Forming 4-Methylcatechol as the Dominant Bioavailable Metabolite of Intraruminal Rutin Inhibits p-Cresol Production in Dairy Cows
Abstract
:1. Introduction
2. Results
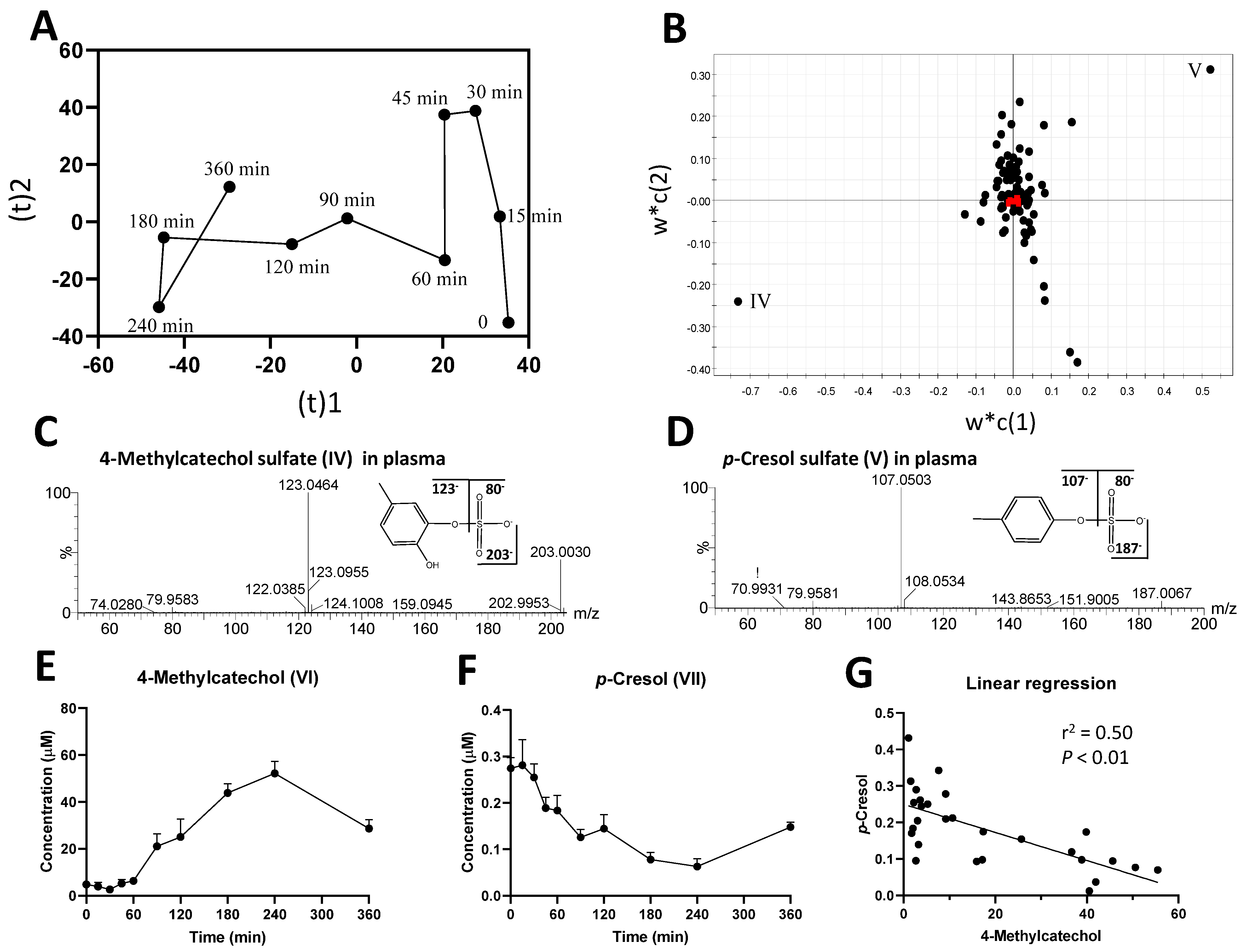
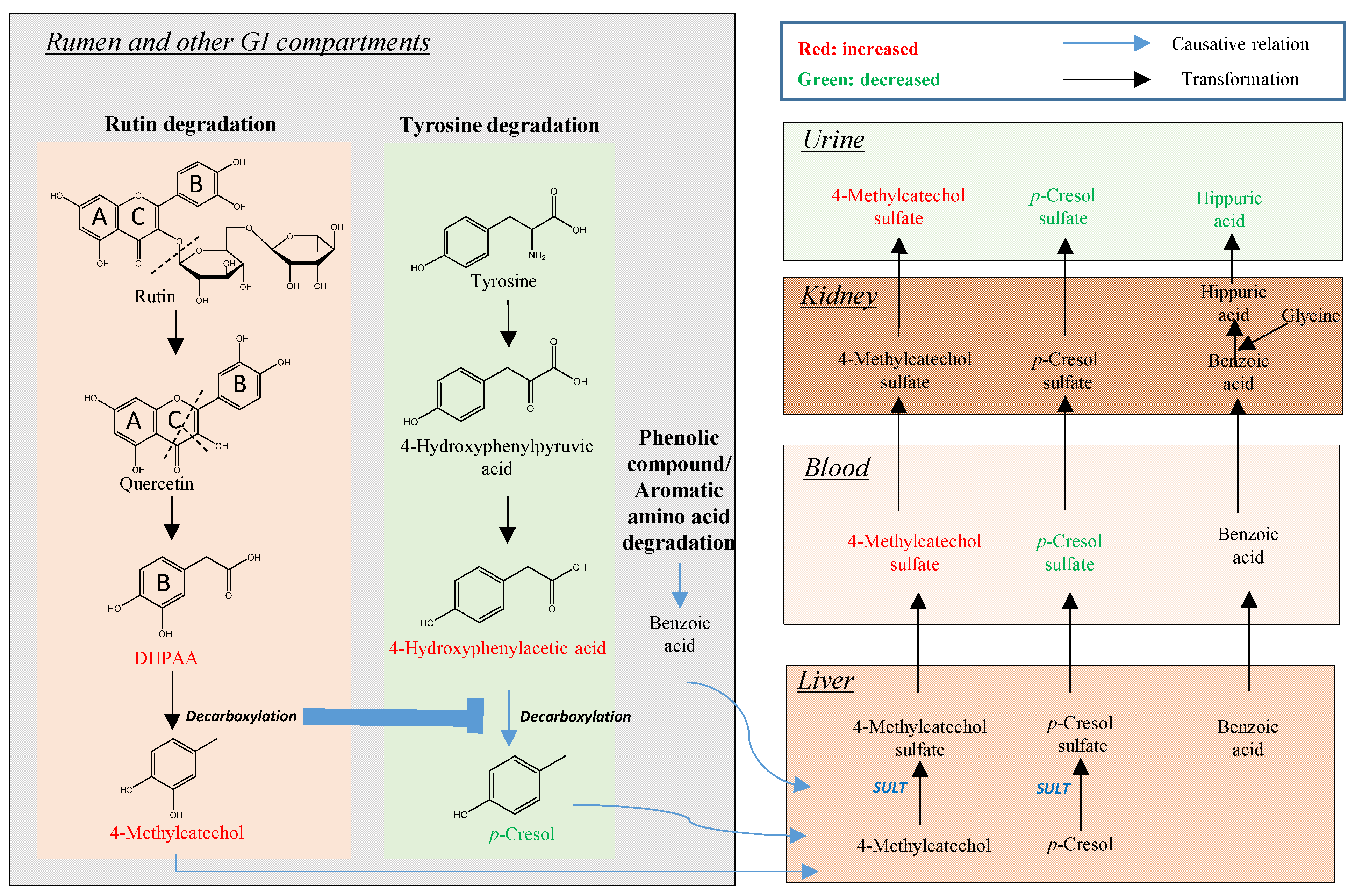
2.1. Identification of Plasma Metabolites Affected by Intraruminal Rutin
2.2. Identification of Urinary Metabolites Affected by Intraruminal Rutin
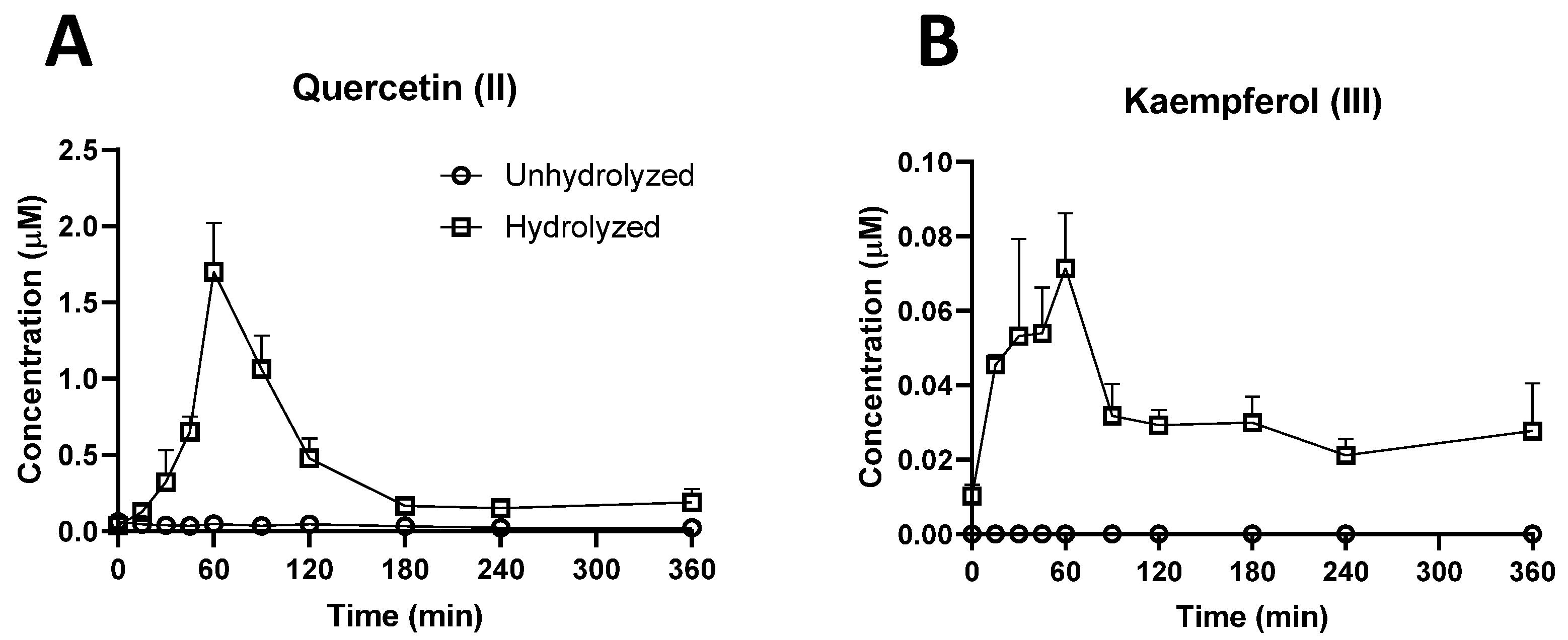
2.3. Investigation of Ruminal Degradation of Rutin and Quercetin
2.4. Influence of Rutin on Ruminal Tyrosine Metabolism
2.5. Ruminal Short-Chain Fatty Acids (SCFAs)
3. Discussion
3.1. 4-Methylcatechol as the Most Bioavailable Metabolite of Rutin and its Significance
3.1.1. Bioavailability of Rutin, Quercetin, and their Derivatives
3.1.2. Significance of 4-Methylcatechol as the Dominant Bioavailable Rutin Metabolite
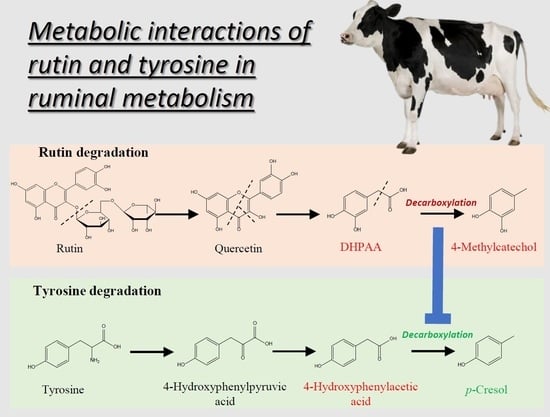
3.2. Rutin Degradation Pathway and its Interactions with Tyrosine Degradation in the Rumen
3.2.1. Rutin and the Bacteria Responsible for Rutin Degradation
3.2.2. Influence of Rutin Degradation on p-Cresol Production and its Potential Mechanism
3.2.3. Significance of Inhibiting p-Cresol Production
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animals, Experimental Design, and Sample Collection
4.3. Metabolites Extraction
4.4. Acid Hydrolysis of Conjugated Metabolites
4.5. Chemical Derivatization
4.6. LC-MS Analysis
4.7. Targeted/Quantitative Analysis
4.8. Untargeted Multivariate Data Analysis and Marker Characterization
4.9. Kinetic Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Middleton, E., Jr.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar]
- Erlund, I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Cui, K.; Guo, X.D.; Tu, Y.; Zhang, N.F.; Ma, T.; Diao, Q.Y. Effect of dietary supplementation of rutin on lactation performance, ruminal fermentation and metabolism in dairy cows. J. Anim. Physiol. Anim. Nutr. 2015, 99, 1065–1073. [Google Scholar] [CrossRef]
