Defining Blood Plasma and Serum Metabolome by GC-MS
Abstract
1. Introduction
2. Serum and Plasma Metabolome as a “Snapshot” of a Human Biochemistry
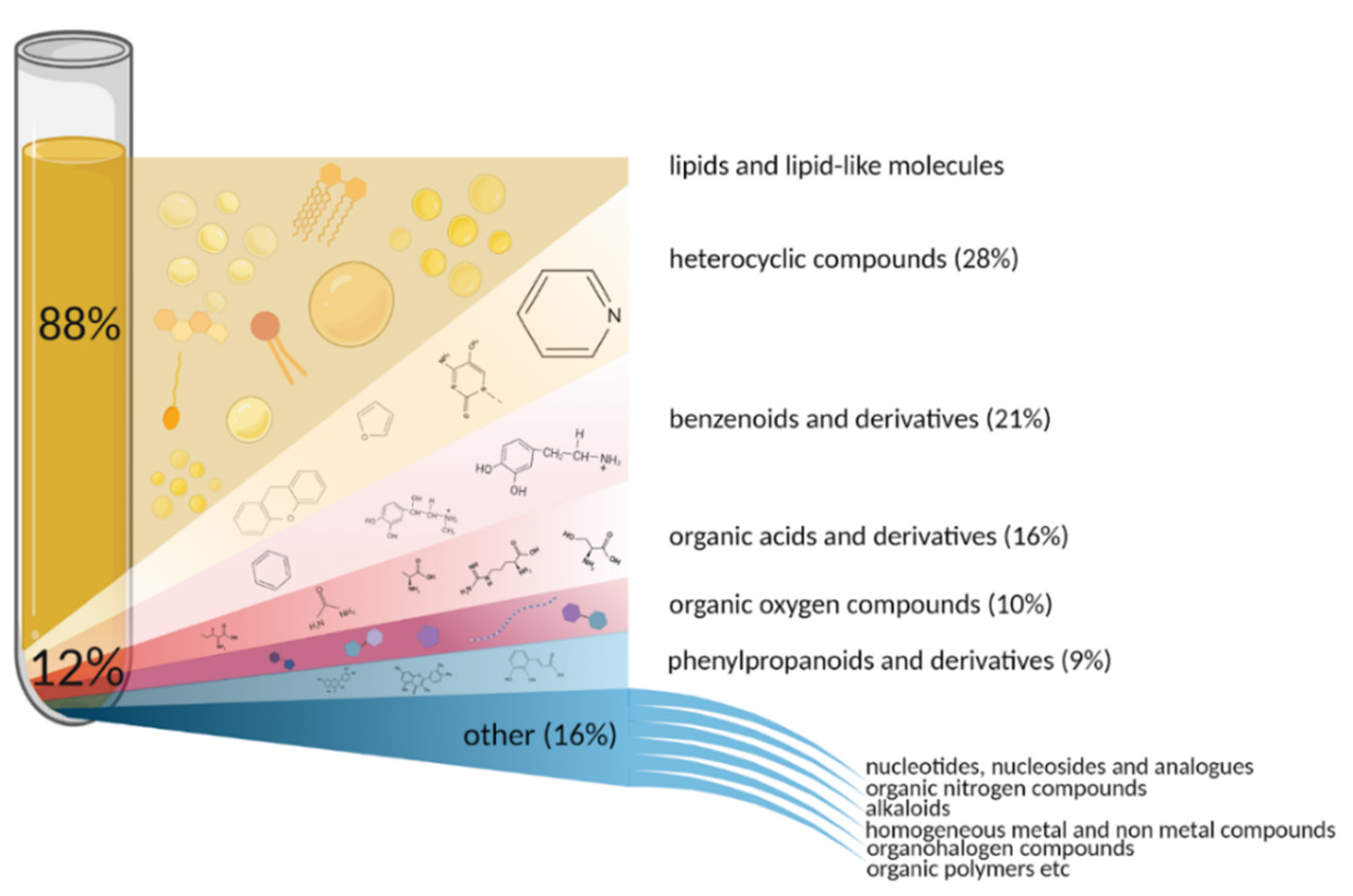
3. How Many Blood Metabolites Are There?
4. Approaches of Metabolome Exploration
4.1. NMR
4.2. Tandem of Chromatography and Mass Spectrometry
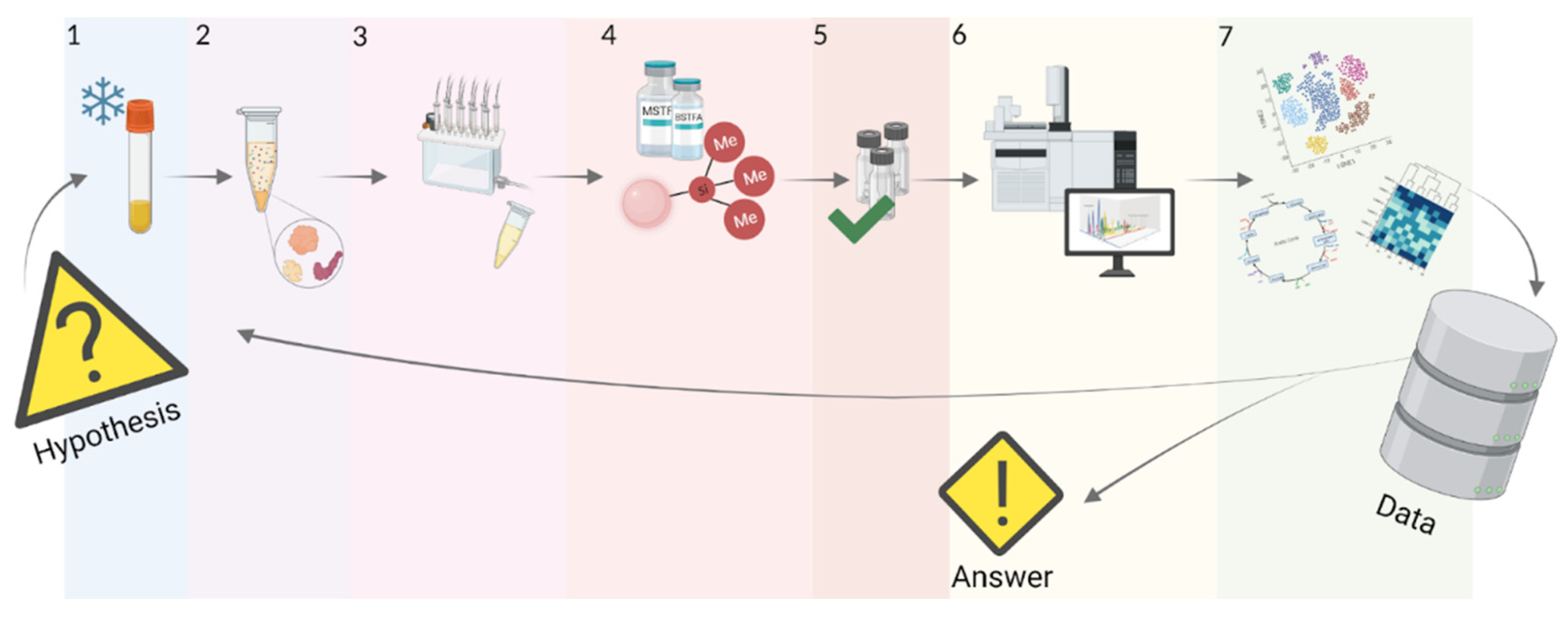
5. Workflow of GC-MS Analysis of Blood Metabolome
5.1. Sample Preparation
5.1.1. Quenching
5.1.2. Protein Cleanup
5.1.3. Extraction
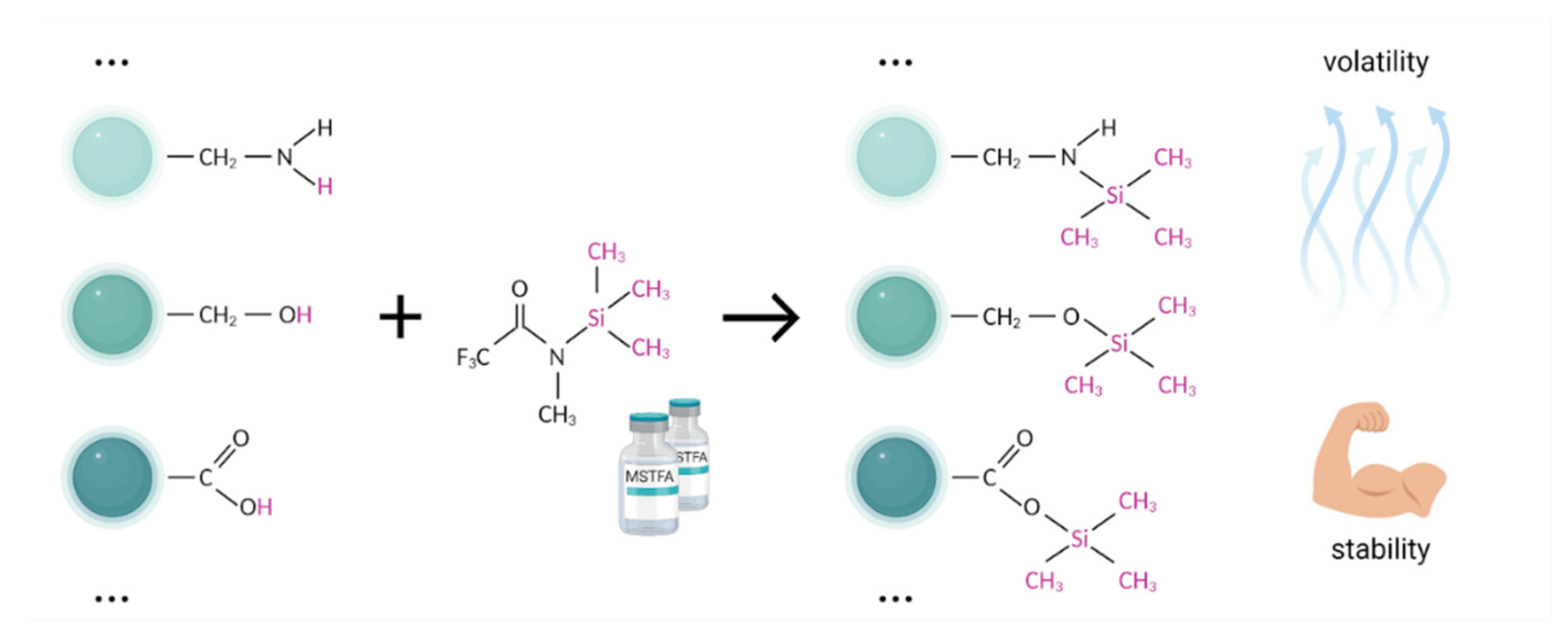
5.1.4. Derivatization
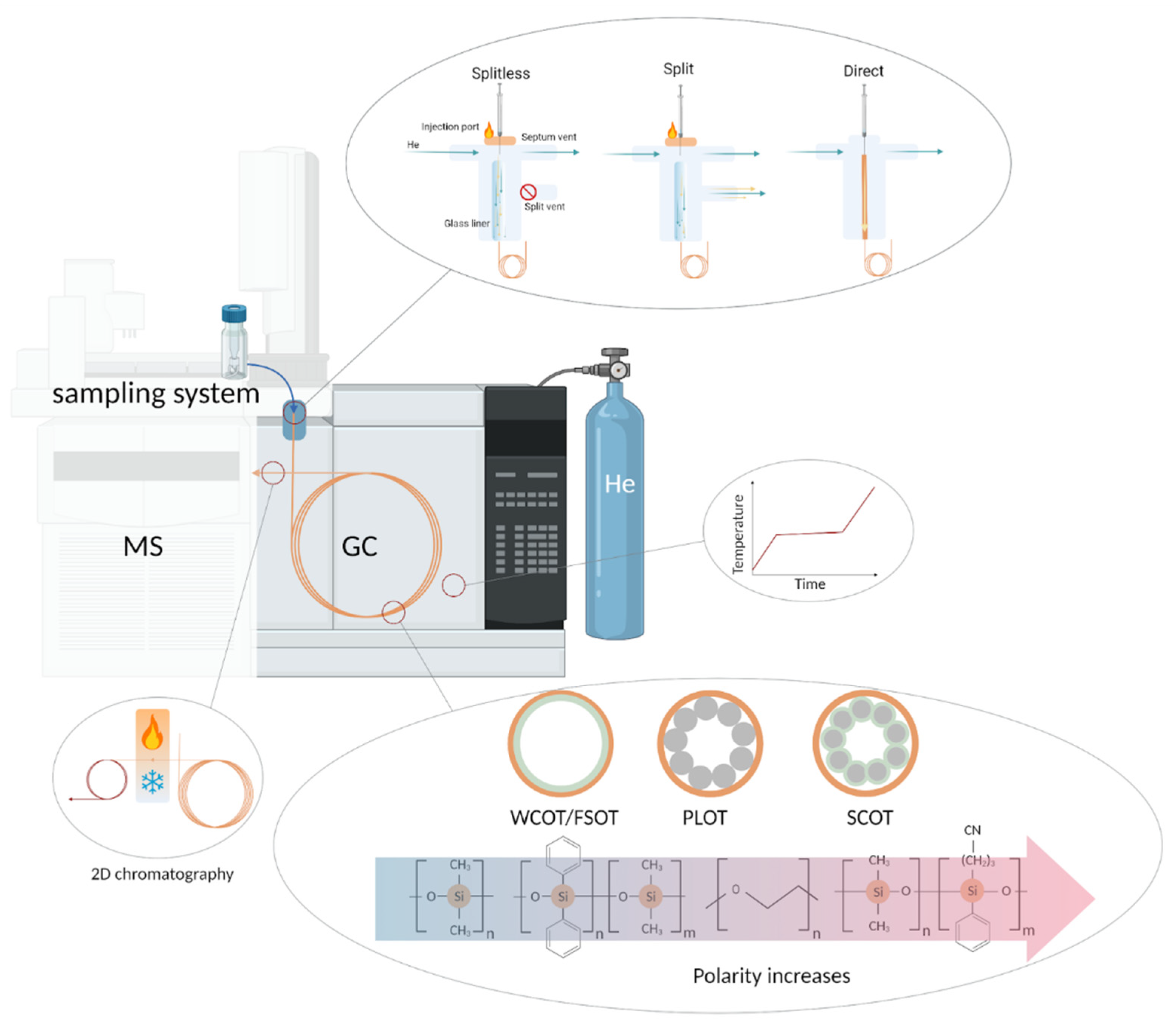
5.2. Gas Chromatography
5.2.1. Injection
5.2.2. Thermal Conditions
5.2.3. Solid and Liquid Stationary Phases
5.2.4. Retention Times and Indices
5.2.5. Multidimensional Chromatography
5.3. Mass Spectrometry
5.3.1. Ionization
5.3.2. Mass Analyzers
5.4. Data Processing
6. Current Challenges and Prospects in Measuring Metabolites
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goodacre, R. Metabolomics of a superorganism. J. Nutr. 2007, 137, 259S–266S. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, S.H.; Kim, J.H.; Hwang, S.; Yoo, H.J. Understanding Metabolomics in Biomedical Research. Endocrinol. Metab. 2016, 31, 7. [Google Scholar] [CrossRef]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Guasch-Ferre, M.; Bhupathiraju, S.N.; Hu, F.B. Use of Metabolomics in Improving Assessment of Dietary Intake. Clin. Chem. 2018, 64, 82–98. [Google Scholar] [CrossRef]
- Li, J.; Guasch-Ferré, M.; Chung, W.; Ruiz-Canela, M.; Toledo, E.; Corella, D.; Bhupathiraju, S.N.; Tobias, D.K.; Tabung, F.K.; Hu, J.; et al. The Mediterranean diet, plasma metabolome, and cardiovascular disease risk. Eur. Heart J. 2020, 41, 2645–2656. [Google Scholar] [CrossRef]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward personalized medicine. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef]
- Heaney, L.M.; Deighton, K.; Suzuki, T. Non-targeted metabolomics in sport and exercise science. J. Sports Sci. 2019, 37, 959–967. [Google Scholar] [CrossRef]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Lima-Oliveira, G.; Monneret, D.; Guerber, F.; Guidi, G.C. Sample management for clinical biochemistry assays: Are serum and plasma interchangeable specimens? Crit. Rev. Clin. Lab. Sci. 2018, 55, 480–500. [Google Scholar] [CrossRef]
- Yasumoto, A.; Tokuoka, S.M.; Kita, Y.; Shimizu, T.; Yatomi, Y. Multiplex quantitative analysis of eicosanoid mediators in human plasma and serum: Possible introduction into clinical testing. J. Chromatogr. B 2017, 1068–1069, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Brown, M.; Davey, H.M.; Dunn, W.B.; Spasic, I.; Oliver, S.G. Metabolic footprinting and systems biology: The medium is the message. Nat. Rev. Microbiol. 2005, 3, 557–565. [Google Scholar] [CrossRef]
- Shah, N.J.; Sureshkumar, S.; Shewade, D.G. Metabolomics: A Tool Ahead for Understanding Molecular Mechanisms of Drugs and Diseases. Indian J. Clin. Biochem. 2015, 30, 247–254. [Google Scholar] [CrossRef]
- Mitro, S.D.; Wu, J.; Rahman, M.L.; Cao, Y.; Zhu, Y.; Chen, Z.; Chen, L.; Li, M.; Hinkle, S.N.; Bremer, A.A.; et al. Longitudinal Plasma Metabolomics Profile in Pregnancy-A Study in an Ethnically Diverse U.S. Pregnancy Cohort. Nutrients 2021, 13, 3080. [Google Scholar] [CrossRef]
- Harville, E.W.; Li, Y.Y.; Pan, K.; McRitchie, S.; Pathmasiri, W.; Sumner, S. Untargeted analysis of first trimester serum to reveal biomarkers of pregnancy complications: A case–control discovery phase study. Sci. Rep. 2021, 11, 3468. [Google Scholar] [CrossRef]
- Darst, B.F.; Koscik, R.L.; Hogan, K.J.; Johnson, S.C.; Engelman, C.D. Longitudinal plasma metabolomics of aging and sex. Aging 2019, 11, 1262–1282. [Google Scholar] [CrossRef]
- Thompson, D.S.; Bourdon, C.; Massara, P.; Boyne, M.S.; Forrester, T.E.; Gonzales, G.B.; Bandsma, R.H.J. Childhood severe acute malnutrition is associated with metabolic changes in adulthood. JCI Insight 2020, 5, e141316. [Google Scholar] [CrossRef] [PubMed]
- Aderemi, A.V.; Ayeleso, A.O.; Oyedapo, O.O.; Mukwevho, E. Metabolomics: A Scoping Review of Its Role as a Tool for Disease Biomarker Discovery in Selected Non-Communicable Diseases. Metabolites 2021, 11, 418. [Google Scholar] [CrossRef] [PubMed]
- Sindelar, M.; Stancliffe, E.; Schwaiger-Haber, M.; Anbukumar, D.S.; Adkins-Travis, K.; Goss, C.W.; O’Halloran, J.A.; Mudd, P.A.; Liu, W.C.; Albrecht, R.A.; et al. Longitudinal metabolomics of human plasma reveals prognostic markers of COVID-19 disease severity. Cell Rep. Med. 2021, 2, 100369. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Shahjaman, M.; Mollah, M.N.H.; Islam, S.M.S.; Hoque, M.A. Serum and Plasma Metabolomic Biomarkers for Lung Cancer. Bioinformation 2017, 13, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, C.; Xu, S.; Gao, L.; Blaženović, I.; Ji, J.; Wang, J.; Sun, X. Untargeted metabolomics analysis by gas chromatography/time-of-flight mass spectrometry of human serum from methamphetamine abusers. Addict. Biol. 2021, 26, e13062. [Google Scholar] [CrossRef]
- Paris, A.; Labrador, B.; Lejeune, F.X.; Canlet, C.; Molina, J.; Guinot, M.; Mégret, A.; Rieu, M.; Thalabard, J.C.; Le Bouc, Y. Metabolomic signatures in elite cyclists: Differential characterization of a seeming normal endocrine status regarding three serum hormones. Metabolomics 2021, 17, 67. [Google Scholar] [CrossRef]
- Tebani, A.; Bekri, S. Paving the Way to Precision Nutrition Through Metabolomics. Front. Nutr. 2019, 6, 41. [Google Scholar] [CrossRef]
- Rafiq, T.; Azab, S.M.; Teo, K.K.; Thabane, L.; Anand, S.S.; Morrison, K.M.; de Souza, R.J.; Britz-McKibbin, P. Nutritional Metabolomics and the Classification of Dietary Biomarker Candidates: A Critical Review. Adv. Nutr. 2021, 12, 2333–2357. [Google Scholar] [CrossRef]
- Stekovic, S.; Hofer, S.J.; Tripolt, N.; Aon, M.A.; Royer, P.; Pein, L.; Stadler, J.T.; Pendl, T.; Prietl, B.; Url, J.; et al. Alternate Day Fasting Improves Physiological and Molecular Markers of Aging in Healthy, Non-obese Humans. Cell Metab. 2019, 30, 462–476.e5. [Google Scholar] [CrossRef]
