Plasma Metabolomic Profiling in 1391 Subjects with Overweight and Obesity from the SPHERE Study
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. Metabolite Levels
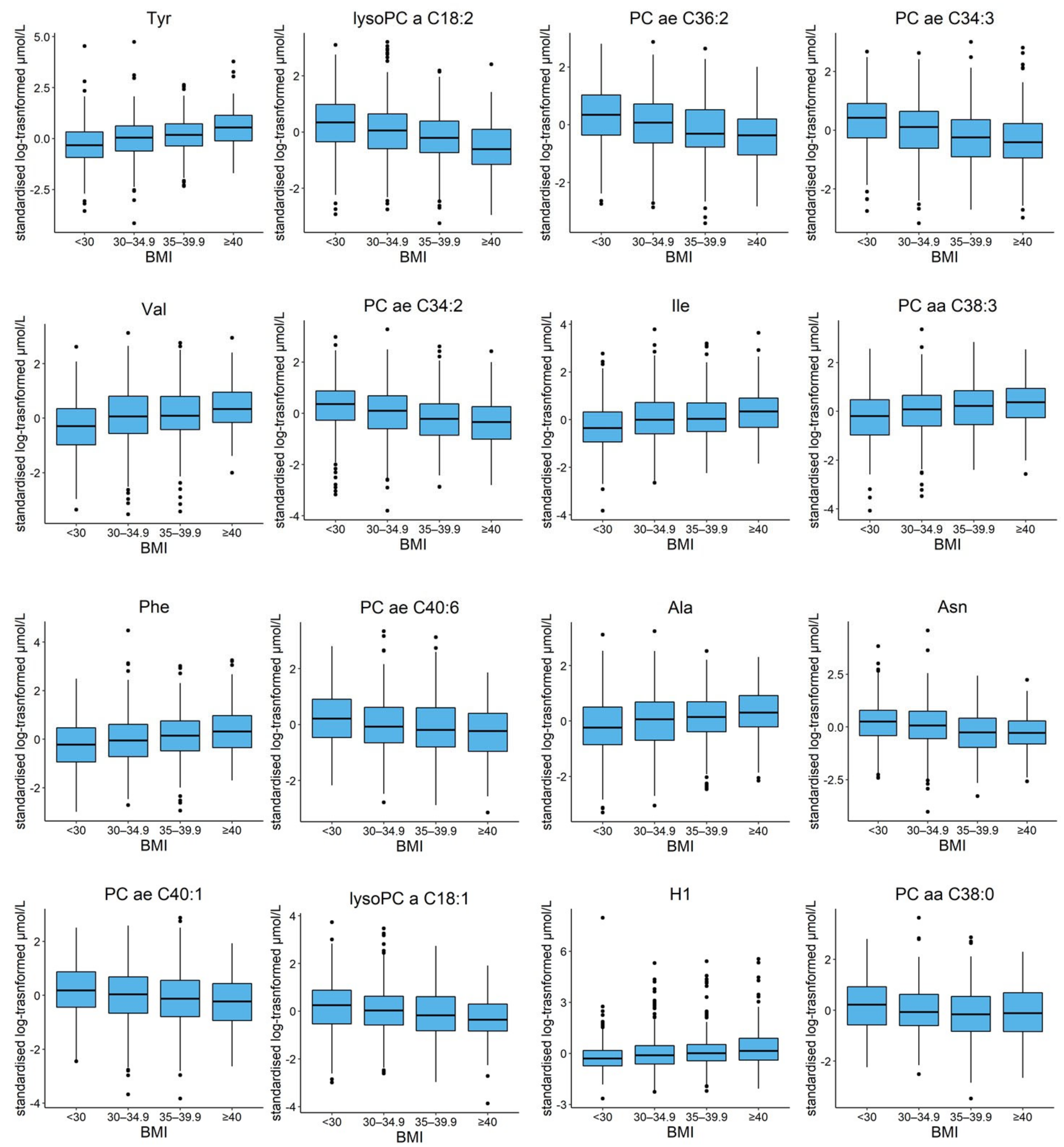