- Gruse, J.; Görs, S.; Tuchscherer, A.; Otten, W.; Weitzel, J.M.; Metges, C.C.; Wolffram, S.; Hammon, H.M. The Effects of Oral Quercetin Supplementation on Splanchnic Glucose Metabolism in 1-Week-Old Calves Depend on Diet after Birth. J. Nutr. 2015, 145, 2486–2495. [Google Scholar] [CrossRef] [Green Version]
- Stoldt, A.K.; Derno, M.; Nürnberg, G.; Weitzel, J.M.; Otten, W.; Starke, A.; Wolffram, S.; Metges, C.C. Effects of a 6-wk intraduodenal supplementation with quercetin on energy metabolism and indicators of liver damage in periparturient dairy cows. J. Dairy Sci. 2015, 98, 4509–4520. [Google Scholar] [CrossRef] [Green Version]
- Oskoueian, E.; Abdullah, N.; Oskoueian, A. Effects of Flavonoids on Rumen Fermentation Activity, Methane Production, and Microbial Population. BioMed Res. Int. 2013, 2013, 349129. [Google Scholar] [CrossRef] [Green Version]
- Maciej, J.; Schäff, C.T.; Kanitz, E.; Tuchscherer, A.; Bruckmaier, R.M.; Wolffram, S.; Hammon, H.M. Short communication: Effects of oral flavonoid supplementation on the metabolic and antioxidative status of newborn dairy calves. J. Dairy Sci. 2016, 99, 805–811. [Google Scholar] [CrossRef] [Green Version]
- Gohlke, A.; Ingelmann, C.J.; Nürnberg, G.; Weitzel, J.M.; Hammon, H.M.; Görs, S.; Starke, A.; Wolffram, S.; Metges, C.C. Influence of 4-week intraduodenal supplementation of quercetin on performance, glucose metabolism, and mRNA abundance of genes related to glucose metabolism and antioxidative status in dairy cows. J. Dairy Sci. 2013, 96, 6986–7000. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.M.; Hwang, M.K.; Lee, D.E.; Lee, K.W.; Lee, H.J. Protective Effect of Quercetin against Arsenite-Induced COX-2 Expression by Targeting PI3K in Rat Liver Epithelial Cells. J. Agric. Food Chem. 2010, 58, 5815–5820. [Google Scholar] [CrossRef]