- Tenori, L.; Oakman, C.; Claudino, W.M.; Bernini, P.; Cappadona, S.; Nepi, S.; Biganzoli, L.; Arbushites, M.C.; Luchinat, C.; Bertini, I.; et al. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: A pilot study. Mol. Oncol. 2012, 6, 437–444. [Google Scholar] [CrossRef]
- Misra, B. Individualized metabolomics: Opportunities and challenges. Clin. Chem. Lab. Med. 2020, 58, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Bar, N.; Korem, T.; Weissbrod, O.; Zeevi, D.; Rothschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weinberger, A.; Le Roy, C.I.; et al. A reference map of potential determinants for the human serum metabolome. Nature 2020, 588, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Lin, W.; Broadhurst, D.; Begley, P.; Brown, M.; Zelena, E.; Vaughan, A.A.; Halsall, A.; Harding, N.; Knowles, J.D.; et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics 2015, 11, 9–26. [Google Scholar] [CrossRef]
- Sethi, S.; Hayashi, M.A.F.; Barbosa, B.S.; Pontes, J.G.M.; Tasic, L.; Brietzke, E. Metabolomics: From Fundamentals to Clinical Applications; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2017; Volume 965, ISBN 978-3-319-47655-1. [Google Scholar]
- Ossola, R.; Schiess, R.; Picotti, P.; Rinner, O.; Reiter, L.; Aebersold, R. Biomarker validation in blood specimens by selected reaction monitoring mass spectrometry of N-glycosites. Methods Mol. Biol. 2011, 728, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Plebani, M.; Banfi, G.; Bernardini, S.; Bondanini, F.; Conti, L.; Dorizzi, R.; Ferrara, F.E.; Mancini, R.; Trenti, T. Serum or plasma? An old question looking for new answers. Clin. Chem. Lab. Med. 2020, 58, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Tuck, M.K.; Chan, D.W.; Chia, D.; Godwin, A.K.; Grizzle, W.E.; Krueger, K.E.; Rom, W.; Sanda, M.; Sorbara, L.; Stass, S.; et al. Standard Operating Procedures for Serum and Plasma Collection: Early Detection Research Network Consensus Statement Standard Operating Procedure Integration Working Group. J. Proteome Res. 2010, 8, 113–117. [Google Scholar] [CrossRef]
- Wedge, D.C.; Allwood, J.W.; Dunn, W.; Vaughan, A.A.; Simpson, K.; Brown, M.; Priest, L.; Blackhall, F.H.; Whetton, A.D.; Dive, C.; et al. Is serum or plasma more appropriate for intersubject comparisons in metabolomic studies? An assessment in patients with small-cell lung cancer. Anal. Chem. 2011, 83, 6689–6697. [Google Scholar] [CrossRef]
- Liu, X.; Hoene, M.; Wang, X.; Yin, P.; Häring, H.U.; Xu, G.; Lehmann, R. Serum or plasma, what is the difference? Investigations to facilitate the sample material selection decision making process for metabolomics studies and beyond. Anal. Chim. Acta 2018, 1037, 293–300. [Google Scholar] [CrossRef]
- Dettmer, K.; Almstetter, M.F.; Appel, I.J.; Nürnberger, N.; Schlamberger, G.; Gronwald, W.; Meyer, H.H.D.; Oefner, P.J. Comparison of serum versus plasma collection in gas chromatography—Mass spectrometry-based metabolomics. Electrophoresis 2010, 31, 2365–2373. [Google Scholar] [CrossRef]
- Kaluarachchi, M.; Boulangé, C.L.; Karaman, I.; Lindon, J.C.; Ebbels, T.M.D.; Elliott, P.; Tracy, R.P.; Olson, N.C. A comparison of human serum and plasma metabolites using untargeted 1 H NMR spectroscopy and UPLC-MS. Metabolomics 2018, 14, 32. [Google Scholar] [CrossRef]
- Sotelo-Orozco, J.; Chen, S.Y.; Hertz-Picciotto, I.; Slupsky, C.M. A Comparison of Serum and Plasma Blood Collection Tubes for the Integration of Epidemiological and Metabolomics Data. Front. Mol. Biosci. 2021, 8, 650. [Google Scholar] [CrossRef]
- Sandlers, Y. Amino Acids Profiling for the Diagnosis of Metabolic Disorders. In Biochemical Testing—Clinical Correlation and Diagnosis; InTech Open: London, UK, 2019. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The Human Serum Metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef]
- Yu, Z.; Kastenmüller, G.; He, Y.; Belcredi, P.; Möller, G.; Prehn, C.; Mendes, J.; Wahl, S.; Roemisch-Margl, W.; Ceglarek, U.; et al. Differences between human plasma and serum metabolite profiles. PLoS ONE 2011, 6, e21230. [Google Scholar] [CrossRef] [PubMed]
- Peironcely, J.E.; Reijmers, T.; Coulier, L.; Bender, A.; Hankemeier, T. Understanding and classifying metabolite space and metabolite-likeness. PLoS ONE 2011, 6, e28966. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef]
- Quehenberger, O.; Dennis, E.A. The human plasma lipidome. N. Engl. J. Med. 2011, 365, 1812–1823. [Google Scholar] [CrossRef]
- O’Donnell, V.B.; Ekroos, K.; Liebisch, G.; Wakelam, M. Lipidomics: Current state of the art in a fast moving field. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1466. [Google Scholar] [CrossRef]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef]
- Giraudeau, P. NMR-based metabolomics and fluxomics: Developments and future prospects. Analyst 2020, 145, 2457–2472. [Google Scholar] [CrossRef]
- Kirwan, J.A.; Broadhurst, D.I.; Davidson, R.L.; Viant, M.R. Characterising and correcting batch variation in an automated direct infusion mass spectrometry (DIMS) metabolomics workflow. Anal. Bioanal. Chem. 2013, 405, 5147–5157. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.; Zhou, Y.; Zhao, C.; Lu, X.; Xu, G. A rapid GC method coupled with quadrupole or time of flight mass spectrometry for metabolomics analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020, 1160, 122355. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-based metabolic phenotyping (metabonomics/metabolomics): The state of the art. J. Chromatogr. B 2019, 1117, 136–147. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Nagana Gowda, G.A.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Martias, C.; Baroukh, N.; Mavel, S.; Blasco, H.; Lefèvre, A.; Roch, L.; Montigny, F.; Gatien, J.; Schibler, L.; Dufour-Rainfray, D.; et al. Optimization of Sample Preparation for Metabolomics Exploration of Urine, Feces, Blood and Saliva in Humans Using Combined NMR and UHPLC-HRMS Platforms. Molecules 2021, 26, 4111. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Can NMR solve some significant challenges in metabolomics? J. Magn. Reson. 2015, 260, 144. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Hu, Y.; Li, P.; Wan, J.B. Current state of the art of mass spectrometry-based metabolomics studies—A review focusing on wide coverage, high throughput and easy identification. RSC Adv. 2015, 5, 78728–78737. [Google Scholar] [CrossRef]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef] [PubMed]