2.3. Impact of BMI on the Metabolomic Profile
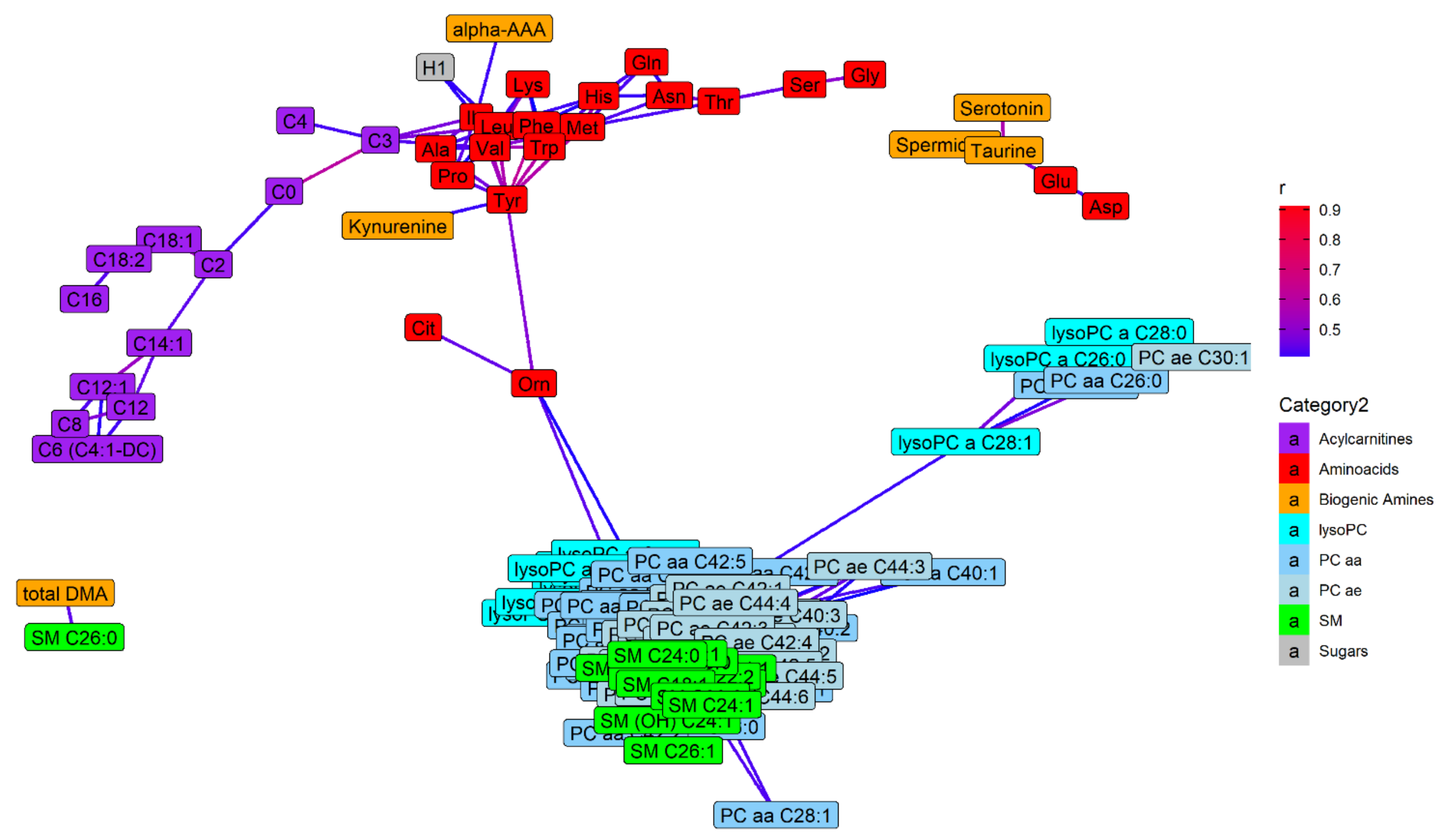
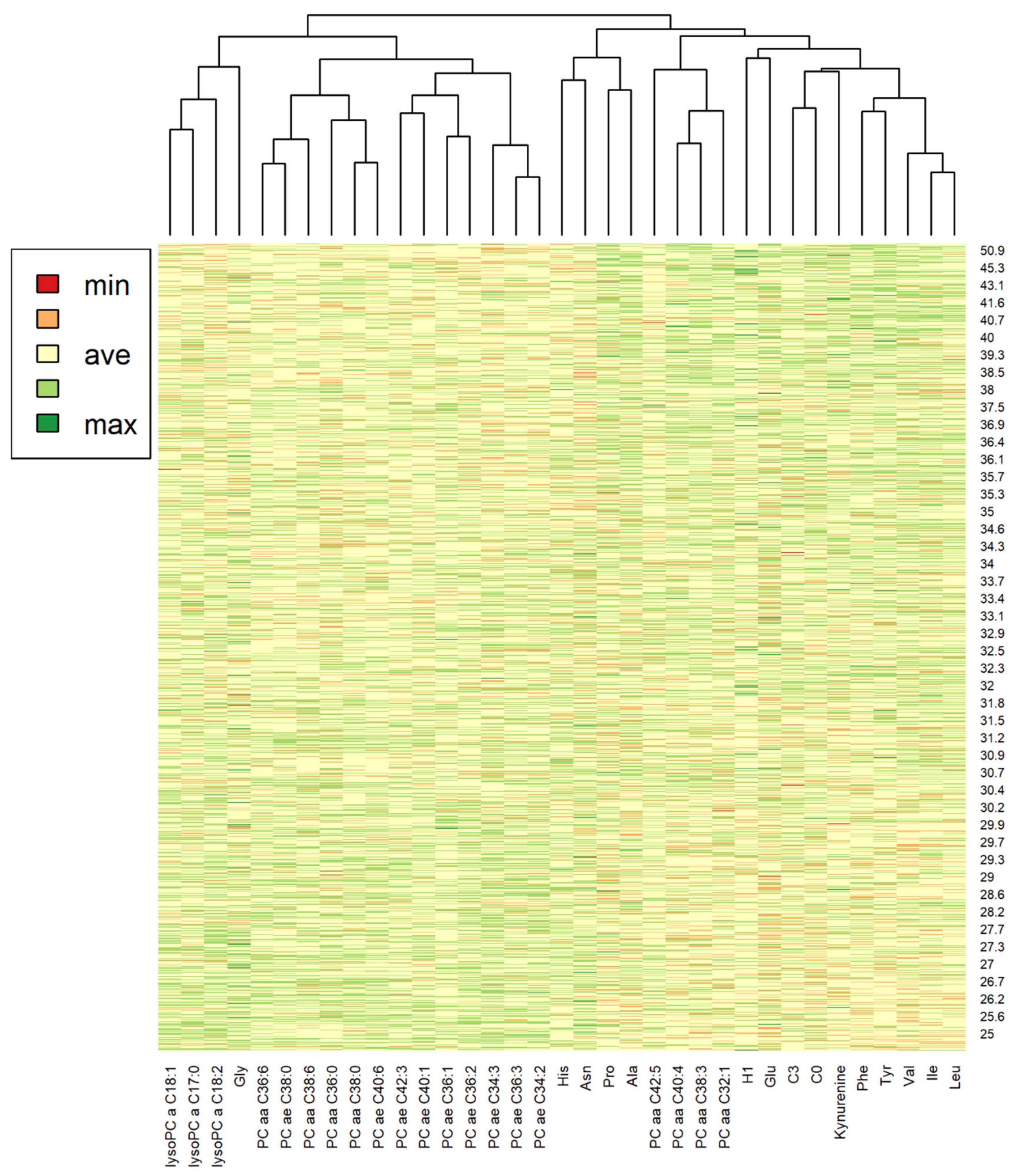
2.4. Metabolite Distributions and Correlations
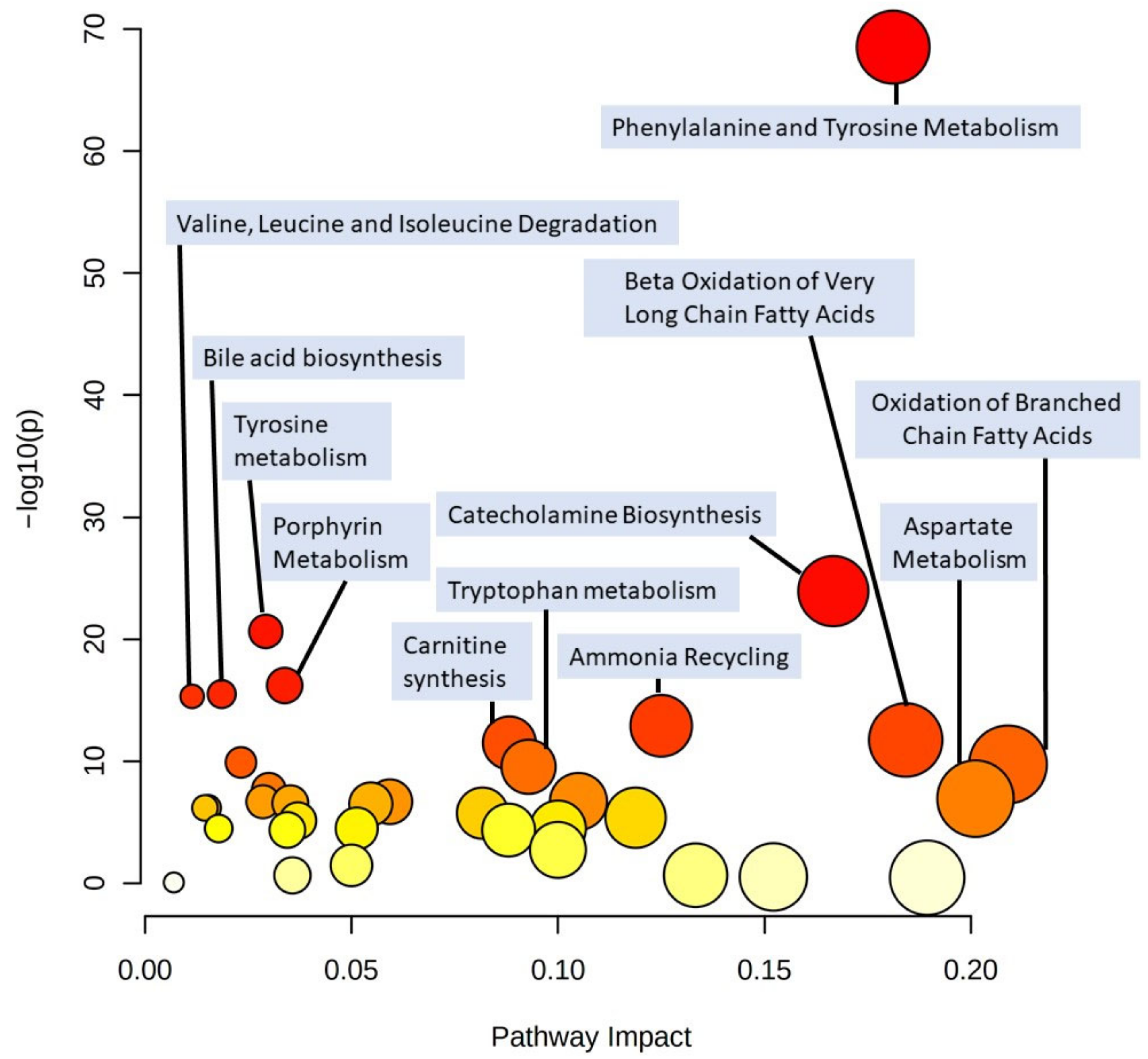
2.5. Possible Involved Biochemical Pathways
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Plasma Sample Collection
4.3. Metabolomic Analyses
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 17 December 2020).
