Fish Skin and Gill Mucus: A Source of Metabolites for Non-Invasive Health Monitoring and Research
Abstract
:1. Introduction
2. Results
2.1. Untargeted Analyses
2.1.1. Normalization of Data from Skin and Gill Mucus
2.1.2. Metabolic Pathways: Functional Analysis
2.2. AbsoluteIDQ® p400 HR Kit
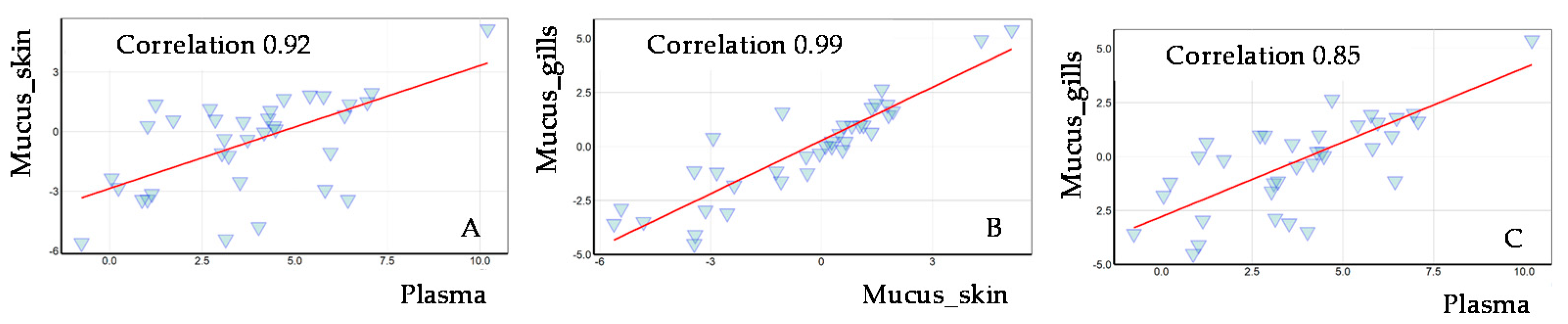
2.2.1. Comparing Mucus and Plasma Samples Using Targeted Metabolomics
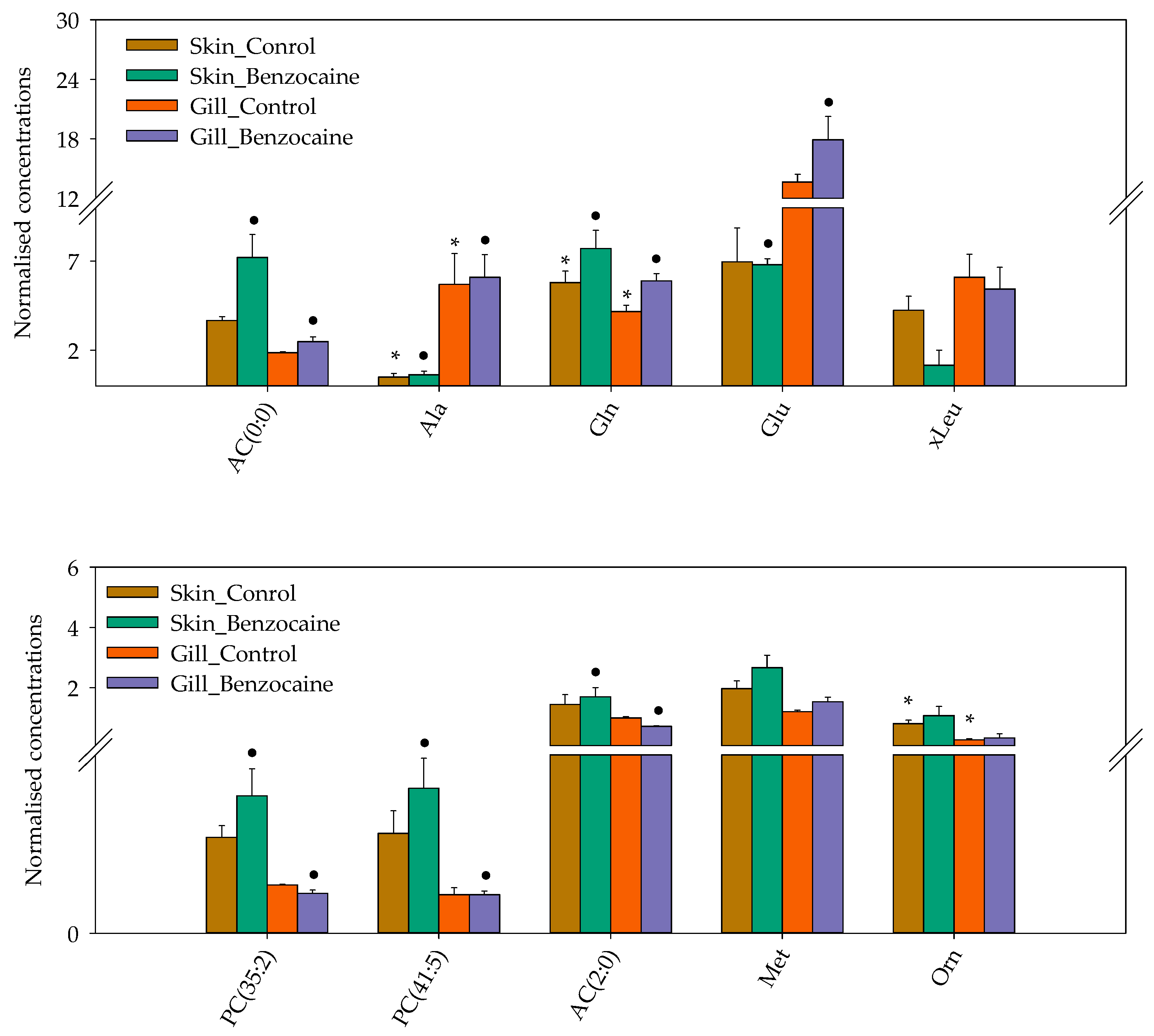
2.2.2. Skin and Gill Mucus: General Considerations
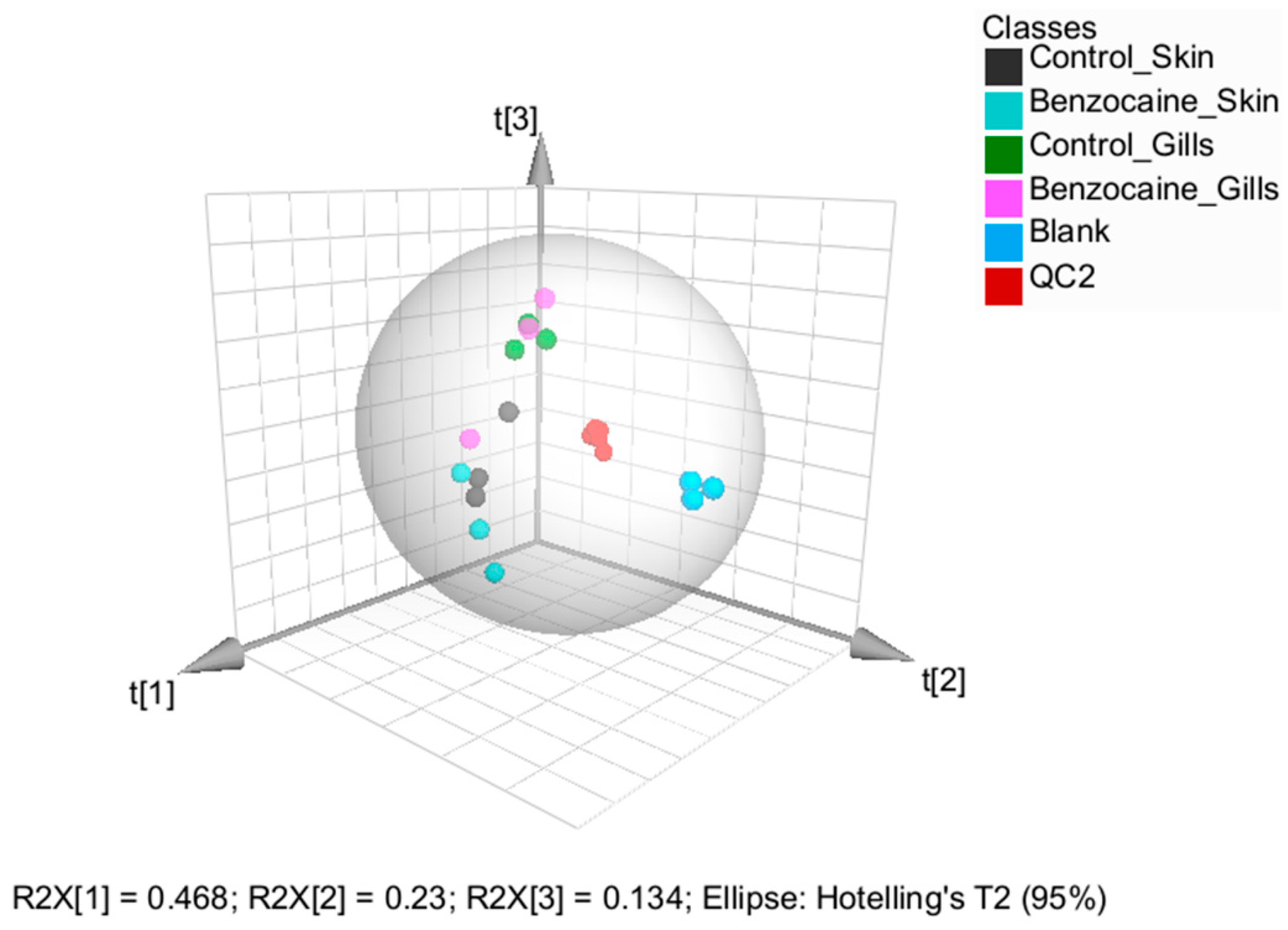
2.2.3. Univariate and Multivariate Analysis: Comparing the Gill and Skin Mucus Metabolome
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. In Vivo Treatments and Fish Sampling
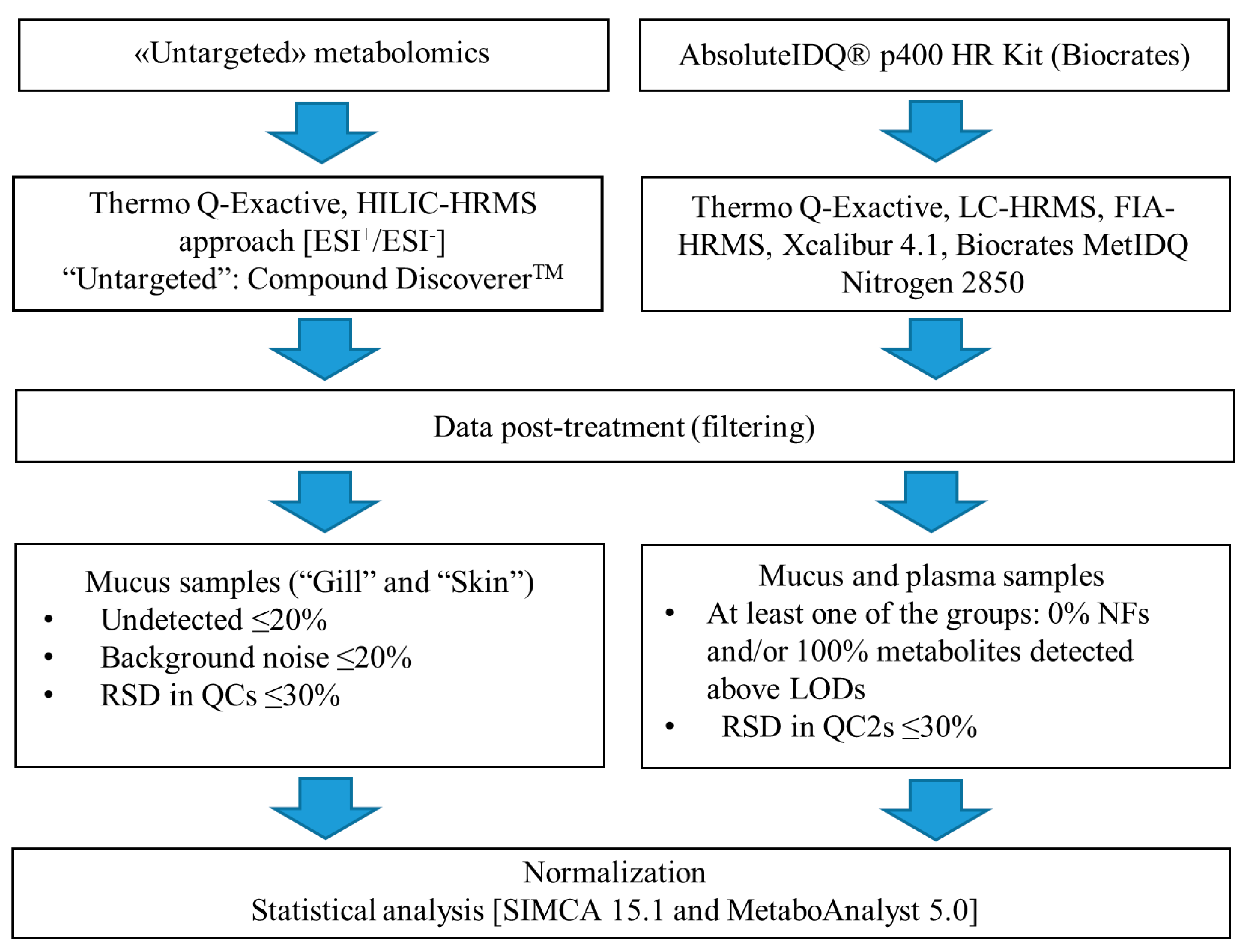
4.3. Targeted and Untargeted Metabolomics
4.3.1. Untargeted Metabolomics
4.3.2. Targeted Metabolomics Using the AbsoluteIDQ® p400 HR Kit
4.3.3. High-Resolution-Mass Spectrometry Analyses (HRMS)
4.4. Data Post-Processing
4.4.1. Targeted Metabolomics: AbsoluteIDQ® p400 HR Kit
4.4.2. Untargted Metabolomics
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alfaro, A.C.; Young, T. Showcasing metabolomic applications in aquaculture: A review. Rev. Aquac. 2016, 10, 135–152. [Google Scholar] [CrossRef]
- Roques, S.; Deborde, C.; Richard, N.; Skiba-Cassy, S.; Moing, A.; Fauconneau, B. Metabolomics and fish nutrition: A review in the context of sustainable feed development. Rev. Aquac. 2020, 12, 261–282. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aru, V.; Khakimov, B.; Sørensen, K.M.; Chikwati, E.M.; Kortner, T.M.; Midtlyng, P.; Krogdahl, A.; Engelsen, S.B. The plasma metabolome of Atlantic salmon as studied by 1H NMR spectroscopy using standard operating procedures: Effect of aquaculture location and growth stage. Metabolomics 2021, 17, 1–13. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Dash, S.; Das, S.; Samal, J.; Thatoi, H.N. Epidermal mucus, a major determinant in fish health: A review. Iran. J. Vet. Res. 2018, 19, 72–81. [Google Scholar]
- Misra, B.B. Data normalization strategies in metabolomics: Current challenges, approaches, and tools. Eur. J. Mass Spectrom. 2020, 26, 165–174. [Google Scholar] [CrossRef]
- Cuevas-Delgado, P.; Dudzik, D.; Miguel, V.; Lamas, S.; Barbas, C. Data-dependent normalization strategies for untargeted metabolomics—A case study. Anal. Bioanal. Chem. 2020, 412, 6391–6405. [Google Scholar] [CrossRef]
- Reverter, M.; Sasal, P.; Banaigs, B.; Lecchini, D.; Lecellier, G.; Tapissier-Bontemps, N. Fish mucus metabolome reveals fish life-history traits. Coral Reefs 2017, 36, 463–475. [Google Scholar] [CrossRef]
- Ekman, D.R.; Skelton, D.M.; Davis, J.M.; Villeneuve, D.L.; Cavallin, J.E.; Schroeder, A.; Jensen, K.M.; Ankley, G.T.; Collette, T. Metabolite Profiling of Fish Skin Mucus: A novel approach for minimally-invasive environmental exposure monitoring and surveillance. Environ. Sci. Technol. 2015, 49, 3091–3100. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Alacid, L.; Sanahuja, I.; Ordóñez-Grande, B.; Sánchez-Nuño, S.; Viscor, G.; Gisbert, E.; Herrera, M.; Ibarz, A. Skin mucus metabolites in response to physiological challenges: A valuable non-invasive method to study teleost marine species. Sci. Total Environ. 2018, 644, 1323–1335. [Google Scholar] [CrossRef]