- Higgins Keppler, E.A.; Jenkins, C.L.; Davis, T.J.; Bean, H.D. Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. TrAC—Trends Anal. Chem. 2018, 109, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Moros, G.; Chatziioannou, A.C.; Gika, H.G.; Raikos, N.; Theodoridis, G. Investigation of the derivatization conditions for GC-MS metabolomics of biological samples. Bioanalysis 2017, 9, 53–65. [Google Scholar] [CrossRef]
- Wu, Z.; Shon, J.C.; Liu, K.-H. Mass Spectrometry-based Lipidomics and Its Application to Biomedical Research. J. Lifestyle Med. 2014, 4, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Chetwynd, A.J.; Dunn, W.B.; Rodriguez-Blanco, G.; Chetwynd, A.J.; Dunn, W.B.; Rodriguez-Blanco, G. Collection and Preparation of Clinical Samples for Metabolomics. Adv. Exp. Med. Biol. 2017, 965, 19–44. [Google Scholar] [CrossRef]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef]
- Hernández Bort, J.A.; Shanmukam, V.; Pabst, M.; Windwarder, M.; Neumann, L.; Alchalabi, A.; Krebiehl, G.; Koellensperger, G.; Hann, S.; Sonntag, D.; et al. Reduced quenching and extraction time for mammalian cells using filtration and syringe extraction. J. Biotechnol. 2014, 182–183, 97–103. [Google Scholar] [CrossRef]
- Castro-Perez, J.; Prakash, C. Recent advances in mass spectrometric and other analytical techniques for the identification of drug metabolites. In Identification and Quantification of Drugs, Metabolites, Drug Metabolizing Enzymes, and Transporters; Elsevier: Amsterdam, The Netherlands, 2020; pp. 39–71. [Google Scholar] [CrossRef]
- Jiye, A.; Trygg, J.; Gullberg, J.; Johansson, A.I.; Jonsson, P.; Antti, H.; Marklund, S.L.; Moritz, T. Extraction and GC/MS analysis of the human blood plasma metabolome. Anal. Chem. 2005, 77, 8086–8094. [Google Scholar] [CrossRef]
- Want, E.J.; O’Maille, G.; Smith, C.A.; Brandon, T.R.; Uritboonthai, W.; Qin, C.; Trauger, S.A.; Siuzdak, G. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal. Chem. 2006, 78, 743–752. [Google Scholar] [CrossRef]
- Wawrzyniak, R.; Kosnowska, A.; Macioszek, S.; Bartoszewski, R.; Markuszewski, M.J. New plasma preparation approach to enrich metabolome coverage in untargeted metabolomics: Plasma protein bound hydrophobic metabolite release with proteinase K. Sci. Rep. 2018, 8, 9541. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cruickshank, C.; Armstrong, M.; Mahaffey, S.; Reisdorph, R.; Reisdorph, N. New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. J. Chromatogr. A 2013, 1300, 217–226. [Google Scholar] [CrossRef]
- Lelli, V.; Belardo, A.; Timperio, A.M. From Targeted Quantification to Untargeted Metabolomics. In Metabolomics—Methodology and Applications in Medical Sciences and Life Sciences; InTech Open: London, UK, 2021. [Google Scholar] [CrossRef]
- He, Z.; Liu, Z.; Gong, L. Systematic evaluation of sample preparation strategy for GC-MS-based plasma metabolomics and its application in osteoarthritis. Anal. Biochem. 2021, 621, 114153. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Reed, D. Gas Chromatography in Metabolomics Study. In Advances in Gas Chromatography; InTech Open: London, UK, 2014. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, T.; Qiu, Y.; Cheng, Y.; Cao, Y.; Zhao, A.; Jia, W. An ultrasonication-assisted extraction and derivatization protocol for GC/TOFMS-based metabolite profiling. Anal. Bioanal. Chem. 2011, 400, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Sana, T.; Fischer, S. Maximizing Metabolite Extraction for Comprehensive Metabolomics Studies of Erythrocytes. Available online: https://www.agilent.com/cs/library/applications/5989-7407EN.pdf (accessed on 23 November 2021).
- Tulipani, S.; Llorach, R.; Urpi-Sarda, M.; Andres-Lacueva, C. Comparative analysis of sample preparation methods to handle the complexity of the blood fluid metabolome: When less is more. Anal. Chem. 2013, 85, 341–348. [Google Scholar] [CrossRef]
- Sitnikov, D.G.; Monnin, C.S.; Vuckovic, D. Systematic Assessment of Seven Solvent and Solid-Phase Extraction Methods for Metabolomics Analysis of Human Plasma by LC-MS. Sci. Rep. 2016, 6, 38885. [Google Scholar] [CrossRef]
- Michopoulos, F.; Lai, L.; Gika, H.; Theodoridis, G.; Wilson, I. UPLC-MS-Based Analysis of Human Plasma for Metabonomics Using Solvent Precipitation or Solid Phase Extraction. J. Proteome Res. 2009, 8, 2114–2121. [Google Scholar] [CrossRef]
- Rico, E.; González, O.; Blanco, M.E.; Alonso, R.M. Evaluation of human plasma sample preparation protocols for untargeted metabolic profiles analyzed by UHPLC-ESI-TOF-MS. Anal. Bioanal. Chem. 2014, 406, 7641–7652. [Google Scholar] [CrossRef]
- Bojko, B.; Reyes-Garcés, N.; Bessonneau, V.; Goryński, K.; Mousavi, F.; Souza Silva, E.A.; Pawliszyn, J. Solid-phase microextraction in metabolomics. TrAC—Trends Anal. Chem. 2014, 61, 168–180. [Google Scholar] [CrossRef]
- Musharraf, S.G.; Mazhar, S.; Siddiqui, A.J.; Choudhary, M.I. Metabolite profiling of human plasma by different extraction methods through gas chromatography–mass spectrometry—An objective comparison. Anal. Chim. Acta 2013, 804, 180–189. [Google Scholar] [CrossRef]
- Caldeira, M.; Barros, A.S.; Bilelo, M.J.; Parada, A.; Câmara, J.S.; Rocha, S.M. Profiling allergic asthma volatile metabolic patterns using a headspace-solid phase microextraction/gas chromatography based methodology. J. Chromatogr. A 2011, 1218, 3771–3780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Raftery, D. Headspace SPME-GC-MS Metabolomics Analysis of Urinary Volatile Organic Compounds (VOCs). Methods Mol. Biol. 2014, 1198, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Rocha, S.M.; Caldeira, M.; Carrola, J.; Santos, M.; Cruz, N.; Duarte, I.F. Exploring the human urine metabolomic potentialities by comprehensive two-dimensional gas chromatography coupled to time of flight mass spectrometry. J. Chromatogr. A 2012, 1252, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Dixon, E.; Clubb, C.; Pittman, S.; Ammann, L.; Rasheed, Z.; Kazmi, N.; Keshavarzian, A.; Gillevet, P.; Rangwala, H.; Couch, R.D. Solid-Phase Microextraction and the Human Fecal VOC Metabolome. PLoS ONE 2011, 6, e18471. [Google Scholar] [CrossRef]