- WHO. Body Mass Index-BMI. Available online: https://www.euro.who.int/en/health-topics/disease-prevention/nutrition/a-healthy-lifestyle/body-mass-index-bmi (accessed on 17 December 2020).
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Rangel-Huerta, O.D.; Pastor-Villaescusa, B.; Gil, A. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics 2019, 15, 93. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Iodice, S.; Favero, C.; Angelici, L.; Albetti, B.; Cacace, R.; Cantone, L.; Carugno, M.; Cavalleri, T.; De Giorgio, B.; et al. Susceptibility to particle health effects, miRNA and exosomes: Rationale and study protocol of the SPHERE study. BMC Public Health 2014, 14, 1137. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Lin, W.; Broadhurst, D.; Begley, P.; Brown, M.; Zelena, E.; Vaughan, A.A.; Halsall, A.; Harding, N.; Knowles, J.D.; et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics 2015, 11, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, J.Y.; Kim, O.Y.; Ham, B.M.; Kim, H.J.; Kwon, D.Y.; Jang, Y.; Lee, J.H. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS). J. Proteome Res. 2010, 9, 4368–4375. [Google Scholar] [CrossRef]
- Bagheri, M.; Farzadfar, F.; Qi, L.; Yekaninejad, M.S.; Chamari, M.; Zeleznik, O.A.; Kalantar, Z.; Ebrahimi, Z.; Sheidaie, A.; Koletzko, B.; et al. Obesity-related metabolomic profiles and discrimination of metabolically unhealthy obesity. J. Proteome Res. 2018, 17, 1452–1462. [Google Scholar] [CrossRef]
- Bagheri, M.; Djazayery, A.; Farzadfar, F.; Qi, L.; Yekaninejad, M.S.; Aslibekyan, S.; Chamari, M.; Hassani, H.; Koletzko, B.; Uhl, O. Plasma metabolomic profiling of amino acids and polar lipids in Iranian obese adults. Lipids Health Dis 2019, 18, 94. [Google Scholar] [CrossRef]
- Libert, D.M.; Nowacki, A.S.; Natowicz, M.R. Metabolomic analysis of obesity, metabolic syndrome, and type 2 diabetes: Amino acid and acylcarnitine levels change along a spectrum of metabolic wellness. PeerJ 2018, 6, e5410. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Guo, L.; Leon Swisher, C.; Shah, N.; Huang, L.; Napier, L.A.; Kirkness, E.F.; Spector, T.D.; Caskey, C.T.; Thorens, B.; et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019, 29, 488–500.e2. [Google Scholar] [CrossRef]
- Ho, J.E.; Larson, M.G.; Ghorbani, A.; Cheng, S.; Chen, M.H.; Keyes, M.; Rhee, E.P.; Clish, C.B.; Vasan, R.S.; Gerszten, R.E.; et al. Metabolomic profiles of body mass index in the framingham heart study reveal distinct cardiometabolic phenotypes. PLoS ONE 2016, 11, e0148361. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Ma, X.; Zhao, A.; Wang, C.; Zhang, Y.; Nieman, D.; Nicholson, J.K.; Jia, W.; Bao, Y. The metabolite profiles of the obese population are gender-dependent. J. Proteome Res. 2014, 13, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- Wang, S.M.; Yang, R.Y.; Wang, M.; Ji, F.S.; Li, H.X.; Tang, Y.M.; Chen, W.X.; Dong, J. Identification of serum metabolites associated with obesity and traditional risk factors for metabolic disease in Chinese adults. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Han, Q.; Liu, Y.; Sun, C.; Gang, X.; Wang, G. The relationship between branched-chain amino acid related metabolomic signature and insulin resistance: A systematic review. J. Diabetes Res. 2016, 2016, 2794591. [Google Scholar] [CrossRef]
- Newgard, C.B. Metabolomics and metabolic diseases: Where do we stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Van Horn, C.; Reid, T.; Hutson, S.M.; Cooney, R.N.; Lynch, C.J. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1552–E1563. [Google Scholar] [CrossRef]