- Fernández-Alacid, L.; Sanahuja, I.; Grande, B.O.; Sánchez-Nuño, S.; Herrera, M.; Ibarz, A. Skin mucus metabolites and cortisol in meagre fed acute stress-attenuating diets: Correlations between plasma and mucus. Aquaculture 2019, 499, 185–194. [Google Scholar] [CrossRef]
- Milligan, C.L. The role of cortisol in amino acid mobilization and metabolism following exhaustive exercise in rainbow trout (Oncorhynchus mykiss Walbaum). Fish Physiol. Biochem. 1997, 16, 119–128. [Google Scholar] [CrossRef]
- Easy, R.H.; Ross, N.W. Changes in Atlantic salmon Salmo salar mucus components following short- and long-term handling stress. J. Fish Biol. 2010, 77, 1616–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwama, G.K.; McGeer, J.C.; Pawluk, M.P. The effects of five fish anaesthetics on acid–base balance, hematocrit, blood gases, cortisol, and adrenaline in rainbow trout. Can. J. Zool. 1989, 67, 2065–2073. [Google Scholar] [CrossRef]
- Li, P.; Mai, K.; Trushenski, J.; Wu, G. New developments in fish amino acid nutrition: Towards functional and environmentally oriented aquafeeds. Amino Acids 2009, 37, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Erben, V.; Poschet, G.; Schrotz-King, P.; Brenner, H. Evaluation of different stool extraction methods for metabolomics measurements in human faecal samples. BMJ Nutr. Prev. Health 2021. [Google Scholar] [CrossRef]
- Reverter, M.; Tapissier-Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Biological and ecological roles of external fish wucus: A review. Fishes 2018, 3, 41. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, L.; Tartor, H.; Grove, S.; Kristoffersen, A.B.; Uhlig, S. Workflow for the targeted and untargeted detection of small etabolites in fish skin mucus. Fishes 2018, 3, 21. [Google Scholar] [CrossRef] [Green Version]