- Endale, M.; Park, S.C.; Kim, S.; Kim, S.H.; Yang, Y.; Cho, J.Y.; Rhee, M.H. Quercetin disrupts tyrosine-phosphorylated phosphatidylinositol 3-kinase and myeloid differentiation factor-88 association, and inhibits MAPK/AP-1 and IKK/NF-κB-induced inflammatory mediators production in RAW 264.7 cells. Immunobiology 2013, 218, 1452–1467. [Google Scholar] [CrossRef]
- Muthian, G.; Bright, J.J. Quercetin, a flavonoid phytoestrogen, ameliorates experimental allergic encephalomyelitis by blocking IL-12 signaling through JAK-STAT pathway in T lymphocyte. J. Clin. Immunol. 2004, 24, 542–552. [Google Scholar] [CrossRef]
- Zhang, M.; Swarts, S.G.; Yin, L.; Liu, C.; Tian, Y.; Cao, Y.; Swarts, M.; Yang, S.; Zhang, S.B.; Zhang, K.; et al. Antioxidant properties of quercetin. Adv. Exp. Med. Biol. 2011, 701, 283–289. [Google Scholar] [CrossRef]
- Xu, D.; Hu, M.-J.; Wang, Y.-Q.; Cui, Y.-L. Antioxidant Activities of Quercetin and Its Complexes for Medicinal Application. Molecules 2019, 24, 1123. [Google Scholar] [CrossRef] [Green Version]
- Ader, P.; Wessmann, A.; Wolffram, S. Bioavailability and metabolism of the flavonol quercetin in the pig. Free. Radic. Biol. Med. 2000, 28, 1056–1067. [Google Scholar] [CrossRef]
- Reinboth, M.; Wolffram, S.; Abraham, G.; Ungemach, F.R.; Cermak, R. Oral bioavailability of quercetin from different quercetin glycosides in dogs. Br. J. Nutr. 2010, 104, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Bokkenheuser, V.D.; Shackleton, C.H.; Winter, J. Hydrolysis of dietary flavonoid glycosides by strains of intestinal Bacteroides from humans. Biochem. J. 1987, 248, 953–956. [Google Scholar] [CrossRef] [Green Version]
- Shin, K.-C.; Nam, H.-K.; Oh, D.-K. Hydrolysis of Flavanone Glycosides by β-Glucosidase from Pyrococcus furiosus and Its Application to the Production of Flavanone Aglycones from Citrus Extracts. J. Agric. Food Chem. 2013, 61, 11532–11540. [Google Scholar] [CrossRef]