- Adams, S.H. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv. Nutr. 2011, 2, 445–456. [Google Scholar] [CrossRef]
- Siddik, M.A.B.; Shin, A.C. Recent Progress on branched-chain amino acids in obesity, diabetes, and beyond. Endocrinol. Metab. 2019, 34, 234–246. [Google Scholar] [CrossRef]
- Melnik, B.C. Leucine signaling in the pathogenesis of type 2 diabetes and obesity. World J. Diabetes 2012, 3, 38–53. [Google Scholar] [CrossRef]
- Lent-Schochet, D.; McLaughlin, M.; Ramakrishnan, N.; Jialal, I. Exploratory metabolomics of metabolic syndrome: A status report. World J. Diabetes 2019, 10, 23–36. [Google Scholar] [CrossRef]
- Yu, H.T.; Fu, X.Y.; Xu, B.; Zuo, L.L.; Ma, H.B.; Wang, S.R. Untargeted metabolomics approach (UPLC-Q-TOF-MS) explores the biomarkers of serum and urine in overweight/obese young men. Asia Pac. J. Clin. Nutr. 2018, 27, 1067–1076. [Google Scholar] [CrossRef]
- Carayol, M.; Leitzmann, M.F.; Ferrari, P.; Zamora-Ros, R.; Achaintre, D.; Stepien, M.; Schmidt, J.A.; Travis, R.C.; Overvad, K.; Tjønneland, A.; et al. Blood metabolic signatures of body mass index: A targeted metabolomics study in the EPIC cohort. J. Proteome Res. 2017, 16, 3137–3146. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhu, Y.; Best, L.G.; Umans, J.G.; Uppal, K.; Tran, V.T.; Jones, D.P.; Lee, E.T.; Howard, B.V.; Zhao, J. Metabolic profiles of obesity in American Indians: The strong heart family study. PLoS ONE 2016, 11, e0159548. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dadvar, S.; Ferreira, D.M.S.; Cervenka, I.; Ruas, J.L. The weight of nutrients: Kynurenine metabolites in obesity and exercise. J. Intern. Med. 2018, 284, 519–533. [Google Scholar] [CrossRef]
- Maltais-Payette, I.; Boulet, M.M.; Prehn, C.; Adamski, J.; Tchernof, A. Circulating glutamate concentration as a biomarker of visceral obesity and associated metabolic alterations. Nutr. Metab. 2018, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Oberbach, A.; Blüher, M.; Wirth, H.; Till, H.; Kovacs, P.; Kullnick, Y.; Schlichting, N.; Tomm, J.M.; Rolle-Kampczyk, U.; Murugaiyan, J.; et al. Combined proteomic and metabolomic profiling of serum reveals association of the complement system with obesity and identifies novel markers of body fat mass changes. J. Proteome Res. 2011, 10, 4769–4788. [Google Scholar] [CrossRef] [PubMed]
- Gurung, P.; Jialal, I. Plasma Glucose; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Kim, O.Y.; Lee, J.H.; Sweeney, G. Metabolomic profiling as a useful tool for diagnosis and treatment of chronic disease: Focus on obesity, diabetes and cardiovascular diseases. Expert Rev. Cardiovasc. Ther. 2013, 11, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Van Hove, J.L.; Zhang, W.; Kahler, S.G.; Roe, C.R.; Chen, Y.T.; Terada, N.; Chace, D.H.; Iafolla, A.K.; Ding, J.H.; Millington, D.S. Medium-chain acyl-CoA dehydrogenase (MCAD) deficiency: Diagnosis by acylcarnitine analysis in blood. Am. J. Hum. Genet. 1993, 52, 958–966. [Google Scholar]
- Xie, B.; Waters, M.J.; Schirra, H.J. Investigating potential mechanisms of obesity by metabolomics. J. Biomed. Biotechnol. 2012, 2012, 805683. [Google Scholar] [CrossRef]