- De Fátima Alpendurada, M. Solid-phase microextraction: A promising technique for sample preparation in environmental analysis. J. Chromatogr. A 2000, 889, 3–14. [Google Scholar] [CrossRef]
- Silva, C.L.; Passos, M.; Câmara, J.S. Solid phase microextraction, mass spectrometry and metabolomic approaches for detection of potential urinary cancer biomarkers--a powerful strategy for breast cancer diagnosis. Talanta 2012, 89, 360–368. [Google Scholar] [CrossRef]
- Silva, C.L.; Passos, M.; Cmara, J.S. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br. J. Cancer 2011, 105, 1894–1904. [Google Scholar] [CrossRef]
- Halket, J.M.; Zaikin, V.G. Derivatization in Mass Spectrometry—7. On-Line Derivatization/Degradation. Eur. J. Mass Spectr. 2017, 12, 1–13. [Google Scholar] [CrossRef]
- Kanani, H.; Chrysanthopoulos, P.K.; Klapa, M.I. Standardizing GC-MS metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 871, 191–201. [Google Scholar] [CrossRef]
- Adebo, O.A.; Oyeyinka, S.A.; Adebiyi, J.A.; Feng, X.; Wilkin, J.D.; Kewuyemi, Y.O.; Abrahams, A.M.; Tugizimana, F. Application of gas chromatography–mass spectrometry (GC-MS)-based metabolomics for the study of fermented cereal and legume foods: A review. Int. J. Food Sci. Technol. 2021, 56, 1514–1534. [Google Scholar] [CrossRef]
- Orata, F. Derivatization Reactions and Reagents for Gas Chromatography Analysis. In Advanced Gas Chromatography—Progress in Agricultural, Biomedical and Industrial Applications; InTech: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef]
- Schummer, C.; Delhomme, O.; Appenzeller, B.M.R.; Wennig, R.; Millet, M. Comparison of MTBSTFA and BSTFA in derivatization reactions of polar compounds prior to GC/MS analysis. Talanta 2009, 77, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Matute, A.I.; Hernández-Hernández, O.; Rodríguez-Sánchez, S.; Sanz, M.L.; Martínez-Castro, I. Derivatization of carbohydrates for GC and GC-MS analyses. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 1226–1240. [Google Scholar] [CrossRef] [PubMed]
- Moldoveanu, S.C.; David, V. Derivatization Methods in GC and GC/MS. In Gas Chromatography—Derivatization, Sample Preparation, Application; InTech Open: London, UK, 2018. [Google Scholar] [CrossRef]
- Little, J.L. Artifacts in trimethylsilyl derivatization reactions and ways to avoid them. J. Chromatogr. A 1999, 844, 1–22. [Google Scholar] [CrossRef]
- Gullberg, J.; Jonsson, P.; Nordström, A.; Sjöström, M.; Moritz, T. Design of experiments: An efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal. Biochem. 2004, 331, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Liang, Y.; Xu, C.; Zhang, J.; Hu, M.; Shi, Q. Polycyclic aromatic hydrocarbons (PAHs) in ambient aerosols from Beijing: Characterization of low volatile pahs by positive-ion atmospheric pressure photoionization (APPI) coupled with fourier transform ion cyclotron resonance. Environ. Sci. Technol. 2014, 48, 4716–4723. [Google Scholar] [CrossRef]
- Miyagawa, H.; Bamba, T. Comparison of sequential derivatization with concurrent methods for GC/MS-based metabolomics. J. Biosci. Bioeng. 2019, 127, 160–168. [Google Scholar] [CrossRef]
- Hyötyläinen, T. Sample Collection, Storage and Preparation. In Chromatographic Methods in Metabolomics; RSC Chromatography Monographs; Royal Society of Chemistry: London, UK, 2013; Chapter 2; pp. 11–42. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Smart, K.F.; Sivakumaran, S.; Lane, G.A. Alkylation or Silylation for Analysis of Amino and Non-Amino Organic Acids by GC-MS? Metabolism 2011, 1, 3–20. [Google Scholar] [CrossRef]
- Ferreira, A.M.C.; Laespada, M.E.F.; Pavón, J.L.P.; Cordero, B.M. In situ aqueous derivatization as sample preparation technique for gas chromatographic determinations. J. Chromatogr. A 2013, 1296, 70–83. [Google Scholar] [CrossRef]
- Qiu, Y.; Su, M.; Liu, Y.; Chen, M.; Gu, J.; Zhang, J.; Jia, W. Application of ethyl chloroformate derivatization for gas chromatography-mass spectrometry based metabonomic profiling. Anal. Chim. Acta 2007, 583, 277–283. [Google Scholar] [CrossRef]
- Hušek, P.; Matucha, P.; Vránková, A.; Šimek, P. Simple plasma work-up for a fast chromatographic analysis of homocysteine, cysteine, methionine and aromatic amino acids. J. Chromatogr. B 2003, 789, 311–322. [Google Scholar] [CrossRef]
- Tao, X.; Liu, Y.; Wang, Y.; Qiu, Y.; Lin, J.; Zhao, A.; Su, M.; Jia, W. GC-MS with ethyl chloroformate derivatization for comprehensive analysis of metabolites in serum and its application to human uremia. Anal. Bioanal. Chem. 2008, 391, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Husek, P.; Simek, P. Alkyl Chloroformates in Sample Derivatization Strategies for GC Analysis. Review on a Decade Use of the Reagents as Esterifying Agents. Curr. Pharm. Anal. 2006, 2, 23–43. [Google Scholar] [CrossRef]
- Hušek, P. Chloroformates in gas chromatography as general purpose derivatizing agents. J. Chromatogr. B Biomed. Sci. Appl. 1998, 717, 57–91. [Google Scholar] [CrossRef]
- McNair, H.M.; Miller, J.M.; Snow, N.H. Temperature Programming. In Basic Gas Chromatography; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 87–98. [Google Scholar] [CrossRef]
- Blumberg, L.M.; Klee, M.S. Quantitative comparison of performance of isothermal and temperature-programmed gas chromatography. J. Chromatogr. A 2001, 933, 13–26. [Google Scholar] [CrossRef]
- Blumberg, L.M. Metrics of separation performance in chromatography: Part 2. Separation performance of a heating ramp in temperature-programmed gas chromatography. J. Chromatogr. A 2012, 1244, 148–160. [Google Scholar] [CrossRef]
- Jespers, S.; Roeleveld, K.; Lynen, F.; Broeckhoven, K.; Desmet, G. Kinetic plots for programmed temperature gas chromatography. J. Chromatogr. A 2016, 1450, 94–100. [Google Scholar] [CrossRef]