- Berger, L.M.; Wein, S.; Blank, R.; Metges, C.C.; Wolffram, S. Bioavailability of the flavonol quercetin in cows after intraruminal application of quercetin aglycone and rutin. J. Dairy Sci. 2012, 95, 5047–5055. [Google Scholar] [CrossRef]
- Berger, L.M.; Blank, R.; Zorn, F.; Wein, S.; Metges, C.C.; Wolffram, S. Ruminal degradation of quercetin and its influence on fermentation in ruminants. J. Dairy Sci. 2015, 98, 5688–5698. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Li, Y.; Brobbey Oppong, M.; Qiu, F. Insights into the intestinal bacterial metabolism of flavonoids and the bioactivities of their microbe-derived ring cleavage metabolites. Drug Metab. Rev. 2018, 50, 343–356. [Google Scholar] [CrossRef]
- Feliciano, R.P.; Boeres, A.; Massacessi, L.; Istas, G.; Ventura, M.R.; Nunes dos Santos, C.; Heiss, C.; Rodriguez-Mateos, A. Identification and quantification of novel cranberry-derived plasma and urinary (poly)phenols. Arch. Biochem. Biophys. 2016, 599, 31–41. [Google Scholar] [CrossRef]
- Gläßer, G.; Graefe, E.U.; Struck, F.; Veit, M.; Gebhardt, R. Comparison of antioxidative capacities and inhibitory effects on cholesterol biosynthesis of quercetin and potential metabolites. Phytomedicine 2002, 9, 33–40. [Google Scholar] [CrossRef]
- Boots, A.W.; Haenen, G.R.M.M.; den Hartog, G.J.M.; Bast, A. Oxidative damage shifts from lipid peroxidation to thiol arylation by catechol-containing antioxidants. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2002, 1583, 279–284. [Google Scholar] [CrossRef]
- Bobe, G.; Young, J.W.; Beitz, D.C. Invited Review: Pathology, Etiology, Prevention, and Treatment of Fatty Liver in Dairy Cows. J. Dairy Sci. 2004, 87, 3105–3124. [Google Scholar] [CrossRef] [Green Version]
- Pourová, J.; Najmanová, I.; Vopršalová, M.; Migkos, T.; Pilařová, V.; Applová, L.; Nováková, L.; Mladěnka, P. Two flavonoid metabolites, 3,4-dihydroxyphenylacetic acid and 4-methylcatechol, relax arteries ex vivo and decrease blood pressure in vivo. Vascul. Pharmacol. 2018, 111, 36–43. [Google Scholar] [CrossRef]