- Tulipani, S.; Palau-Rodriguez, M.; Miñarro Alonso, A.; Cardona, F.; Marco-Ramell, A.; Zonja, B.; de Alda, M.; Muñoz-Garach, A.; Sanchez-Pla, A.; Tinahones, F.J.; et al. Biomarkers of morbid obesity and prediabetes by metabolomic profiling of human discordant phenotypes. Clin. Chim. Acta 2016, 463, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.A.; Herling, A.W.; Stemmer, K.; Lechner, A.; Tschöp, M.H. Chip-based sensing for release of unprocessed cell surface proteins in vitro and in serum and its (patho)physiological relevance. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E212–E233. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.A.; Tschöp, M.H.; Müller, T.D. Upregulated phospholipase D activity toward glycosylphosphatidylinositol-anchored proteins in micelle-like serum complexes in metabolically deranged rats and humans. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E462–E479. [Google Scholar] [CrossRef] [PubMed]
- Trabado, S.; Al-Salameh, A.; Croixmarie, V.; Masson, P.; Corruble, E.; Fève, B.; Colle, R.; Ripoll, L.; Walther, B.; Boursier-Neyret, C.; et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE 2017, 12, e0173615. [Google Scholar] [CrossRef] [PubMed]
- Fucho, R.; Casals, N.; Serra, D.; Herrero, L. Ceramides and mitochondrial fatty acid oxidation in obesity. FASEB J. 2017, 31, 1263–1272. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Siskos, A.P.; Jain, P.; Römisch-Margl, W.; Bennett, M.; Achaintre, D.; Asad, Y.; Marney, L.; Richardson, L.; Koulman, A.; Griffin, J.L.; et al. Interlaboratory reproducibility of a targeted metabolomics platform for analysis of human serum and plasma. Anal. Chem. 2017, 89, 656–665. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Age and sex effects on plasma metabolite association networks in healthy subjects. J. Proteome Res. 2018, 17, 97–107. [Google Scholar] [CrossRef]
- Ramos Salas, X.; Alberga, A.S.; Cameron, E.; Estey, L.; Forhan, M.; Kirk, S.F.L.; Russell-Mayhew, S.; Sharma, A.M. Addressing weight bias and discrimination: Moving beyond raising awareness to creating change. Obes. Rev. 2017, 18, 1323–1335. [Google Scholar] [CrossRef]
- Rubino, F.; Puhl, R.M.; Cummings, D.E.; Eckel, R.H.; Ryan, D.H.; Mechanick, J.I.; Nadglowski, J.; Ramos Salas, X.; Schauer, P.R.; Twenefour, D.; et al. Joint international consensus statement for ending stigma of obesity. Nat. Med. 2020, 26, 485–497. [Google Scholar] [CrossRef]
- Albury, C.; Strain, W.D.; Brocq, S.L.; Logue, J.; Lloyd, C.; Tahrani, A. The importance of language in engagement between health-care professionals and people living with obesity: A joint consensus statement. Lancet Diabetes Endocrinol. 2020, 8, 447–455. [Google Scholar] [CrossRef]
- Biocrates-Life-Sciences-AG AbsoluteIDQ® p180 Kit. Available online: https://biocrates.com/absoluteidq-p180-kit/ (accessed on 31 August 2020).
- Altman, N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am. Stat. 1992, 46, 175–185. [Google Scholar]
- Tobin, J. Estimation of relationship for limited dependent variables. Econometrica 1958, 26, 24–36. [Google Scholar] [CrossRef]
- McDonald, J.F.; Robert, A.M. The uses of Tobit analysis. Rev. Econ. Stat. 1980, 62, 318–321. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 14 December 2020).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Pedersen, T.L. tidygraph: A Tidy API for Graph Manipulation. Available online: https://cran.r-project.org/package=tidygraph (accessed on 31 August 2020).
- Pedersen, T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. Available online: https://cran.r-project.org/package=ggraph (accessed on 31 August 2020).
