Drosophila INDY and Mammalian INDY: Major Differences in Transport Mechanism and Structural Features despite Mostly Similar Biological Functions
Abstract
1. Introduction
2. Identification of Drosophila INDY and Biological Consequences of Its Loss of Function
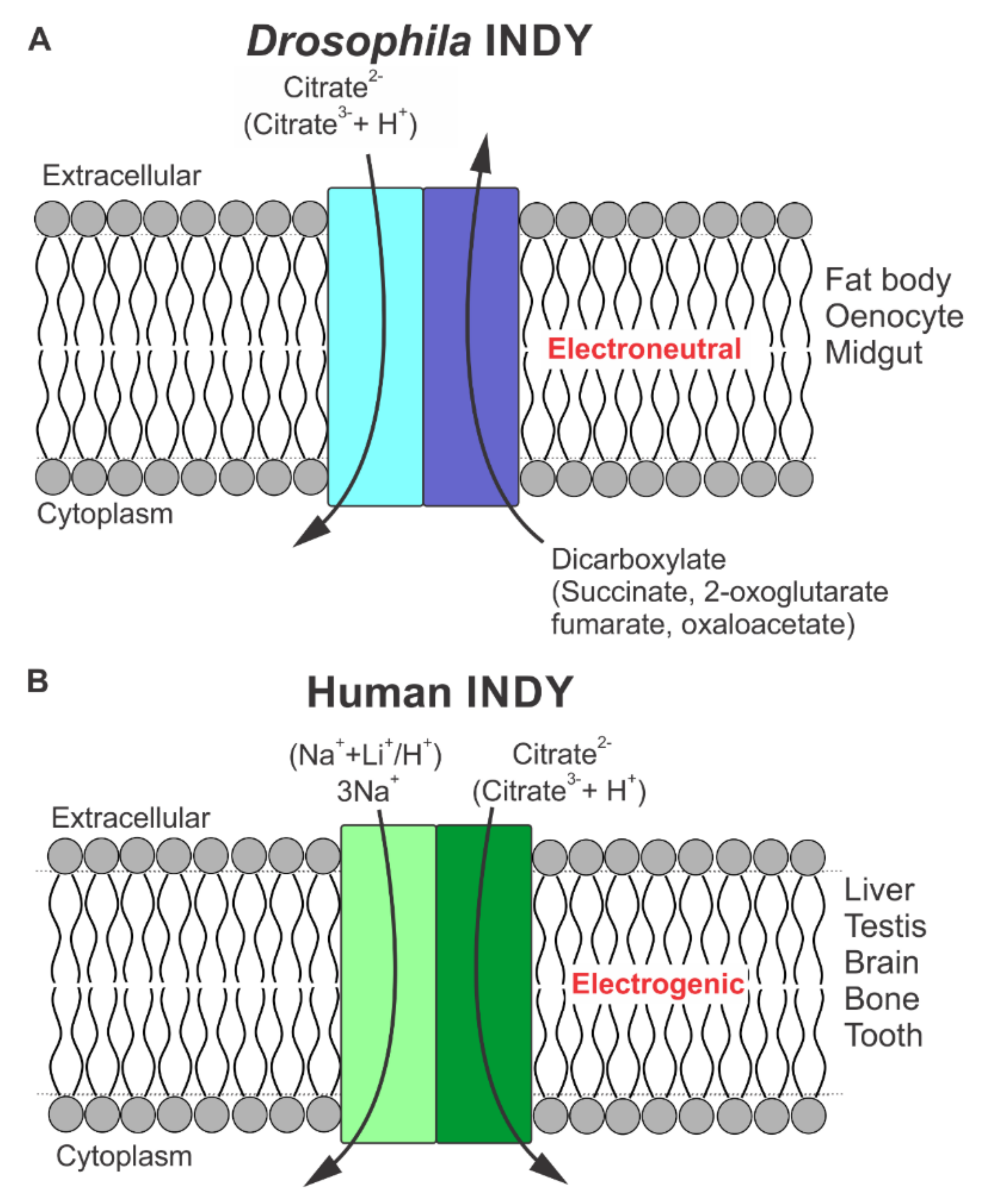
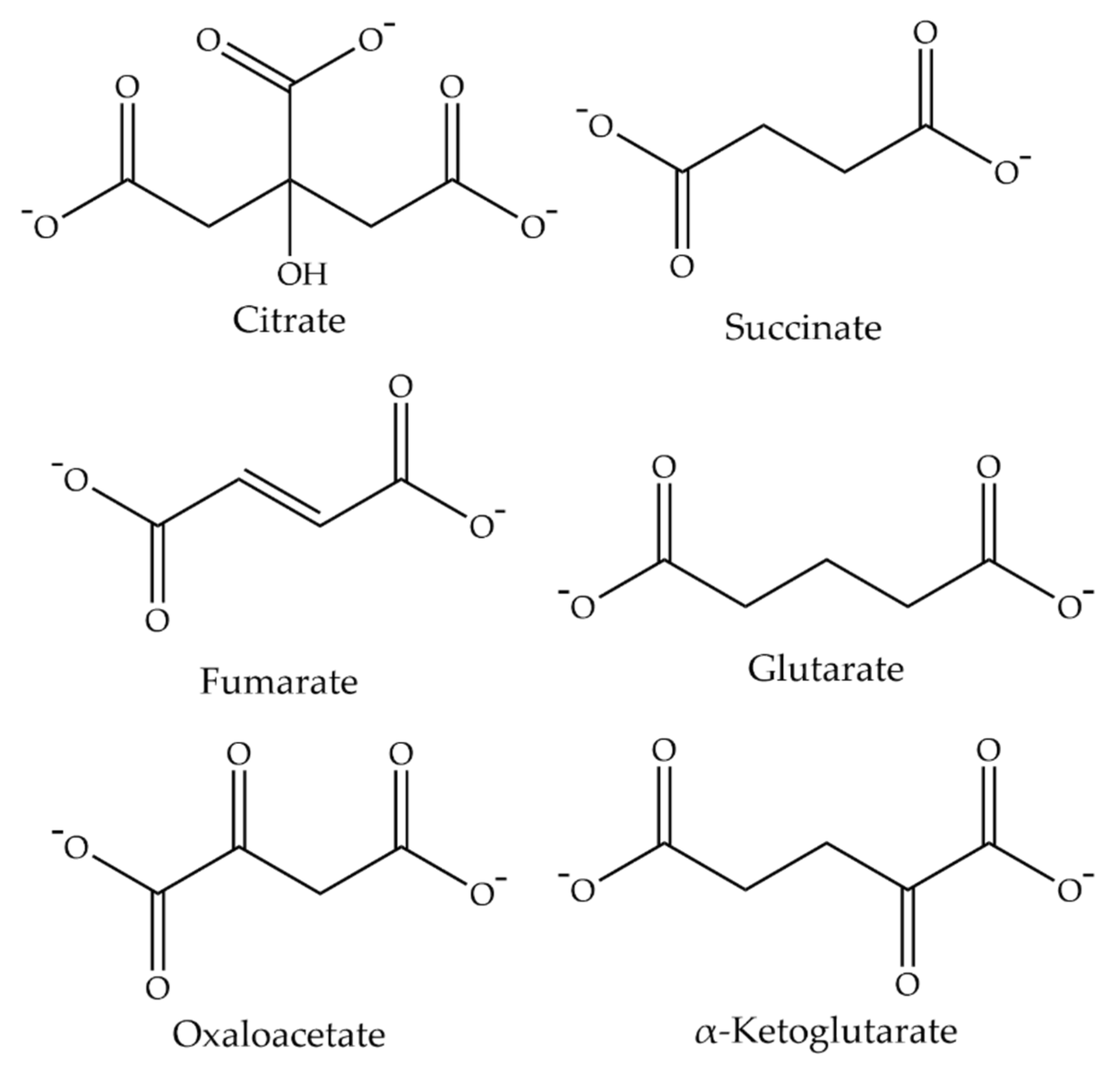
Functional Features of Drosophila INDY as a Transporter
3. Discovery of Mammalian INDY
3.1. Functional Identity of SLC13A5 as a Citrate Transporter
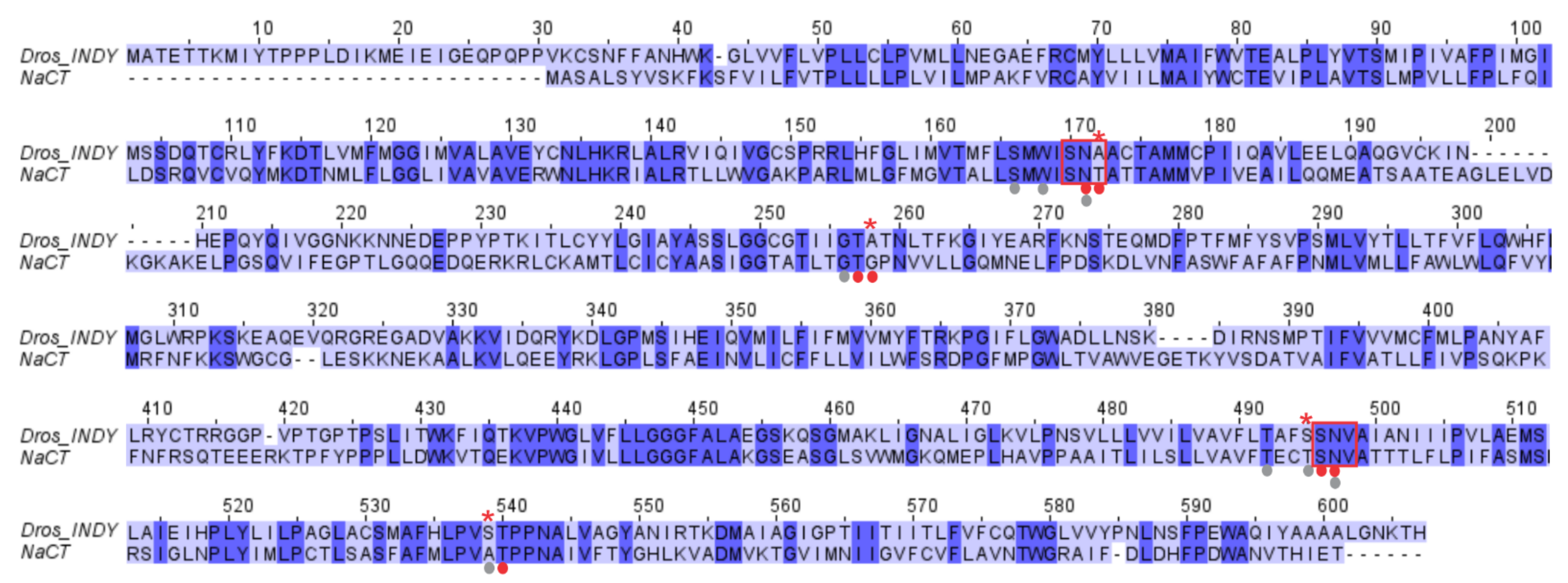
3.2. Species-Specific Functional Differences in SLC13A5 between Primates and Non-Primates
3.3. Biological Consequences of SLC13A5 Deficiency in Mammals, including Humans
3.4. Functions of SLC13A5 in the Liver Versus Brain
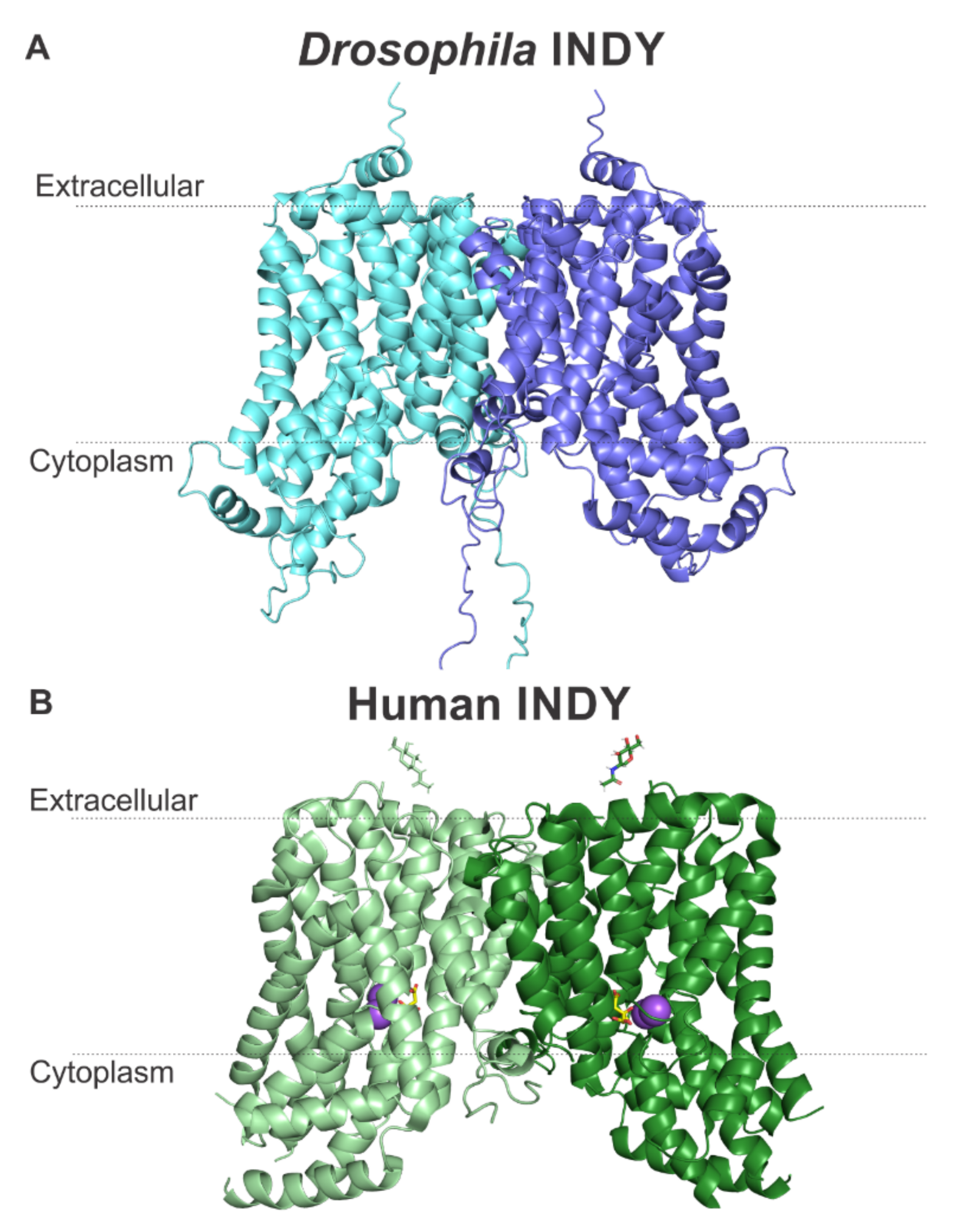