- Applová, L.; Karlíčková, J.; Warncke, P.; Macáková, K.; Hrubša, M.; Macháček, M.; Tvrdý, V.; Fischer, D.; Mladěnka, P. 4-Methylcatechol, a Flavonoid Metabolite with Potent Antiplatelet Effects. Mol. Nutr. Food Res. 2019, 63, 1900261. [Google Scholar] [CrossRef]
- Yasuoka, M.M.; Monteiro, B.M.; Fantinato-Neto, P.; Paiano, R.B.; Fantoni, D.T.; Otsuki, D.A.; Birgel Junior, E.H. Transient Pulmonary Artery Hypertension in Holstein Neonate Calves. Animals 2020, 10, 2277. [Google Scholar] [CrossRef]
- Lübke, T.; Damme, M. Lysosomal sulfatases: A growing family*. Biochem. J. 2020, 477, 3963–3983. [Google Scholar] [CrossRef]
- Cheng, K.-J.; Jones, G.A.; Simpson, F.J.; Bryant, M.P. Isolation and identification of rumen bacteria capable of anaerobic rutin degradation. Can. J. Microbiol. 1969, 15, 1365–1371. [Google Scholar] [CrossRef]
- Bryant, M.P.; Small, N. The anaerobic monotrichous butyric acid-producing curved rod-shaped bacteria of the rumen. J. Bacteriol. 1956, 72, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurty, H.G.; Cheng, K.J.; Jones, G.A.; Simpson, F.J.; Watkin, J.E. Identification of products produced by the anaerobic degradation of rutin and related flavonoids by Butyrivibrio sp. C3. Can. J. Microbiol. 1970, 16, 759–767. [Google Scholar] [CrossRef]
- Winter, J.; Moore, L.H.; Dowell, V.R., Jr.; Bokkenheuser, V.D. C-ring cleavage of flavonoids by human intestinal bacteria. Appl. Environ. Microbiol. 1989, 55, 1203–1208. [Google Scholar] [CrossRef] [Green Version]
- Krumholz, L.R.; Crawford, R.L.; Hemling, M.E.; Bryant, M.P. Metabolism of gallate and phloroglucinol in Eubacterium oxidoreducens via 3-hydroxy-5-oxohexanoate. J. Bacteriol. 1987, 169, 1886–1890. [Google Scholar] [CrossRef] [Green Version]
- Simpson, F.J.; Jones, G.A.; Wolin, E.A. Anaerobic degradation of some bioflavonoids by microflora of the rumen. Can. J. Microbiol. 1969, 15, 972–974. [Google Scholar] [CrossRef]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 2018, 94, fiy125. [Google Scholar] [CrossRef]