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified interface to visualize statistical result of popular R packages. R J. 2016, 8, 478–489. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Jewison, T.; Su, Y.; Disfany, F.M.; Liang, Y.; Knox, C.; MacIejewski, A.; Poelzer, J.; Huynh, J.; Zhou, Y.; Arndt, D.; et al. SMPDB 2.0: Big improvements to the small molecule pathway database. Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef]


| All | BMI <30 | BMI 30–34.9 | BMI 35–39.9 | BMI ≥40 | ||
|---|---|---|---|---|---|---|
| N | 1391 | 397 | 530 | 304 | 160 | |
| Age, years (mean ± SD) | 51.8 ± 13.5 | 50.3 ± 13.6 | 52.3 ± 13.6 | 52.8 ± 13.6 | 52.3 ± 12.6 | |
| Ages, category | 18–29 | 101 (7.3%) | 39 (9.8%) | 34 (6.4%) | 18 (5.9%) | 10 (6.2%) |
| 30–49 | 486 (34.9%) | 150 (37.8%) | 180 (34.0%) | 106 (34.9%) | 50 (31.2%) | |
| 50–69 | 689 (49.5%) | 187 (47.1%) | 260 (49.1%) | 150 (49.3%) | 92 (57.5%) | |
| 70–89 | 115 (8.3%) | 21 (5.3%) | 56 (10.6%) | 30 (9.9%) | 8 (5.0%) | |
| Gender | Males | 250 (18.0%) | 52 (13.1%) | 112 (21.1%) | 61 (20.1%) | 25 (15.6%) |
| Females | 1141 (82.0%) | 345 (86.9%) | 418 (78.9%) | 243 (79.9%) | 135 (84.4%) | |
| Smoking status | Never smoker | 705 (50.7%) | 207 (52.1%) | 259 (48.9%) | 155 (51.0%) | 84 (52.5%) |
| Former smoker | 476 (34.2%) | 130 (32.8%) | 182 (34.3%) | 108 (35.5%) | 56 (35.0%) | |
| Current smoker | 201 (14.4%) | 57 (14.3%) | 85 (16.0%) | 41 (13.5%) | 18 (11.3%) | |
| N.A. | 9 (0.7%) | 3 (0.8%) | 4 (0.8%) | - | 2 (1.2%) | |
| Occupation | Employee | 830 (59.7%) | 244 (62.5%) | 329 (62.1%) | 177 (58.2%) | 80 (50.0%) |
| Unemployed | 125 (9.0%) | 39 (9.8%) | 40 (7.5%) | 24 (7.9%) | 22 (13.8%) | |
| Pensioner | 309 (22.2%) | 81 (20.4%) | 118 (22.3%) | 75 (24.7%) | 35 (21.9%) | |
| Homemaker | 111 (8.0%) | 29 (7.3%) | 38 (7.2%) | 22 (7.2%) | 22 (13.8%) | |
| N.A. | 16 (1.1%) | 4 (1.0%) | 5 (0.9%) | 6 (2.0%) | 1 (0.6%) | |
| Positively Associated | |||
| Category | Metabolite | % Variation | FDR p-Value |
| Aminoacids | Tyrosine (Tyr) | 29.8 | 1.01 × 10−22 |
| Aminoacids | Valine (Val) | 23.2 | 1.12 × 10−14 |
| Aminoacids | Isoleucine (Ile) | 20.0 | 2.59 × 10−12 |
| PC aa | PC aa C38:3 | 21.1 | 4.02 × 10−12 |
| Aminoacids | Phenylalanine (Phe) | 20.4 | 1.35 × 10−11 |
| Aminoacids | Alanine (Ala) | 18.9 | 5.14 × 10−10 |
| Sugars | Sum of hexose (H1) | 17.8 | 1.31 × 10−9 |
| Aminoacids | Proline (Pro) | 18.1 | 1.62 × 10−9 |
| Aminoacids | Glutamic acid (Glu) | 17.1 | 1.18 × 10−8 |
| Biogenic Amines | Kynurenine | 16.1 | 4.64 × 10−8 |
| Aminoacids | Leucine (Leu) | 15.0 | 2.03 × 10−7 |
| PC aa | PC aa C40:4 | 14.3 | 4.71 × 10−6 |
| Acylcarnitines | Carnitine (C0) | 12.6 | 1.5 × 10−5 |
| Acylcarnitines | Propionylcarnitine (C3) | 12.2 | 4.61 × 10−5 |
| PC aa | PC aa C32:1 | 12.4 | 4.61 × 10−5 |
| Biogenic Amines | Aminoadipic acid (alpha-AAA) | 18.5 | 7.02 × 10−4 |
| Aminoacids | Ornithine (Orn) | 9.2 | 0.002 |
| Acylcarnitines | Acetylcarnitine (C2) | 9.1 | 0.003 |
| SM | SM C18:1 | 8.3 | 0.006 |
| Biogenic Amines | 4-Hydroxyproline (t4-OH-Pro) | 8.0 | 0.012 |
| PC aa | PC aa C38:4 | 7.6 | 0.015 |
| SM | SM C16:1 | 7.0 | 0.019 |
| lysoPC | lysoPC a C16:1 | 7.3 | 0.021 |
| Aminoacids | Lysine (Lys) | 7.1 | 0.026 |
| PC aa | PC aa C40:5 | 6.9 | 0.028 |
| Acylcarnitines | Valerylcarnitine (C5) | 6.5 | 0.037 |
| Negatively Associated | |||
| Category | Metabolite | % Variation | FDR p-Value |
| lysoPC | lysoPC a C18:2 | −23.3 | 1.28 × 10−22 |
| PC ae | PC ae C36:2 | −20.5 | 3.11 × 10−17 |
| PC ae | PC ae C34:3 | −20.2 | 1.36 × 10−16 |
| PC ae | PC ae C34:2 | −18.2 | 2.18 × 10−13 |
| PC ae | PC ae C40:6 | −16.1 | 3.50 × 10−10 |
| Aminoacids | Asparagine (Asn) | −16.1 | 5.24 × 10−10 |
| PC ae | PC ae C40:1 | −16.0 | 5.40 × 10−10 |