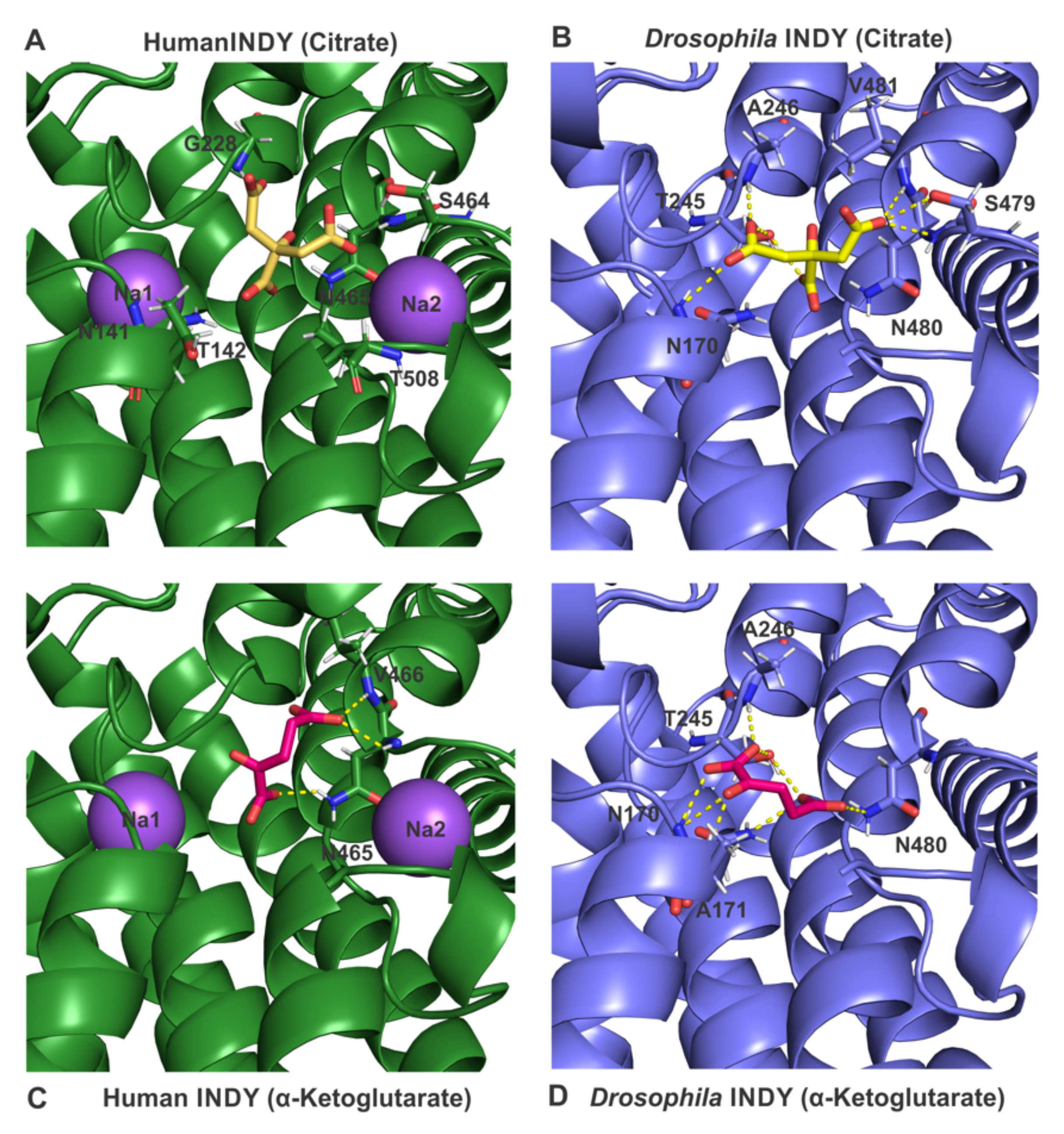
4. Structural Differences between Drosophila INDY and Human INDY
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rogina, B.; Reenan, R.A.; Nilsen, S.P.; Helfand, S.L. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 2000, 290, 2137–2140. [Google Scholar] [CrossRef]
- Wang, P.Y.; Neretti, N.; Whitaker, R.; Hosier, S.; Chang, C.; Lu, D.; Rogina, B.; Helfand, S.L. Long-lived Indy and calorie restriction interact to extend life span. Proc. Natl. Acad. Sci. USA 2009, 106, 9262–9267. [Google Scholar] [CrossRef]
- Rogina, B.; Helfand, S.L. Indy mutations and Drosophila longevity. Front. Genet. 2013, 4, 47. [Google Scholar] [CrossRef]