- Ross, N.W.; Firth, K.J.; Wang, A.; Burka, J.F.; Johnson, S.C. Changes in hydrolytic enzyme activities of naïve Atlantic salmon Salmo salar skin mucus due to infection with the salmon louse Lepeophtheirus salmonis and cortisol implantation. Dis. Aquat. Org. 2000, 41, 43–51. [Google Scholar] [CrossRef]
- Fæste, C.K.; Tartor, H.; Moen, A.; Kristoffersen, A.B.; Dhanasiri, A.K.S.; Anonsen, J.H.; Furmanek, T.; Grove, S. Proteomic profiling of salmon skin mucus for the comparison of sampling methods. J. Chtomatogr. B 2020, 121965. [Google Scholar] [CrossRef]
- Fernández-Montero, A.; Torrecillas, S.; Tort, L.; Ginés, R.; Acosta, F.; Izquierdo, M.; Montero, D. Stress response and skin mucus production of greater amberjack (Seriola dumerili) under different rearing conditions. Aquaculture 2020, 520, 735005. [Google Scholar] [CrossRef]
- Jiang, W.; Tian, X.; Fang, Z.; Li, L.; Dong, S.; Li, H.; Zhao, K. Metabolic responses in the gills of tongue sole (Cynoglossus semilaevis) exposed to salinity stress using NMR-based metabolomics. Sci. Total Environ. 2019, 653, 465–474. [Google Scholar] [CrossRef]
- Ip, A.Y.K. Ammonia production, excretion, toxicity, and defense in fish: A review. Front. Physiol. 2010, 1, 134. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.-C.; Liu, T.-Y.; Hu, M.; Casties, I.; Tseng, Y.-C. Energy and nitrogenous waste from glutamate/glutamine catabolism facilitates acute osmotic adjustment in non-neuroectodermal branchial cells. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-L.; Wang, G.-Y.; Zhang, Z.-H.; Xie, Y.-Y.; Jin, H.; Dong, Z.-R. Partial amino acid metabolism and glutamine synthesis as the ammonia defensive strategies during aerial exposure in Chinese loach Paramisgurnus dabryanus. Front. Physiol. 2019, 10, 14. [Google Scholar] [CrossRef]
- Clark, T.C.; Tinsley, J.; Macqueen, D.J.; Martin, S.A.M. Rainbow trout (Oncorhynchus mykiss) urea cycle and polyamine synthesis gene families show dynamic expression responses to inflammation. Fish Shellfish Immunol. 2019, 89, 290–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, T.; Walker, S.P.; Alfaro, A.C.; Fletcher, L.M.; Murray, J.S.; Lulijwa, R.; Symonds, J. Impact of acute handling stress, anaesthesia, and euthanasia on fish plasma biochemistry: Implications for veterinary screening and metabolomic sampling. Fish Physiol. Biochem. 2019, 45, 1485–1494. [Google Scholar] [CrossRef]
- Van De Vis, H.; Kestin, S.; Robb, D.; Oehlenschläger, J.; Lambooij, B.; Münkner, W.; Kuhlmann, H.; Kloosterboer, K.; Tejada, M.; Huidobro, A. Is humane slaughter of fish possible for industry? Aquac. Res. 2003, 34, 211–220. [Google Scholar] [CrossRef]
- Tartor, H.; Monjane, A.L.; Grove, S. Quantification of defensive proteins in skin mucus of Atlantic salmon using minimally invasive sampling and high-sensitivity ELISA. Animals 2020, 10, 1374. [Google Scholar] [CrossRef]
- Thompson, J.W.; Adams, K.J.; Adamski, J.; Asad, Y.; Borts, D.; Bowden, J.A.; Byram, G.; Dang, V.D.; Dunn, W.B.; Fernandez, F.M. International ring trial of a high resolution targeted metabolomics and lipidomics platform for serum and plasma analysis. Anal. Chem. 2019, 91, 14407–14416. [Google Scholar] [CrossRef] [PubMed]