| lysoPC | lysoPC a C18:1 | −15.9 | 6.19 × 10−10 |
| PC ae | PC ae C38:0 | −15.2 | 1.59 × 10−9 |
| lysoPC | lysoPC a C17:0 | −14.3 | 7.32 × 10−8 |
| Aminoacids | Glycine (Gly) | −13.6 | 1.98 × 10−7 |
| PC aa | PC aa C38:6 | −12.9 | 1.31 × 10−6 |
| PC ae | PC ae C36:3 | −12.3 | 4.71 × 10−6 |
| PC aa | PC aa C38:0 | −11.7 | 1.52 × 10−5 |
| PC ae | PC ae C42:3 | −11.9 | 1.52 × 10−5 |
| Aminoacids | Histidine (His) | −11.8 | 1.61 × 10−5 |
| PC aa | PC aa C36:0 | −11.7 | 1.61 × 10−5 |
| PC aa | PC aa C42:5 | −11.6 | 3.00 × 10−5 |
| PC ae | PC ae C36:1 | −10.6 | 8.42 × 10−5 |
| PC aa | PC aa C36:6 | −10.6 | 9.45 × 10−5 |
| PC ae | PC ae C30:0 | −10.6 | 1.45 × 10−4 |
| SM | SM C24:1 | −10.5 | 1.51 × 10−4 |
| PC ae | PC ae C40:5 | −10.5 | 1.69 × 10−4 |
| PC aa | PC aa C34:2 | −10.5 | 1.91 × 10−4 |
| PC ae | PC ae C42:2 | −10.1 | 2.17 × 10−4 |
| PC ae | PC ae C44:6 | −10.3 | 2.32 × 10−4 |
| Aminoacids | Serine (Ser) | −10.0 | 4.03 × 10−4 |
| PC aa | PC aa C42:1 | −10.0 | 4.78 × 10−4 |
| PC ae | PC ae C34:1 | −9.5 | 5.05 × 10−4 |
| SM | SM C16:0 | −9.7 | 5.06 × 10−4 |
| PC aa | PC aa C42:6 | −10.5 | 7.24 × 10−4 |
| PC ae | PC ae C38:6 | −9.4 | 7.72 × 10−4 |
| PC ae | PC ae C32:1 | −9.0 | 0.002 |
| SM | SM C26:1 | −8.9 | 0.002 |
| SM | SM (OH) C22:2 | −8.3 | 0.002 |
| PC aa | PC aa C40:3 | −8.8 | 0.002 |
| PC ae | PC ae C42:1 | −8.9 | 0.002 |
| PC aa | PC aa C42:2 | −9.6 | 0.003 |
| PC aa | PC aa C42:0 | −8.4 | 0.003 |
| PC ae | PC ae C38:5 | −8.5 | 0.003 |
| PC ae | PC ae C42:4 | −8.4 | 0.003 |
| PC ae | PC ae C34:0 | −7.8 | 0.006 |
| PC ae | PC ae C36:5 | −7.9 | 0.006 |
| PC ae | PC ae C44:5 | −7.8 | 0.008 |
| PC aa | PC aa C40:2 | −7.3 | 0.015 |
| PC ae | PC ae C42:5 | −7.2 | 0.015 |
| PC ae | PC ae C36:0 | −7.1 | 0.017 |
| Biogenic Amines | Serotonin | −7.4 | 0.018 |
| PC ae | PC ae C32:2 | −6.7 | 0.019 |
| PC ae | PC ae C38:4 | −6.9 | 0.020 |
| SM | SM (OH) C16:1 | −6.7 | 0.021 |
| Biogenic Amines | Creatinine | −6.1 | 0.023 |
| Biogenic Amines | N-Acetylornithine (Ac-Orn) | −9.2 | 0.030 |
| Aminoacids | Citrulline (Cit) | −6.2 | 0.030 |
| Acylcarnitines | Dodecanoylcarnitine (C12) | −24.5 | 0.032 |
| SM | SM (OH) C14:1 | −5.9 | 0.041 |
| PC ae | PC ae C38:2 | −6.1 | 0.043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frigerio, G.; Favero, C.; Savino, D.; Mercadante, R.; Albetti, B.; Dioni, L.; Vigna, L.; Bollati, V.; Pesatori, A.C.; Fustinoni, S. Plasma Metabolomic Profiling in 1391 Subjects with Overweight and Obesity from the SPHERE Study. Metabolites 2021, 11, 194. https://doi.org/10.3390/metabo11040194
Frigerio G, Favero C, Savino D, Mercadante R, Albetti B, Dioni L, Vigna L, Bollati V, Pesatori AC, Fustinoni S. Plasma Metabolomic Profiling in 1391 Subjects with Overweight and Obesity from the SPHERE Study. Metabolites. 2021; 11(4):194. https://doi.org/10.3390/metabo11040194
Chicago/Turabian StyleFrigerio, Gianfranco, Chiara Favero, Diego Savino, Rosa Mercadante, Benedetta Albetti, Laura Dioni, Luisella Vigna, Valentina Bollati, Angela Cecilia Pesatori, and Silvia Fustinoni. 2021. "Plasma Metabolomic Profiling in 1391 Subjects with Overweight and Obesity from the SPHERE Study" Metabolites 11, no. 4: 194. https://doi.org/10.3390/metabo11040194
APA StyleFrigerio, G., Favero, C., Savino, D., Mercadante, R., Albetti, B., Dioni, L., Vigna, L., Bollati, V., Pesatori, A. C., & Fustinoni, S. (2021). Plasma Metabolomic Profiling in 1391 Subjects with Overweight and Obesity from the SPHERE Study. Metabolites, 11(4), 194. https://doi.org/10.3390/metabo11040194