- Zhu, C.T.; Chang, C.; Reenan, R.A.; Helfand, S.L. Indy gene variation in natural populations confers fitness advantage and life span extension through transposon insertion. Aging 2014, 6, 58–69. [Google Scholar] [CrossRef][Green Version]
- Inoue, K.; Zhuang, L.; Maddox, D.M.; Smith, S.B.; Ganapathy, V. Structure, function, and expression of a novel sodium-coupled citrate transporter (NaCT) cloned from mammalian brain. J. Biol. Chem. 2002, 277, 39469–39476. [Google Scholar] [CrossRef]
- Inoue, K.; Zhuang, L.; Ganapathy, V. Human Na+-coupled citrate transporter: Primary structure, genomic organization, and transport function. Biochem. Biophys. Res. Commun. 2002, 299, 465–471. [Google Scholar] [CrossRef]
- Inoue, K.; Fei, Y.J.; Zhuang, L.; Gopal, E.; Miyauchi, S.; Ganapathy, V. Functional features and genomic organization of mouse NaCT, a sodium-coupled transporter for tricarboxylic acid cycle intermediates. Biochem. J. 2004, 378, 949–957. [Google Scholar] [CrossRef]
- Gopal, E.; Babu, E.; Ramachandran, S.; Bhutia, Y.D.; Prasad, P.D.; Ganapathy, V. Species-specific influence of lithium on the activity of SLC13A5 (NaCT): Lithium-induced activation is specific for the transporter in primates. J. Pharmacol. Exp. Ther. 2015, 353, 17–26. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Kopel, J.J.; Lawrence, J.J.; Neugebauer, V.; Ganapathy, V. Plasma membrane Na+-coupled citrate transporter (SLC13A5) and neonatal epileptic encephalopathy. Molecules 2017, 22, 378. [Google Scholar] [CrossRef]
- Rogina, B. INDY – A new link to metabolic regulation in animals and humans. Front. Genet. 2017, 8, 66. [Google Scholar] [CrossRef]
- Willmes, D.M.; Kurzbach, A.; Henke, C.; Schumann, T.; Zahn, G.; Heifetz, A.; Jordan, J.; Helfand, S.L.; Birkenfeld, A.L. The longevity gene INDY (I’m Not Dead Yet) in metabolic control: Potential as pharmacological target. Pharmacol. Ther. 2018, 185, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kopel, J.J.; Bhutia, Y.D.; Sivaprakasam, S.; Ganapathy, V. Consequences of NaCT/SLC13A5/mINDY deficiency: Good versus evil, separated only by the blood-brain barrier. Biochem. J. 2021, 478, 463–486. [Google Scholar] [CrossRef] [PubMed]
- Nye, K.; Porter, B.; Dubey, D. SLC13A5 Epileptic Encephalopathy. Rare Disease Database, National Organization for Rare Disorders. Available online: https://rarediseases.org/rare-diseases/slc13a5-epileptic-encephalopathy (accessed on 5 September 2021).