- Roediger, W.E.; Babidge, W. Human colonocyte detoxification. Gut 1997, 41, 731–734. [Google Scholar] [CrossRef]
- Selmer, T.; Andrei, P.I. p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. Eur. J. Biochem. 2001, 268, 1363–1372. [Google Scholar] [CrossRef]
- Yokoyama, M.T.; Carlson, J.R. Production of Skatole and para-Cresol by a Rumen Lactobacillus sp. Appl. Environ. Microbiol. 1981, 41, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, N.; Arikawa, A.Y.; Chen, C. Inhibitory Effects of Green Tea Polyphenols on Microbial Metabolism of Aromatic Amino Acids in Humans Revealed by Metabolomic Analysis. Metabolites 2019, 9, 96. [Google Scholar] [CrossRef] [Green Version]
- Andriamihaja, M.; Lan, A.; Beaumont, M.; Audebert, M.; Wong, X.; Yamada, K.; Yin, Y.; Tomé, D.; Carrasco-Pozo, C.; Gotteland, M.; et al. The deleterious metabolic and genotoxic effects of the bacterial metabolite p-cresol on colonic epithelial cells. Free. Radic. Biol. Med. 2015, 85, 219–227. [Google Scholar] [CrossRef]
- Shiba, T.; Kawakami, K.; Sasaki, T.; Makino, I.; Kato, I.; Kobayashi, T.; Uchida, K.; Kaneko, K. Effects of intestinal bacteria-derived p-cresyl sulfate on Th1-type immune response in vivo and in vitro. Toxicol. Appl. Pharmacol. 2014, 274, 191–199. [Google Scholar] [CrossRef]
- Meijers, B.K.; Van Kerckhoven, S.; Verbeke, K.; Dehaen, W.; Vanrenterghem, Y.; Hoylaerts, M.F.; Evenepoel, P. The uremic retention solute p-cresyl sulfate and markers of endothelial damage. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2009, 54, 891–901. [Google Scholar] [CrossRef]
- Borhan, M.S.; Capareda, S.; Mukhtar, S.; Faulkner, W.B.; McGee, R.; Parnell, C.B. Comparison of seasonal phenol and p-cresol emissions from ground-level area sources in a dairy operation in central Texas. J. Air Waste Manag. Assoc. 2012, 62, 381–392. [Google Scholar] [CrossRef]
