tong, X.S.; Wang, J.; Zheng, S.; Pivnichny, J.V.; Griffin, P.R.; Shen, X.; Donnelly, M.; Vakerich, K.; Nunes, C.; Fenyk-Melody, J. Effect of signal interference from dosing excipients on pharmacokinetic screening of drug candidates by liquid chromatography/mass spectrometry. Anal. Chem. 2002, 74, 6305–6313. [Google Scholar] [CrossRef] [PubMed]
- Lucio, M.; Fekete, A.; Frommberger, M.; Schmitt-Kopplin, P. Metabolomics: High-resolution tools offer to follow bacterial growth on a molecular level. In Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches; De Brujin, F.J., Ed.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2011; Chapter 72. [Google Scholar]
- Green, N.W.; Perdue, E.M. Fast Graphically Inspired Algorithm for Assignment of Molecular Formulae in Ultrahigh Resolution Mass Spectrometry. Anal. Chem. 2015, 87, 5086–5094. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007, 8, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertkorn, N.; Frommberger, M.; Witt, M.; Koch, B.P.; Schimitt-Kopplin, P.; Perdue, E.M. Natural organic matter and the event horizon of mass spectrometry. Anal. Chem. 2008, 80, 8908–8919. [Google Scholar] [CrossRef] [PubMed]


| Bacterial Strain. | Code | Source | Effect on DC a | Effect on KM-H2 b |
|---|---|---|---|---|
| Bifidobacterium animalis subsp.lactis | BB-12 c | Chr. Hansen, Horsholm, Denmark | + | + |
| Bifidobacterium animalis | W53 | Winclove Bioindustries BV, The Netherlands | − | − |
| Bifidobacterium breve | W9 | Winclove Bioindustries BV, The Netherlands | + | + |
| Bifidobacterium breve | W25 | Winclove Bioindustries BV, The Netherlands | − | − |
| Bifidobacterium lactis | BB-420 | Danisco, Niebüll, Germany | + | + |
| Bifidobacterium lactis | W51 | Winclove Bioindustries BV, The Netherlands | − | − |
| Enterococcus faecium | W54 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lactobacillus acidophilus | LA-5 | Chr. Hansen, Horsholm, Denmark | + | + |
| Lactobacillus acidophilus | W12 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lactobacillus acidophilus | W33 | Winclove Bioindustries BV, The Netherlands | + | + |
| Lactobacillus acidophilus | W74 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lacticaseibacillus casei | LC-01 | Chr. Hansen, Horsholm, Denmark | + | + |
| Lacticaseibacillus casei | W20 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lacticaseibacillus casei | W56 | Winclove Bioindustries BV, The Netherlands | + | + |
| Lacticaseibacillus casei | W79 | Winclove Bioindustries BV, The Netherlands | + | + |
| Lactobacillus helveticus | W60 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lacticaseibacillus paracasei | W7 | Winclove Bioindustries BV, The Netherlands | + | + |
| Lactiplantibacillus plantarum | W21 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lactiplantibacillus plantarum | W62 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lacticaseibacillus rhamnosus | LGG | Valio Ltd., Helsinki, Finland | + | + |
| Lacticaseibacillus rhamnosus | W102 | Winclove Bioindustries BV, The Netherlands | − | − |
| Ligilactobacillus salivarius | W24 | Winclove Bioindustries BV, The Netherlands | − | − |
| Lactococcis lactis | W32 | Winclove Bioindustries BV, The Netherlands | − | − |
| Streptococcus salivarius | W122 | Winclove Bioindustries BV, The Netherlands | − | − |
| Streptococcus thermophilus | W69 | Winclove Bioindustries BV, The Netherlands | − | − |
| ESI Mode | Sample Pretreatment Applied | ||
|---|---|---|---|
| Crude Supernatant * | HLB-SPE Extract | CN-E-SPE Extract | |
| Positive ** | 344 | 2658 | 1367 |
| Negative | 1970 | 3932 | 3353 |
| Bioactive Supernatants SPE Extract | Non-Bioactive Supernatants Extract | Metabolite Hit Prediction * | ||
|---|---|---|---|---|
| HLB-SPE/ESI[+] | HLB-SPE/ESI[-] | CN-E-SPE/ESI[-] | HLB-SPE/ESI[-] | |
| C00078 | C00078 | NP | NP | Tryptophan |
| C00331 | C00331 | C00331 | NP | Indolepyruvic acid |
| C00643 | C00643 | C00643 | C00643 | 5-Hydroxy-l-tryptophan |
| C01598 | C01598 | NP | C01598 | Melatonin |
| C00978 | C00978 | NP | NP | N-Acetylserotonin |
| C00780 | C00780 | C00780 | NP | Serotonin |
| C02298 | C02298 | C02298 | C02298 | N-Acetylindoxyl |
| C02700 | C02700 | C02700 | C02700 | l-Formylkynurenine |
| C00328 | C00328 | C00328 | NP | l-Kynurenine |
| C03227 | C03227 | NP | NP | 3-Hydroxy-l-kynurenine |
| C00637 | C00637 | C00637 | C00637 | Indole-3-acetaldehyde |
| C02693 | C02693 | NP | NP | Indole-3-acetamide |
| C00954 | C00954 | C00954 | C00954 | Indole-3-acetic acid |
| C02937 | C02937 | NP | NP | Indole-3-acetaldehyde oxime |
| C03230 | C03230 | C03230 | C03230 | 3-Indoleglycolaldehyde |
| C02043 | C02043 | NP | C02043 | Indolelactate |
| NP | C00955 | NP | C00955 | Indole-3-ethanol |
| C02470 | Xanthurenic acid | |||
| C01987 | 2-Aminophenol | |||
| C01249 | 7,8-Dihydro-7,8-dihydroxykynurenate | |||
| C01717 | Kynurenic acid | |||
| C00398 | Tryptamine | |||
| C02172 | N-Acetylisatin | |||
| C01252 | 4-(2-Aminophenyl)-2,4-dioxobutanoate | |||
| C00463 | Indole; 2,3-Benzopyrrole | |||
| C02775 | Dihydroxyindole | |||
| C02938 | 3-Indoleacetonitrile | |||
| C03574 | 2-Formylaminobenzaldehyde | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, J.R.; Lucio, M.; Harir, M.; Schmitt-Kopplin, P. Mining for Active Molecules in Probiotic Supernatant by Combining Non-Targeted Metabolomics and Immunoregulation Testing. Metabolites 2022, 12, 35. https://doi.org/10.3390/metabo12010035
Fonseca JR, Lucio M, Harir M, Schmitt-Kopplin P. Mining for Active Molecules in Probiotic Supernatant by Combining Non-Targeted Metabolomics and Immunoregulation Testing. Metabolites. 2022; 12(1):35. https://doi.org/10.3390/metabo12010035
Chicago/Turabian StyleFonseca, Juliano Roldan, Marianna Lucio, Mourad Harir, and Philippe Schmitt-Kopplin. 2022. "Mining for Active Molecules in Probiotic Supernatant by Combining Non-Targeted Metabolomics and Immunoregulation Testing" Metabolites 12, no. 1: 35. https://doi.org/10.3390/metabo12010035
APA StyleFonseca, J. R., Lucio, M., Harir, M., & Schmitt-Kopplin, P. (2022). Mining for Active Molecules in Probiotic Supernatant by Combining Non-Targeted Metabolomics and Immunoregulation Testing. Metabolites, 12(1), 35. https://doi.org/10.3390/metabo12010035