- Aigaki, T.; Seong, K.; Matsuo, T. Longevity determination genes in Drosophila melanogaster. Mech. Ageing Dev. 2002, 123, 1531–1541. [Google Scholar] [CrossRef]
- Marden, J.H.; Rogina, B.; Montooth, K.L.; Helfand, S.L. Conditional tradeoffs between aging and organismal performance of Indy long-lived mutant flies. Proc. Natl. Acad. Sci. USA 2003, 100, 3369–3373. [Google Scholar] [CrossRef] [PubMed]
- Pajor, A.M. Sodium-coupled dicarboxylate and citrate transporters from the SLC13 family. Pflugers Arch. 2014, 466, 119–130. [Google Scholar] [CrossRef]
- Inoue, K.; Fei, Y.J.; Huang, W.; Zhuang, L.; Chen, Z.; Ganapathy, V. Functional identity of Drosophila melanogaster Indy as a cation-independent, electroneutral transporter for tricarboxylic acid intermediates. Biochem. J. 2002, 367, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Knauf, F.; Rogina, B.; Jiang, Z.; Aronson, P.S.; Helfand, S.L. Functional characterization and immunolocalization of the transporter encoded by the life-extending gene Indy. Proc. Natl. Acad. Sci. USA 2002, 99, 14315–14319. [Google Scholar] [CrossRef]
- Knauf, F.; Mohebbi, N.; Teichert, C.; Herold, D.; Rogina, B.; Helfand, S.L.; Gollasch, M.; Luft, F.C.; Aronson, P.S. The life-extending gene Indy encodes an exchanger for Krebs-cycle intermediates. Biochem. J. 2006, 397, 25–29. [Google Scholar] [CrossRef]
- Neretti, N.; Wang, P.Y.; Brodsky, A.S.; Nyguyen, H.H.; White, K.P.; Rogina, B.; Helfand, S.L. Long-lived Indy induces reduced mitochondrial reactive oxygen species production and oxidative damage. Proc. Natl. Acad. Sci. USA 2009, 106, 2277–2282. [Google Scholar] [CrossRef]
- Rogers, R.P.; Rogina, B. Increased mitochondrial biogenesis preserves intestinal stem cell homeostasis and contributes to longevity in Indy mutant flies. Aging 2014, 6, 335–350. [Google Scholar] [CrossRef]
- Palmieri, F. The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Mol. Aspects Med. 2013, 34, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.C.; O’Conner, M.B. Systemic activin signaling independently regulates sugar homeostasis, cellular metabolism, and pH balance in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2014, 111, 5729–5734. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.J.; Liu, J.C.; Inoue, K.; Zhuang, L.; Miyake, K.; Miyauchi, S.; Ganapathy, V. Relevance of NAC-2, an Na+-coupled citrate transporter, to life span, body size and fat content in Caenorhabditis elegans. Biochem. J. 2004, 379, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Gopal, E.; Miyauchi, S.; Martin, P.M.; Ananth, S.; Srinivas, S.R.; Smith, S.B.; Prasad, P.D.; Ganapathy, V. Expression and functional features of NaCT, a sodium-coupled citrate transporter, in human and rat livers and cell lines. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G402–G408. [Google Scholar] [CrossRef]
- Inoue, K.; Zhuang, L.; Maddox, D.M.; Smith, S.B.; Ganapathy, V. Human sodium-coupled citrate transporter, the orthologue of Drosophila Indy, as a novel target for lithium action. Biochem. J. 2003, 374, 21–26. [Google Scholar] [CrossRef]
- Higuchi, K.; Kopel, J.J.; Sivaprakasam, S.; Jaramillo-Martinez, V.; Sutton, R.B.; Urbatsch, I.L.; Ganapathy, V. Functional analysis of a species-specific inhibitor selective for human Na+-coupled citrate transporter (NaCT/SLC13A5/mINDY). Biochem. J. 2020, 477, 4149–4165. [Google Scholar] [CrossRef]
- Sauer, D.B.; Song, J.; Hilton, J.K.; Karpowich, N.K.; Mindell, J.A.; Rice, W.J.; Wang, D.N. Structure and inhibition mechanism of the human citrate transporter NaCT. Nature 2021, 591, 157–161. [Google Scholar] [CrossRef]