- Hertog, M.G.L.; Hollman, P.C.H.; Venema, D.P. Optimization of a quantitative HPLC determination of potentially anticarcinogenic flavonoids in vegetables and fruits. J. Agric. Food Chem. 1992, 40, 1591–1598. [Google Scholar] [CrossRef]
- Wang, L.; Urriola, P.E.; Luo, Z.H.; Rambo, Z.J.; Wilson, M.E.; Torrison, J.L.; Shurson, G.C.; Chen, C. Metabolomics revealed diurnal heat stress and zinc supplementation-induced changes in amino acid, lipid, and microbial metabolism. Physiol. Rep. 2016, 4, e12676. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, D.; Chen, C. 2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis. Metabolites 2013, 3, 993–1010. [Google Scholar] [CrossRef]
- Zhang, Y.; Huo, M.; Zhou, J.; Xie, S. PKSolver: An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 2010, 99, 306–314. [Google Scholar] [CrossRef]




| Ion | Sample | Mode of Ion Detection | m/z of Detected Ion | Identity | Formula | Δppm | Database ID |
|---|---|---|---|---|---|---|---|
| I | Plasma Rumen | (M − H)− | 609.1454 | Rutin | C27H30O16 | −0.3 | HMDB0003249 |
| II | Plasma Rumen | (M − H)− | 301.0351 | Quercetin | C15H10O7 | 1 | HMDB0005794 |
| III | Plasma | (M − H)− | 285.0361 | Kaempferol | C15H10O6 | −2 | HMDB0005801 |
| IV | Plasma Urine | (M − H)− | 203.0012 | 4-Methylcatechol sulfate | C7H8O5S | −0.9 | HMDB0240459 |
| V | Plasma Urine | (M − H)− | 187.0064 | p-Cresol sulfate | C7H8O4S | −0.5 | HMDB0011635 |
| VI | Plasma Urine Rumen | (M − H)− | 123.0448 | 4-Methylcatechol | C7H8O2 | −3 | HMDB0000873 |
| VII | Plasma Urine Rumen | (M + DC)− | 342.1166 | p-Cresol | C7H8O | 0.6 | HMDB0001858 |
| VIII | Urine | (M − H)− | 178.0506 | Hippuric acid | C9H9NO3 | 1 | HMDB0000714 |
| IX | Rumen | (M − H)− | 167.0343 | DHPAA | C8H8O4 | −0.6 | HMDB0001336 |
| IX’ | Rumen | (M − H)− | 123.0446 | DHPAA (fragment) | C7H8O2 | 0 | |
| X | Rumen | (M + DC)+ | 386.1061 | 4-Hydroxyphenylacetic acid | C8H8O3 | −0.3 | HMDB0060390 |