- Tolley, H.D.; Tolley, S.E.; Wang, A.; Lee, M.L. Moving thermal gradients in gas chromatography. J. Chromatogr. A 2014, 1374, 189–198. [Google Scholar] [CrossRef]
- Avila, S.; Tolley, H.D.; Iverson, B.D.; Hawkins, A.R.; Johnson, S.L.; Lee, M.L. Comparison of the Dynamic Thermal Gradient to Temperature-Programmed Conditions in Gas Chromatography Using a Stochastic Transport Model. Anal. Chem. 2021, 93, 11785–11791. [Google Scholar] [CrossRef]
- Contreras, J.A.; Wang, A.; Rockwood, A.L.; Tolley, H.D.; Lee, M.L. Dynamic thermal gradient gas chromatography. J. Chromatogr. A 2013, 1302, 143–151. [Google Scholar] [CrossRef]
- Honour, J.W. Gas chromatography-mass spectrometry. Methods Mol. Biol. 2006, 324, 53–74. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.F.; Poole, S.K. Separation characteristics of wall-coated open-tubular columns for gas chromatography. J. Chromatogr. A 2008, 1184, 254–280. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.F.; Poole, S.K. Ionic liquid stationary phases for gas chromatography. J. Sep. Sci. 2011, 34, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Almstetter, M.F.; Oefner, P.J.; Dettmer, K. Comprehensive two-dimensional gas chromatography in metabolomics. Anal. Bioanal. Chem. 2012, 402, 1993–2013. [Google Scholar] [CrossRef] [PubMed]
- Papadimitropoulos, M.E.P.; Vasilopoulou, C.G.; Maga-Nteve, C.; Klapa, M.I. Untargeted GC-MS Metabolomics. Methods Mol. Biol. 2018, 1738, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Dołowy, M.; Pyka, A. Chromatographic methods in the separation of long-chain mono- and polyunsaturated fatty acids. J. Chem. 2015, 2015, 120830. [Google Scholar] [CrossRef]
- Rahman, M.M.; El-Aty, A.M.A.; Choi, J.-H.; Shin, H.-C.; Shin, S.C.; Shim, J.-H. Basic Overview on Gas Chromatography Columns. Anal. Sep. Sci. 2015, 3, 823–834. [Google Scholar] [CrossRef]
- Kováts, E. Gas-chromatographische Charakterisierung organischer Verbindungen. Teil 1: Retentionsindices aliphatischer Halogenide, Alkohole, Aldehyde und Ketone. Helv. Chim. Acta 1958, 41, 1915–1932. [Google Scholar] [CrossRef]
- Kostiainen, R.; Nokeleinen, S. Use of M-series retention index standards in the identification of trichothecenes by electron impact mass spectrometry. J. Chromatogr. 1990, 513, 31–37. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef]
- Trinklein, T.J.; Schöneich, S.; Sudol, P.E.; Warren, C.G.; Gough, D.V.; Synovec, R.E. Total-transfer comprehensive three-dimensional gas chromatography with time-of-flight mass spectrometry. J. Chromatogr. A 2020, 1634, 461654. [Google Scholar] [CrossRef]
- Haggarty, J.; Burgess, K.E. Recent advances in liquid and gas chromatography methodology for extending coverage of the metabolome. Curr. Opin. Biotechnol. 2017, 43, 77–85. [Google Scholar] [CrossRef]
- Yuan, F.; Kim, S.; Yin, X.; Zhang, X.; Kato, I. Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study. Metabolites 2020, 10, 343. [Google Scholar] [CrossRef]
- Beckstrom, A.C.; Tanya, P.; Humston, E.M.; Snyder, L.R.; Synovec, R.E.; Juul, S.E. The perinatal transition of the circulating metabolome in a nonhuman primate. Pediatr. Res. 2012, 71, 338–344. [Google Scholar] [CrossRef]
- Winnike, J.H.; Wei, X.; Knagge, K.J.; Colman, S.D.; Gregory, S.G.; Zhang, X. Comparison of GC-MS and GC×GC-MS in the analysis of human serum samples for biomarker discovery. J. Proteome Res. 2015, 14, 1810–1817. [Google Scholar] [CrossRef]
- Dallüge, J.; Beens, J.; Brinkman, U.A.T. Comprehensive two-dimensional gas chromatography: A powerful and versatile analytical tool. J. Chromatogr. A 2003, 1000, 69–108. [Google Scholar] [CrossRef]
- Górecki, T.; Harynuk, J.; Panić, O. The evolution of comprehensive two-dimensional gas chromatography (GC × GC). J. Sep. Sci. 2004, 27, 359–379. [Google Scholar] [CrossRef]
- Prodhan, M.A.I.; Shi, B.; Song, M.; He, L.; Yuan, F.; Yin, X.; Bohman, P.; McClain, C.J.; Zhang, X. Integrating comprehensive two-dimensional gas chromatography mass spectrometry and parallel two-dimensional liquid chromatography mass spectrometry for untargeted metabolomics. Analyst 2019, 144, 4331–4341. [Google Scholar] [CrossRef]
- Hummel, J.; Strehmel, N.; Bölling, C.; Schmidt, S.; Walther, D.; Kopka, J. Mass Spectral Search and Analysis Using the Golm Metabolome Database. In The Handbook of Plant Metabolomics; Wiley-VCH: Weinheim, Germany, 2013; Chapter 18; pp. 321–343. [Google Scholar] [CrossRef]
- Oberacher, H.; Sasse, M.; Antignac, J.P.; Guitton, Y.; Debrauwer, L.; Jamin, E.L.; Schulze, T.; Krauss, M.; Covaci, A.; Caballero-Casero, N.; et al. A European proposal for quality control and quality assurance of tandem mass spectral libraries. Environ. Sci. Eur. 2020, 32, 43. [Google Scholar] [CrossRef]
- NIST Standard Reference Database 1A. Available online: https://www.nist.gov/srd/nist-standard-reference-database-1a (accessed on 23 November 2021).
- Montenegro-Burke, J.R.; Guijas, C.; Siuzdak, G. METLIN: A Tandem Mass Spectral Library of Standards. Methods Mol. Biol. 2020, 2104, 149–163. [Google Scholar] [CrossRef]
- Van Renterghem, P.; Viaene, W.; Van Gansbeke, W.; Barrabin, J.; Iannone, M.; Polet, M.; T’Sjoen, G.; Deventer, K.; Van Eenoo, P. Validation of an ultra-sensitive detection method for steroid esters in plasma for doping analysis using positive chemical ionization GC-MS/MS. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2020, 1141, 122026. [Google Scholar] [CrossRef]
- Chobanyan, K.; Mitschke, A.; Gutzki, F.M.; Stichtenoth, D.O.; Tsikas, D. Accurate quantification of dimethylamine (DMA) in human plasma and serum by GC-MS and GC-tandem MS as pentafluorobenzamide derivative in the positive-ion chemical ionization mode. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 851, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Špánik, I.; Machyňáková, A. Recent applications of gas chromatography with high-resolution mass spectrometry. J. Sep. Sci. 2018, 41, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D.; Zoerner, A.A. Analysis of eicosanoids by LC-MS/MS and GC-MS/MS: A historical retrospect and a discussion. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014, 964, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D.; Hanff, E.; Kayacelebi, A.A.; Böhmer, A. Gas chromatographic-mass spectrometric analysis of the tripeptide glutathione in the electron-capture negative-ion chemical ionization mode. Amino Acids 2016, 48, 593–598. [Google Scholar] [CrossRef]