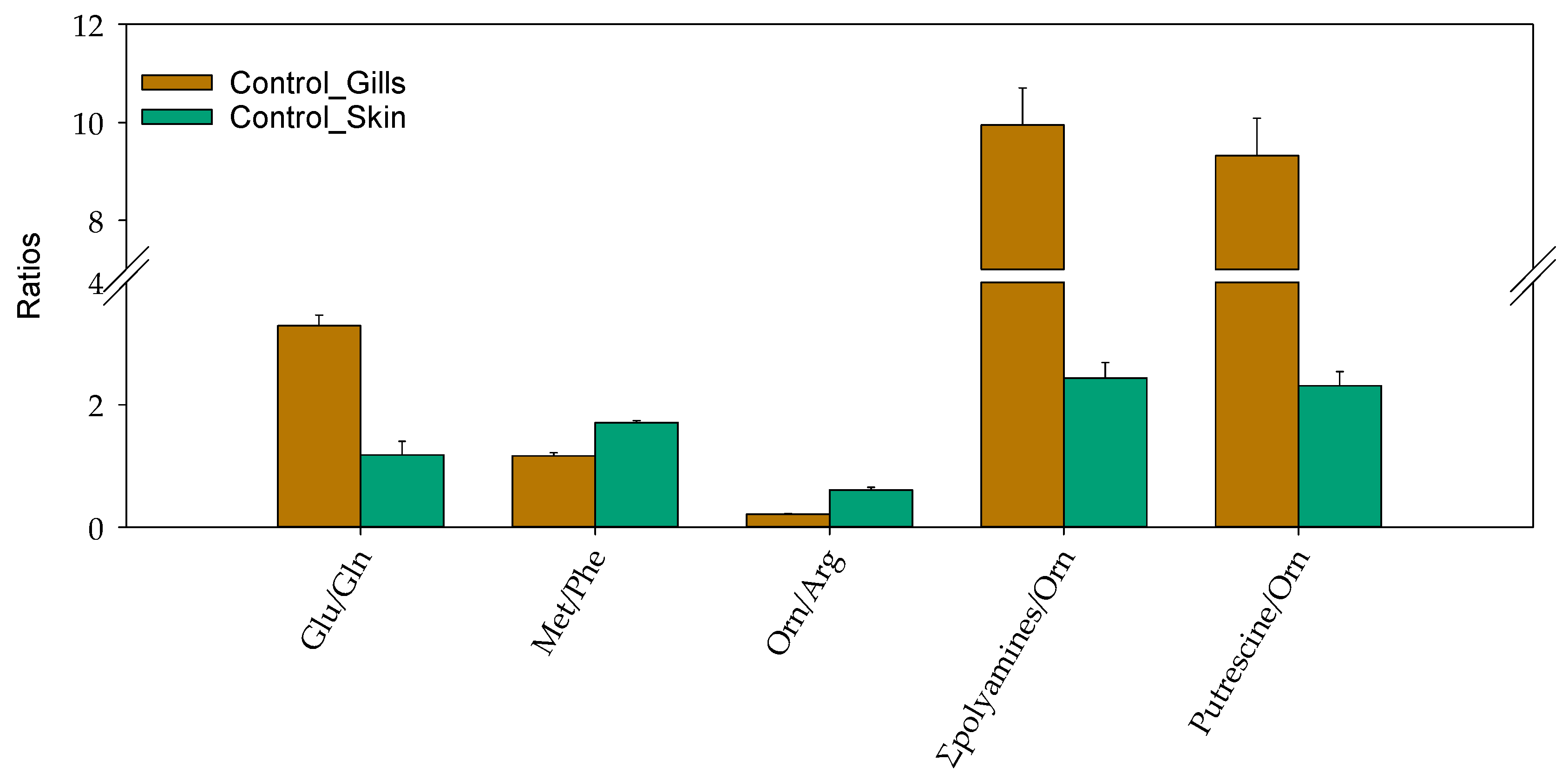
| Normalization Method | N (RSD ≤ 30%) 1 | Median RSD (%) 2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Controls 3 | Benzocain | Controls 3 | Benzocaine | |||||
| Gills | Skin | Gills | Skin | Gills | Skin | Gills | Skin | |
| Non-normalized | 719 | 724 | 811 | 909 | 32 | 33 | 30 | 24 |
| Sum of peak areas | 845 | 807 | 825 | 894 | 28 | 30 | 29 | 27 |
| Protein content | 358 | 170 | 72 | 868 | 40 | 48 | 66 | 28 |
| Median | 853 | 908 | 815 | 932 | 28 | 25 | 30 | 25 |
| Median fold change | 853 | 913 | 838 | 938 | 28 | 25 | 29 | 24 |
| OPLS-DA 1 | R2Y | Q2Y | CV-ANOVA [p Value] | Permutation | |||
|---|---|---|---|---|---|---|---|
| Median | MFC | Median | MFC | Median | MFC | Median/MFC | |
| Control_Skin vs. Benzocaine_Skin | 0.927 | 0.929 | 0.833 | 0.834 | 0.0019 | 0.0019 | Valid |
| Control_Gills vs. Benzocaine_Gills | 0.95 | 0.948 | 0.757 | 0.756 | 0.0071 | 0.0071 | Valid |
| Benzocaine_Skin vs. Benzocaine_Gills | 0.906 | 0.905 | 0.874 | 0.873 | 0.0007 | 0.0007 | Valid |
| Control_Skin vs. Control_Gills | 0.927 | 0.935 | 0.833 | 0.863 | 0.0019 | 0.0010 | Valid |
| Groups Comparison | Metabolic Pathways | p Value | Significant Hits |
|---|---|---|---|
| Control_Skin vs. Benzocaine_Skin | Aminoacyl-tRNA biosynthesis | 0.009 | L-phenylalanine, L-alanine, L-lysine, L-isoleucine, L-aspartate, L-proline |
| Control_Gills vs. Benzocaine_Gills | Fructose and mannose metabolism | 0.021 | D-fructose, D-mannose, D-glyceraldehyde, 6-deoxy-L-galactose, D-glucose, 2-dehydro-3-deoxy-L-fuconate, D-lactic acid |