| XI | Rumen | (M + DC)+ | 648.1838 | Tyrosine | C9H11NO3 | 0.8 | HMDB0000158 |
| Parameter | Quercetin |
|---|---|
| AUC0–t (µmol/L × min) | 143.30 |
| Vd (L/kg) | 129.63 |
| t1/2 (min) | 12.24 |
| Cmax (µmol/L) | 1.70 |
| tmax (min) | 60 |
| Cl (L/min/kg) | 0.97 |
| Rutin | Quercetin | 3,4-dihydroxyphenylacetic Acid | 4-Methylcatechol | |
|---|---|---|---|---|
| t1/2 (min) | 10.89 | 14.62 | 60.65 | 48.22 |
| Cmax (µmol/mL) | 156.42 | 71.16 | 181.21 | 10.09 |
| tmax (min) | 30 | 60 | 120 | 120 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Weber, W.J.; Yao, D.; Caixeta, L.; Zimmerman, N.P.; Thompson, J.; Block, E.; Rehberger, T.G.; Crooker, B.A.; Chen, C. Forming 4-Methylcatechol as the Dominant Bioavailable Metabolite of Intraruminal Rutin Inhibits p-Cresol Production in Dairy Cows. Metabolites 2022, 12, 16. https://doi.org/10.3390/metabo12010016
Guo Y, Weber WJ, Yao D, Caixeta L, Zimmerman NP, Thompson J, Block E, Rehberger TG, Crooker BA, Chen C. Forming 4-Methylcatechol as the Dominant Bioavailable Metabolite of Intraruminal Rutin Inhibits p-Cresol Production in Dairy Cows. Metabolites. 2022; 12(1):16. https://doi.org/10.3390/metabo12010016
Chicago/Turabian StyleGuo, Yue, Wanda J. Weber, Dan Yao, Luciano Caixeta, Noah P. Zimmerman, Jesse Thompson, Elliot Block, Thomas G. Rehberger, Brian A. Crooker, and Chi Chen. 2022. "Forming 4-Methylcatechol as the Dominant Bioavailable Metabolite of Intraruminal Rutin Inhibits p-Cresol Production in Dairy Cows" Metabolites 12, no. 1: 16. https://doi.org/10.3390/metabo12010016
APA StyleGuo, Y., Weber, W. J., Yao, D., Caixeta, L., Zimmerman, N. P., Thompson, J., Block, E., Rehberger, T. G., Crooker, B. A., & Chen, C. (2022). Forming 4-Methylcatechol as the Dominant Bioavailable Metabolite of Intraruminal Rutin Inhibits p-Cresol Production in Dairy Cows. Metabolites, 12(1), 16. https://doi.org/10.3390/metabo12010016