- Warren, C.R. Use of chemical ionization for GC-MS metabolite profiling. Metabolomics 2013, 9, 110–120. [Google Scholar] [CrossRef]
- Lisec, J.; Hoffmann, F.; Schmitt, C.; Jaeger, C. Extending the Dynamic Range in Metabolomics Experiments by Automatic Correction of Peaks Exceeding the Detection Limit. Anal. Chem. 2016, 88, 7487–7492. [Google Scholar] [CrossRef]
- Turner, M.A.; Guallar-Hoyas, C.; Kent, A.L.; Wilson, I.D.; Thomas, C.L. Comparison of metabolomic profiles obtained using chemical ionization and electron ionization MS in exhaled breath. Bioanalysis 2011, 3, 2731–2738. [Google Scholar] [CrossRef]
- Misra, B.B.; Olivier, M. High Resolution GC-Orbitrap-MS Metabolomics Using Both Electron Ionization and Chemical Ionization for Analysis of Human Plasma. J. Proteome Res. 2020, 19, 2717–2731. [Google Scholar] [CrossRef]
- Wachsmuth, C.J.; Hahn, T.A.; Oefner, P.J.; Dettmer, K. Enhanced metabolite profiling using a redesigned atmospheric pressure chemical ionization source for gas chromatography coupled to high-resolution time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2015, 407. [Google Scholar] [CrossRef]
- Roboz, J. A History of Ion Current Detectors for Mass Spectrometry. In Volume 9: Historical Perspectives, Part A: The Development of Mass Spectrometry of The Encyclopedia of Mass Spectrometry; Elsevier: Amsterdam, The Netherlands, 2016; pp. 183–188. [Google Scholar] [CrossRef]
- Kirchner, M.; Matisová, E.; Hrouzková, S.; De Zeeuw, J. Possibilities and limitations of quadrupole mass spectrometric detector in fast gas chromatography. J. Chromatogr. A 2005, 1090, 126–132. [Google Scholar] [CrossRef]
- Purcaro, G.; Tranchida, P.Q.; Ragonese, C.; Conte, L.; Dugo, P.; Dugo, G.; Mondello, L. Evaluation of a rapid-scanning quadrupole mass spectrometer in an apolar × ionic-liquid comprehensive two-dimensional gas chromatography system. Anal. Chem. 2010, 82, 8583–8590. [Google Scholar] [CrossRef] [PubMed]
- Schwaiger-Haber, M.; Stancliffe, E.; Arends, V.; Thyagarajan, B.; Sindelar, M.; Patti, G.J. A Workflow to Perform Targeted Metabolomics at the Untargeted Scale on a Triple Quadrupole Mass Spectrometer. ACS Meas. Sci. Au 2021, 1, 35–45. [Google Scholar] [CrossRef]
- Jayasinghe, N.S.; Mendis, H.; Roessner, U.; Dias, D.A. Quantification of Sugars and Organic Acids in Biological Matrices Using GC-QqQ-MS. Methods Mol. Biol. 2018, 1778, 207–223. [Google Scholar] [CrossRef]
- Szpot, P.; Wachełko, O.; Zawadzki, M. Application of ultra-sensitive GC-QqQ-MS/MS (MRM) method for the determination of diclofenac in whole blood samples without derivatization. J. Chromatogr. B 2021, 1179, 122860. [Google Scholar] [CrossRef]
- Balogh, M.P. Debating Resolutiom and Mass Accuracy. LC-GC Eur. 2004, 17, 152–159. [Google Scholar]
- Toribio-Delgado, A.F.; Maynar-Mariño, M.; Caballero-Loscos, M.J.; Robles-Gil, M.C.; Olcina-Camacho, G.J.; Maynar-Mariño, J.I. Qualification and Quantification of Seventeen Natural Steroids in Plasma by GC–Q-MS and GC-IT–MS/MS. J. Chromatogr. Sci. 2012, 50, 349–357. [Google Scholar] [CrossRef]
- Eliuk, S.; Makarov, A. Evolution of Orbitrap Mass Spectrometry Instrumentation. Annu. Rev. Anal. Chem. 2015, 8, 61–80. [Google Scholar] [CrossRef]
- Makarov, A. Orbitrap journey: Taming the ion rings. Nat. Commun. 2019, 10, 3743. [Google Scholar] [CrossRef] [PubMed]
- Stettin, D.; Poulin, R.X.; Pohnert, G. Metabolomics Benefits from Orbitrap GC-MS-Comparison of Low- and High-Resolution GC-MS. Metabolites 2020, 10, 143. [Google Scholar] [CrossRef]
- Misra, B.B.; Bassey, E.; Bishop, A.C.; Kusel, D.T.; Cox, L.A.; Olivier, M. High-resolution gas chromatography/mass spectrometry metabolomics of non-human primate serum. Rapid Commun. Mass Spectrom. 2018, 32, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, E.N.; Kostyukevich, Y.I.; Vladimirov, G.N. Fourier transform ion cyclotron resonance (FT ICR) mass spectrometry: Theory and simulations. Mass Spectrom. Rev. 2016, 35, 219–258. [Google Scholar] [CrossRef] [PubMed]
- Ghaste, M.; Mistrik, R.; Shulaev, V. Applications of Fourier Transform Ion Cyclotron Resonance (FT-ICR) and Orbitrap Based High Resolution Mass Spectrometry in Metabolomics and Lipidomics. Int. J. Mol. Sci. 2016, 17, 816. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Heffner, C.; Solouki, T. Multidimensional GC-fourier transform ion cyclotron resonance MS analyses: Utilizing gas-phase basicities to characterize multicomponent gasoline samples. J. Chromatogr. Sci. 2009, 47, 75–82. [Google Scholar] [CrossRef][Green Version]
- Liu, T.; Li, R.; Cui, Y.; Yu, Z.; Zhao, Y. Metabonomic analysis of plasma biochemical changes in pyrexia rats after treatment with Gegenqinlian decoction, aspirin and itraconazole by UHPLC-FT-ICR-MS. J. Pharm. Anal. 2020, 10, 581–587. [Google Scholar] [CrossRef]
- Junot, C.; Madalinski, G.; Tabet, J.C.; Ezan, E. Fourier transform mass spectrometry for metabolome analysis. Analyst 2010, 135, 2203–2219. [Google Scholar] [CrossRef]
- Han, J.; Danell, R.M.; Patel, J.R.; Gumerov, D.R.; Scarlett, C.O.; Speir, J.P.; Parker, C.E.; Rusyn, I.; Zeisel, S.; Borchers, C.H. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics 2008, 4, 128–140. [Google Scholar] [CrossRef]
- Macherone, A. The Future of GC/Q-TOF in Environmental Analysis. Compr. Anal. Chem. 2013, 61, 471–490. [Google Scholar] [CrossRef]
- Peterson, A.C.; Balloon, A.J.; Westphall, M.S.; Coon, J.J. Development of a GC/quadrupole-orbitrap mass spectrometer, part II: New approaches for discovery metabolomics. Anal. Chem. 2014, 86, 10044–10051. [Google Scholar] [CrossRef]
- Understanding Your Agilent ChemStation. Available online: https://www.agilent.com/cs/library/usermanuals/Public/G2070-91126_Understanding.pdf (accessed on 24 November 2021).