| beta-Alanine metabolism | 0.022 | glycerone, D-lactate, D-glyceraldehyde, 3-hydroxypropanoate, β-alanine, L-aspartate, 3-aminopropanal, L-alanine | |
| Benzocaine_Skin vs. Benzocaine_Gills | Alanine, aspartate and glutamate metabolism | 0.006 | N-acetyl-L-aspartate, L-aspartate, D-aspartate, L-alanine, L-glutamic acid, 4-aminobutanoate, L-glutamine, fumaric acid, β-citryl-L-glutamate |
| Aminoacyl-tRNA biosynthesis | 0.028 | L-phenylalanine, L-glutamine, glycine, L-aspartate, L-alanine, L-isoleucine, L-leucine, L-threonine, L-proline, L-glutamic acid | |
| Pyrimidine metabolism | 0.033 | L-glutamine, thymine, (R)-3-ureidoisobutyrate, β-alanine, (R)-3-aminoisobutyrate | |
| Glutathione metabolism | 0.033 | glutathione, glycine, L-glutamic acid, 5-oxoproline, L-ornithine | |
| Selenocompound metabolism | 0.041 | L-alanine, β-alanine, sarcosine | |
| Control_Skin vs. Control_Gills | β-Alanine metabolism | 0.024 | 3-hydroxypropanoate, β-alanine, L-aspartate,3-ureidopropionate, dihydrouracil |
| Aminoacyl-tRNA biosynthesis | 0.039 | L-asparagine, L-phenylalanine, glycine, L-aspartate, L-methionine, L-alanine, L-lysine, L-isoleucine, L-leucine, L-glutamic acid |
| Total Number of Metabolites | Detected | Most Abundant | ||||
|---|---|---|---|---|---|---|
| Mucus | Plasma | Gill | Skin | Plasma | ||
| Acylcarnitines [AC (X:Y)] | 55 | 3 | 10 | L-Carnitine | ||
| Amino Acids [AA] | 21 | 19 | 20 | L-Glutamic acid | L-Valine | L-Valine |
| Biogenic Amines [BA] | 21 | 5 | 10 | Taurine | ||
| Lysophosphatidylcholines[LPC (X:Y)] | 24 | - | 12 | ND | ND | LPC (22:6) |