- Jaramillo-Martinez, V.; Urbatsch, I.L.; Ganapathy, V. Functional distinction between human and mouse sodium-coupled citrate transporters and its biological significance: An attempt for structural basis using a homology modeling approach. Chem. Rev. 2021, 121, 5359–5377. [Google Scholar] [CrossRef]
- Jaramillo-Martinez, V.; Ganapathy, V.; Urbatsch, J.L. A home run for human NaCT/SLC13A5/INDY: Cryo-EM structure and homology model to predict transport mechanisms, inhibitor interaction and mutational defects. Biochem. J. 2021, 478, 2051–2057. [Google Scholar] [CrossRef]
- Birkenfeld, A.L.; Lee, H.Y.; Guebre-Egziabher, F.; Alves, T.C.; Jurczak, M.J.; Jornayvaz, F.R.; Zhang, D.; Hsiao, J.J.; Martin-Montalvo, A.; Fischer-Rosinsky, A.; et al. Deletion of the mammalian INDY homolog mimics aspects of dietary restriction and protects against adiposity and insulin resistance in mice. Cell Metab. 2011, 14, 184–195. [Google Scholar] [CrossRef]
- Willmes, D.M.; Daniels, M.; Kurzbach, A.; Lieske, S.; Bechmann, N.; Schumann, T.; Henke, C.; El-Agroudy, N.N.; Goncalves, A.C.D.C.; Peitzsch, M.; et al. The longevity gene mIndy (I’m Not Dead Yet) affects blood pressure through sympathoadrenal mechanisms. JCI Insight 2021, 6, e136083. [Google Scholar] [CrossRef] [PubMed]
- Brachs, S.; Winkel, A.F.; Tang, H.; Birkenfeld, A.L.; Brunner, B.; Jahn-Hofmann, K.; Margerie, D.; Ruetten, H.; Schmoll, D.; Spranger, J. Inhibition of citrate cotransporter Slc13a5.mINDY by RNAi improves hepatic insulin sensitivity and prevents diet-induced non-alcoholic fatty liver disease in mice. Mol. Metab. 2016, 5, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Thevenon, J.; Milh, M.; Feillet, F.; St-Onge, J.; Duffourd, Y.; Juge, C.; Roubertie, A.; Heron, D.; Mignot, C.; Raffo, E.; et al. Mutations in SLC13A5 cause autosomal-recessive epileptic encephalopathy with seizure onset in the first days of life. Am. J. Hum. Genet. 2014, 95, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Kopel, J.J.; Bhutia, Y.D.; Ramachandran, S.; Lawrence, J.J.; Neugebauer, V.; Ganapathy, V. Tooth hypoplasia for differential diagnosis of childhood epilepsy associated with SLC13A5 mutations. Int. J. Neurol. Disord. 2017, 1, 33–37. [Google Scholar]
- Irizarry, A.R.; Yan, G.; Zeng, Q.; Lucchesi, J.; Hamang, M.J.; Ma, Y.L.; Rong, J.X. Defective enamel and bone development in sodium-dependent citrate transporter (NaCT) Slc13a5 deficient mice. PLoS ONE 2017, 12, e0175465. [Google Scholar] [CrossRef] [PubMed]
- Henke, C.; Tollner, K.; van Dijk, R.M.; Miljanovic, N.; Cordes, T.; Twele, F.; Broer, S.; Ziesak, V.; Rohde, M.; Hauck, S.M.; et al. Disruption of the sodium-dependent citrate transporter SLC13A5 in mice causes alterations in brain citrate levels and neuronal network excitability in the hippocampus. Neurobiol. Dis. 2020, 143, 105018. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.Z.; Sung, C.W.; Tsai, Y.H.; Yeh, S.R.; Lin, W.S.; Wang, P.Y. Nervous system deletion of mammalian INDY in mice mimics dietary restriction-induced memory enhancement. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, D.; Choi, E.Y.; Lapidus, R.; Zhang, L.; Huang, S.M.; Shapiro, P.; Wang, H. Silencing of solute carrier family 13 member 5 disrupts energy homeostasis and inhibits proliferation of human hepatocarcinoma cells. J. Biol. Chem. 2017, 292, 13890–13901. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Huang, W.; Li, Z.; Kane, M.A.; Zhang, L.; Huang, S.M.; Wang, H. Comparative proteomic analysis of SLC13A5 knockdown reveals elevated ketogenesis and enhanced cellular toxic response to chemotherapeutic agents in HepG2 cells. Toxicol. Appl. Pharmacol. 2020, 402, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Bhutia, Y.D.; Ramachandran, S.; Ganapathy, V. Cell-surface and nuclear receptors in the colon as targets for bacterial metabolites and its relevance to colon health. Nutrients 2017, 9, 856. [Google Scholar] [CrossRef] [PubMed]
- Ristic, B.; Bhutia, Y.D.; Ganapathy, V. Cell-surface G-protein-coupled receptors for tumor-associated metabolites: A direct link to mitochondrial dysfunction in cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Sauer, D.B.; Trebesch, N.; Marden, J.J.; Cocco, N.; Song, J.; Koide, A.; Koide, S.; Tajkhorshid, E.; Wang, D.N. Structural basis for the reaction cycle of DASS dicarboxylate transporters. eLife 2020, 9, e61350. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mancusso, R.; Gregorio, G.G.; Liu, Q.; Wang, D.N. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature 2012, 491, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Nie, R.; Stark, S.; Symersky, J.; Kaplan, R.S.; Lu, M. Structure and function of the divalent anion/Na+ symporter from Vibrio cholerae and a humanized variant. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Bolla, J.R.; Su, C.-C.; Delmar, J.A.; Radhakrishnan, A.; Kumar, N.; Chou, T.-H.; Long, F.; Rajashankar, K.R.; Edward, W.Y. Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Su, C.-C.; Bolla, J.R.; Kumar, N.; Radhakrishnan, A.; Long, F.; Delmar, J.A.; Chou, T.H.; Rajashankar, K.R.; Shafer, W.M.; Edward, W.Y. Structure and function of Neisseria gonorrhoeae MtrF illuminates a class of antimetabolite efflux pumps. Cell Rep. 2015, 11, 61–70. [Google Scholar] [CrossRef] [PubMed]
| Drosophila INDY | Drosophila INDY | Human INDY | Human INDY | |
|---|---|---|---|---|
| Molecule | Computed Binding Affinity(kcal/mol) | Difference Compared to Citrate(kcal/mol) | Computed Binding Affinity(kcal/mol) | Difference Compared to Citrate(kcal/mol) |
| Citrate | −5.7 | 0 | −5.5 | 0 |
| Oxaloacetate | −5.1 | 0.6 | −4.4 | 1.1 |
| α-Ketoglutarate | −5.3 | 0.4 | −4.5 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaramillo-Martinez, V.; Sivaprakasam, S.; Ganapathy, V.; Urbatsch, I.L. Drosophila INDY and Mammalian INDY: Major Differences in Transport Mechanism and Structural Features despite Mostly Similar Biological Functions. Metabolites 2021, 11, 669. https://doi.org/10.3390/metabo11100669
Jaramillo-Martinez V, Sivaprakasam S, Ganapathy V, Urbatsch IL. Drosophila INDY and Mammalian INDY: Major Differences in Transport Mechanism and Structural Features despite Mostly Similar Biological Functions. Metabolites. 2021; 11(10):669. https://doi.org/10.3390/metabo11100669
Chicago/Turabian StyleJaramillo-Martinez, Valeria, Sathish Sivaprakasam, Vadivel Ganapathy, and Ina L. Urbatsch. 2021. "Drosophila INDY and Mammalian INDY: Major Differences in Transport Mechanism and Structural Features despite Mostly Similar Biological Functions" Metabolites 11, no. 10: 669. https://doi.org/10.3390/metabo11100669
APA StyleJaramillo-Martinez, V., Sivaprakasam, S., Ganapathy, V., & Urbatsch, I. L. (2021). Drosophila INDY and Mammalian INDY: Major Differences in Transport Mechanism and Structural Features despite Mostly Similar Biological Functions. Metabolites, 11(10), 669. https://doi.org/10.3390/metabo11100669