- MassLynx Mass Spectrometry Software. Available online: https://www.waters.com/waters/en_US/MassLynx-Mass-Spectrometry-Software-/nav.htm?locale=en_US&cid=513164 (accessed on 23 November 2021).
- ChromaTOF® Software. Available online: https://www.leco.com/product/chromatof-software (accessed on 23 November 2021).
- Compound Discoverer Software. Available online: https://www.thermofisher.com/ru/ru/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/compound-discoverer-software.html (accessed on 23 November 2021).
- O’Callaghan, S.; De Souza, D.P.; Isaac, A.; Wang, Q.; Hodkinson, L.; Olshansky, M.; Erwin, T.; Appelbe, B.; Tull, D.L.; Roessner, U.; et al. PyMS: A Python toolkit for processing of gas chromatography-mass spectrometry (GC-MS) data. Application and comparative study of selected tools. BMC Bioinform. 2012, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 1999, 10, 770–781. [Google Scholar] [CrossRef]
- Aggio, R.; Villas-Bôas, S.G.; Ruggiero, K. Metab: An R package for high-throughput analysis of metabolomics data generated by GC-MS. Bioinformatics 2011, 27, 2316–2318. [Google Scholar] [CrossRef]
- Behrends, V.; Tredwell, G.D.; Bundy, J.G. A software complement to AMDIS for processing GC-MS metabolomic data. Anal. Biochem. 2011, 415, 206–208. [Google Scholar] [CrossRef]
- Grapp, M.; Maurer, H.H.; Desel, H. Systematic forensic toxicological analysis by GC-MS in serum using automated mass spectral deconvolution and identification system. Drug Test. Anal. 2016, 8, 816–825. [Google Scholar] [CrossRef]
- Hiller, K.; Hangebrauk, J.; Jäger, C.; Spura, J.; Schreiber, K.; Schomburg, D. MetaboliteDetector: Comprehensive Analysis Tool for Targeted and Nontargeted GC/MS Based Metabolome Analysis. Anal. Chem. 2009, 81, 3429–3439. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Almenara, X.; Siuzdak, G. Metabolomics Data Processing Using XCMS. Methods Mol. Biol. 2020, 2104, 11–24. [Google Scholar] [CrossRef]
- Wehrens, R.; Weingart, G.; Mattivi, F. metaMS: An open-source pipeline for GC-MS-based untargeted metabolomics. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2014, 966, 109–116. [Google Scholar] [CrossRef]
- Lapierre, N.; Alser, M.; Eskin, E.; Koslicki, D.; Mangul, S. Metalign: Efficient alignment-based metagenomic profiling via containment min hash. Genome Biol. 2020, 21, 242. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Yao, L.; Sheflin, A.M.; Broeckling, C.D.; Prenni, J.E. Data Processing for GC-MS- and LC-MS-Based Untargeted Metabolomics. Methods Mol. Biol. 2019, 1978, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Balashova, E.E.; Lokhov, P.G.; Ponomarenko, E.A.; Markin, S.S.; Lisitsa, A.V.; Archakov, A.I. Metabolomic diagnostics and human digital image. Pers. Med. 2019, 16, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010, 2, 23–60. [Google Scholar] [CrossRef] [PubMed]
- Boccard, J.; Veuthey, J.L.; Rudaz, S. Knowledge discovery in metabolomics: An overview of MS data handling. J. Sep. Sci. 2010, 33, 290–304. [Google Scholar] [CrossRef]
- Manfredi, M.; Conte, E.; Barberis, E.; Buzzi, A.; Robotti, E.; Caneparo, V.; Cecconi, D.; Brandi, J.; Vanni, E.; Finocchiaro, M.; et al. Integrated serum proteins and fatty acids analysis for putative biomarker discovery in inflammatory bowel disease. J. Proteomics 2019, 195, 138–149. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 11. [Google Scholar] [CrossRef]
- Canzler, S.; Hackermüller, J. multiGSEA: A GSEA-based pathway enrichment analysis for multi-omics data. BMC Bioinform. 2020, 21, 561. [Google Scholar] [CrossRef]
- Jahagirdar, S.; Saccenti, E. Evaluation of Single Sample Network Inference Methods for Metabolomics-Based Systems Medicine. J. Proteome Res. 2020, 20, 932–949. [Google Scholar] [CrossRef]
- Pomyen, Y.; Wanichthanarak, K.; Poungsombat, P.; Fahrmann, J.; Grapov, D.; Khoomrung, S. Deep metabolome: Applications of deep learning in metabolomics. Comput. Struct. Biotechnol. J. 2020, 18, 2818–2825. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Pujos-Guillot, E.; Comte, B.; De Miranda, J.L.; Spiwok, V.; Chorbev, I.; Castiglione, F.; Tieri, P.; Watterson, S.; McAllister, R.; et al. Deep learning in systems medicine. Brief. Bioinform. 2021, 22, 1543–1559. [Google Scholar] [CrossRef]
- Kantz, E.D.; Tiwari, S.; Watrous, J.D.; Cheng, S.; Jain, M. Deep Neural Networks for Classification of LC-MS Spectral Peaks. Anal. Chem. 2019, 91, 12407–12413. [Google Scholar] [CrossRef]
- Mendez, K.M.; Broadhurst, D.I.; Reinke, S.N. The application of artificial neural networks in metabolomics: A historical perspective. Metabolomics 2019, 15, 142. [Google Scholar] [CrossRef]
- Da Silva, R.R.; Dorrestein, P.C.; Quinn, R.A. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.A.H. Illuminating the dark metabolome to advance the molecular characterisation of biological systems. Metabolomics 2018, 14, 101. [Google Scholar] [CrossRef]
- Ji, H.; Xu, Y.; Lu, H.; Zhang, Z. Deep MS/MS-Aided Structural-Similarity Scoring for Unknown Metabolite Identification. Anal. Chem. 2019, 91, 5629–5637. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, R.; Xiong, X.; Yin, Y.; Cai, Y.; Ma, Z.; Liu, N.; Zhu, Z.J. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 2019, 10, 1516. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef]
- Wishart, D.S. Proteomics and the Human Metabolome Project. Expert Rev. Proteomics 2014, 4, 333–335. [Google Scholar] [CrossRef]
- Wishart, D. Systems Biology Resources Arising from the Human Metabolome Project. In Genetics Meets Metabolomics; Springer: New York, NY, USA, 2012; pp. 157–175. [Google Scholar] [CrossRef]
- Costa dos Santos, G.; Renovato-Martins, M.; de Brito, N.M. The remodel of the “central dogma”: A metabolomics interaction perspective. Metabolomics 2021, 17, 48. [Google Scholar] [CrossRef]
- Suhre, K.; Zaghlool, S. Connecting the epigenome, metabolome and proteome for a deeper understanding of disease. J. Intern. Med. 2021, 290, 527–548. [Google Scholar] [CrossRef] [PubMed]
- A Table of All Published GWAS with Metabolomics—Human Metabolic Individuality. Available online: http://www.metabolomix.com/list-of-all-published-gwas-with-metabolomics/ (accessed on 23 November 2021).