| ConfirmedPhosphatidylcholines [PC (X:Y)] | 172 | 8 | 54 | PC (38:6) | PC (34:2) | PC (38:6) |
| Ceramides [Cer (X:Y)] | 9 | - | 1 | ND | ND | Cer (42:2) |
| Sphingomyelins [SM (X:Y)] | 31 | 1 | 10 | SM (42:3) | SM (42:3) | SM (42:2) |
| Sum hexoses [including glucose] | 1 | 1 | 1 | Sum hexoses | ||
| Cholesteryl Esters [CE (X:Y)] | 14 | - | 7 | ND | ND | CE (22:6) |
| Diglycerides [DG (X:Y)] | 18 | - | 10 | ND | ND | DG (36:2) |
| Triglycerides [TG (X:Y)] | 42 | 1 | 28 | TG (52:7) | TG (52:7) | TG (56:7) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, L.; Rangel-Huerta, O.D.; Tartor, H.; Gjessing, M.C.; Dahle, M.K.; Uhlig, S. Fish Skin and Gill Mucus: A Source of Metabolites for Non-Invasive Health Monitoring and Research. Metabolites 2022, 12, 28. https://doi.org/10.3390/metabo12010028
Ivanova L, Rangel-Huerta OD, Tartor H, Gjessing MC, Dahle MK, Uhlig S. Fish Skin and Gill Mucus: A Source of Metabolites for Non-Invasive Health Monitoring and Research. Metabolites. 2022; 12(1):28. https://doi.org/10.3390/metabo12010028
Chicago/Turabian StyleIvanova, Lada, Oscar D. Rangel-Huerta, Haitham Tartor, Mona C. Gjessing, Maria K. Dahle, and Silvio Uhlig. 2022. "Fish Skin and Gill Mucus: A Source of Metabolites for Non-Invasive Health Monitoring and Research" Metabolites 12, no. 1: 28. https://doi.org/10.3390/metabo12010028
APA StyleIvanova, L., Rangel-Huerta, O. D., Tartor, H., Gjessing, M. C., Dahle, M. K., & Uhlig, S. (2022). Fish Skin and Gill Mucus: A Source of Metabolites for Non-Invasive Health Monitoring and Research. Metabolites, 12(1), 28. https://doi.org/10.3390/metabo12010028