- Koshiba, S.; Motoike, I.; Saigusa, D.; Inoue, J.; Shirota, M.; Katoh, Y.; Katsuoka, F.; Danjoh, I.; Hozawa, A.; Kuriyama, S.; et al. Omics research project on prospective cohort studies from the Tohoku Medical Megabank Project. Genes Cells 2018, 23, 406–417. [Google Scholar] [CrossRef]
- Tsepilov, Y.A.; Sharapov, S.Z.; Zaytseva, O.O.; Krumsek, J.; Prehn, C.; Adamski, J.; Kastenmüller, G.; Wang-Sattler, R.; Strauch, K.; Gieger, C.; et al. A network-based conditional genetic association analysis of the human metabolome. Gigascience 2018, 7. [Google Scholar] [CrossRef]
- Cheng, Y.; Schlosser, P.; Hertel, J.; Sekula, P.; Oefner, P.J.; Spiekerkoetter, U.; Mielke, J.; Freitag, D.F.; Schmidts, M.; Oefner, P.J.; et al. Rare genetic variants affecting urine metabolite levels link population variation to inborn errors of metabolism. Nat. Commun. 2021, 12, 964. [Google Scholar] [CrossRef] [PubMed]
- Grassin-Delyle, S.; Roquencourt, C.; Moine, P.; Saffroy, G.; Carn, S.; Heming, N.; Fleuriet, J.; Salvator, H.; Naline, E.; Couderc, L.J.; et al. Metabolomics of exhaled breath in critically ill COVID-19 patients: A pilot study. EBioMedicine 2021, 63, 103154. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef] [PubMed]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems Metabolic Engineering Strategies: Integrating Systems and Synthetic Biology with Metabolic Engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [CrossRef]
- Mahajan, U.M.; Alnatsha, A.; Li, Q.; Oehrle, B.; Weiss, F.U.; Sendler, M.; Distler, M.; Uhl, W.; Fahlbusch, T.; Goni, E.; et al. Plasma Metabolome Profiling Identifies Metabolic Subtypes of Pancreatic Ductal Adenocarcinoma. Cells 2021, 10, 1821. [Google Scholar] [CrossRef] [PubMed]
- Dörr, J.R.; Yu, Y.; Milanovic, M.; Beuster, G.; Zasada, C.; Däbritz, J.H.M.; Lisec, J.; Lenze, D.; Gerhardt, A.; Schleicher, K.; et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 2013, 501, 421–425. [Google Scholar] [CrossRef]
- Smith, A.M.; Natowicz, M.R.; Braas, D.; Ludwig, M.A.; Ney, D.M.; Donley, E.L.R.; Burrier, R.E.; Amaral, D.G. A Metabolomics Approach to Screening for Autism Risk in the Children’s Autism Metabolome Project. Autism Res. 2020, 13, 1270–1285. [Google Scholar] [CrossRef]
- Perng, W.; Rifas-Shiman, S.L.; Sordillo, J.; Hivert, M.F.; Oken, E. Metabolomic Profiles of Overweight/Obesity Phenotypes During Adolescence: A Cross-Sectional Study in Project Viva. Obesity 2020, 28, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Considine, E.C. The Search for Clinically Useful Biomarkers of Complex Disease: A Data Analysis Perspective. Metabolites 2019, 9, 126. [Google Scholar] [CrossRef]
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.D.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Jarmusch, A.K.; Wang, M.; Aceves, C.M.; Advani, R.S.; Aguirre, S.; Aksenov, A.A.; Aleti, G.; Aron, A.T.; Bauermeister, A.; Bolleddu, S.; et al. ReDU: A framework to find and reanalyze public mass spectrometry data. Nat. Methods 2020, 17, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Spicer, R.A.; Salek, R.; Steinbeck, C. A decade after the metabolomics standards initiative it’s time for a revision. Sci. Data 2017, 4, 170138. [Google Scholar] [CrossRef]
- Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Mass spectrometry-based metabolomics diagnostics—Myth or reality? Expert Rev. Proteom. 2021, 18, 7–12. [Google Scholar] [CrossRef]
- Shishkova, E.; Hebert, A.S.; Coon, J.J. Now, More Than Ever, Proteomics Needs Better Chromatography. Cell Syst. 2016, 3, 321–324. [Google Scholar] [CrossRef]
- Metabolon—Enlightening Life. Available online: https://www.metabolon.com/ (accessed on 23 November 2021).
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Jendoubi, T. Approaches to Integrating Metabolomics and Multi-Omics Data: A Primer. Metabolites 2021, 11, 184. [Google Scholar] [CrossRef] [PubMed]


| Technique | Major Strengths | Major Limitations | Major Detectable Compounds |
|---|---|---|---|
| GC-MS | Efficient and reproducible chromatography separation Comprehensive mass spectral libraries | Labor-intensive, time-consuming and varying sample preparation procedure Complicated identification of unknown compounds | Volatile and thermo-stable compound Carbohydrates Esters Sterols Steroids Eicosanoids Fatty acids Aminoacids Organic acids Nucleotides and nucleosides Lipids Non-volatile and thermo-labile compounds |
| LC-MS | Broad range of compounds (including polar, bulky, and thermo-labile metabolites) can be analyzed without derivatization High throughput | Possibility of ion aberrations resulting from a sample matrix Lack of spectral libraries for identification of metabolites | |
| NMR | Non-destructive analysis High reproducibility Simple or even absent sample preparation | Low sensitivity Relatively high sample volume High cost of apparatus | Carbohydrates Amines Aminoacids and organic acids Bulky molecules |
| Mass Analyzer | Resolution | Mass Range (Da) | Acquisition Speed | Major Benefits | Major Limitations |
|---|---|---|---|---|---|
| Quadrupole | ~1000 | 50–6000 | Medium | Highly selective Well suited for pairing with GC Relatively cheap Compact | Low resolution Narrow mass range |
| Ion trap | ~1000 | 50–4000 | Medium | Compact Relatively cheap Highly sensitive | Narrow dynamic range Limited resolution Requires pulsed introduction to MS |
| FT-ICR | over 1,000,000 | 10–10,000 | Slow | High sensitivity High reproducibility High resolving power Wide dynamic range | Expensive and bulky Slow scanning Specific coupling with chromatography systems |
| Orbitrap | up to 240,000 | 40–4000 | Slow | High resolution Compact and elegant solution | Narrow mass range |
| TOF | up to 60,000 | 20–500,000 | Fast | Highly sensitive Wide mass range Fast scanning Well suited for pairing with GC | Requires pulsed introduction to MS Requires fast solutions for data acquisition |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiseleva, O.; Kurbatov, I.; Ilgisonis, E.; Poverennaya, E. Defining Blood Plasma and Serum Metabolome by GC-MS. Metabolites 2022, 12, 15. https://doi.org/10.3390/metabo12010015
Kiseleva O, Kurbatov I, Ilgisonis E, Poverennaya E. Defining Blood Plasma and Serum Metabolome by GC-MS. Metabolites. 2022; 12(1):15. https://doi.org/10.3390/metabo12010015
Chicago/Turabian StyleKiseleva, Olga, Ilya Kurbatov, Ekaterina Ilgisonis, and Ekaterina Poverennaya. 2022. "Defining Blood Plasma and Serum Metabolome by GC-MS" Metabolites 12, no. 1: 15. https://doi.org/10.3390/metabo12010015
APA StyleKiseleva, O., Kurbatov, I., Ilgisonis, E., & Poverennaya, E. (2022). Defining Blood Plasma and Serum Metabolome by GC-MS. Metabolites, 12(1), 15. https://doi.org/10.3390/metabo12010015







