Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions
Abstract
1. Introduction
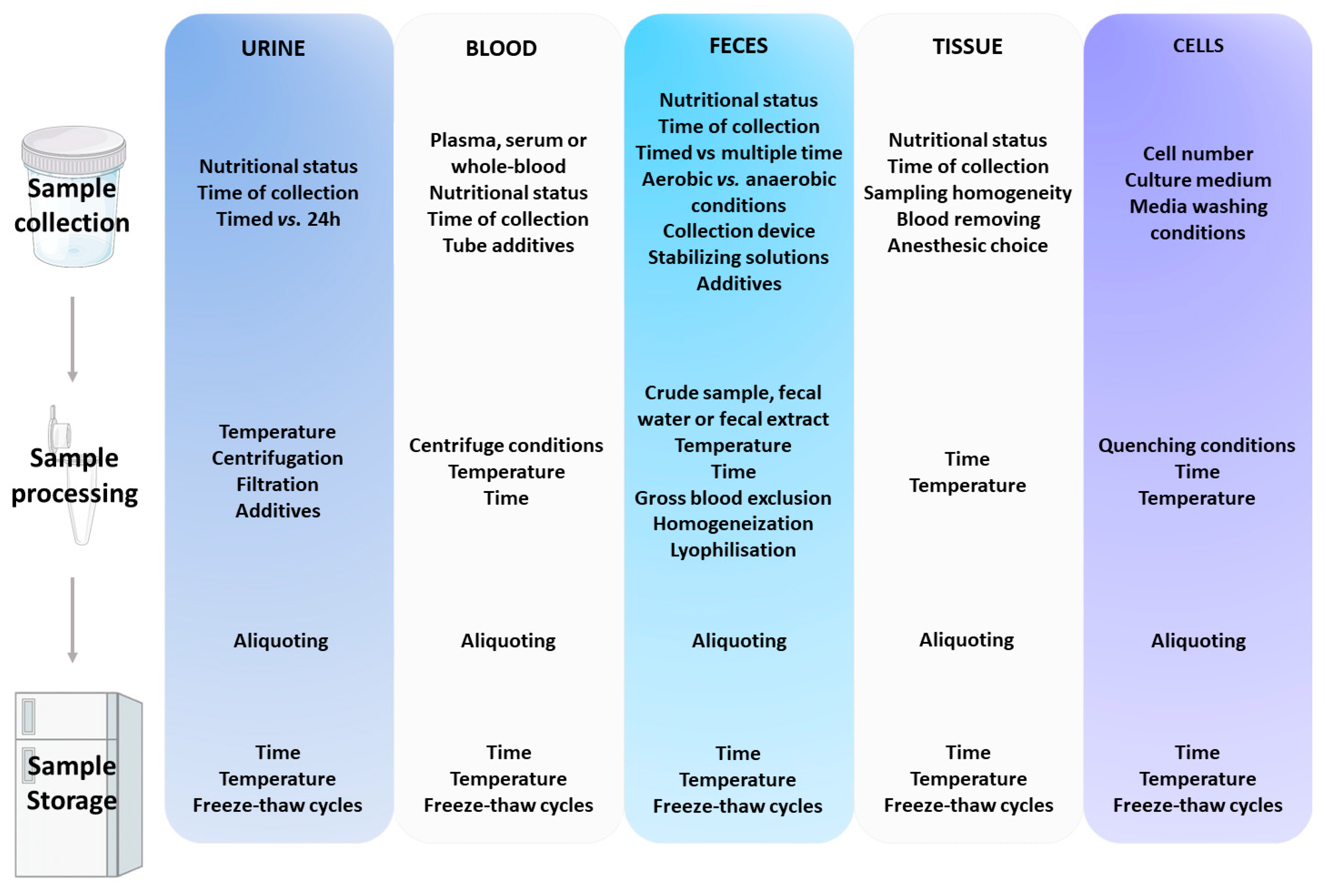
2. Overview of the Pre-Analytical Handling Procedures of the Most Commonly Used Biological Matrices in Metabolomics
2.1. Time of Collection
2.1.1. Considering Nutritional Status
2.1.2. Considering Circadian Rhythm
2.2. Common Sources of Variation in Pre-Analytical Handling of Main Biological Matrices
2.3. Urine
2.3.1. Timed vs. 24-Hour Collection
2.3.2. Sample Collection
2.3.3. Sample Storage
2.4. Blood
2.4.1. Sample Collection
2.4.2. Sample Preparation
2.4.3. Sample Storage
2.5. Feces
2.5.1. Sample Selection
2.5.2. Sample Collection
2.5.3. Sample Storage
2.6. Tissue
2.6.1. Sample Collection
2.6.2. Sample Storage
2.7. Cells
2.7.1. Sample Preparation
2.7.2. Sample Storage
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J. Past, present and future perspectives in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 377–386. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metab Clin. Exp. 2011, 60, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; Fernández-Real, J.-M.; Federici, M.; Serino, M.; Abbott, J.; Charpentier, J.; Heymes, C.; Luque, J.L.; Anthony, E.; Barton, R.H.; et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018, 24, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Moreau, R.; Clària, J.; Aguilar, F.; Fenaille, F.; Lozano, J.; Junot, C.; Colsch, B.; Caraceni, P.; Trebicka, J.; Pavesi, M.; et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J. Hepatol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.-L.; Yang, S.-S. Metabolic Signature of Hepatic Fibrosis: From Individual Pathways to Systems Biology. Cells 2019, 8, 1423. [Google Scholar] [CrossRef]
- Dumas, M.-E.; Barton, R.H.; Toye, A.; Cloarec, O.; Blancher, C.; Rothwell, A.; Fearnside, J.; Tatoud, R.; Blanc, V.; Lindon, J.C.; et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. PNAS 2006, 103, 12511–12516. [Google Scholar] [CrossRef]
- Soltis, A.R.; Kennedy, N.J.; Xin, X.; Zhou, F.; Ficarro, S.B.; Yap, Y.S.; Matthews, B.J.; Lauffenburger, D.A.; White, F.M.; Marto, J.A.; et al. Hepatic Dysfunction Caused by Consumption of a High-Fat Diet. Cell Rep. 2017, 21, 3317–3328. [Google Scholar] [CrossRef]
- Torre Della, S.; Mitro, N.; Meda, C.; Lolli, F.; Pedretti, S.; Barcella, M.; Ottobrini, L.; Metzger, D.; Caruso, D.; Maggi, A. Short-Term Fasting Reveals Amino Acid Metabolism as a Major Sex-Discriminating Factor in the Liver. Cell Metab. 2018, 28, 256–267. [Google Scholar] [CrossRef]
- Li, F.; Patterson, A.D.; Krausz, K.W.; Tanaka, N.; Gonzalez, F.J. Metabolomics reveals an essential role for peroxisome proliferator-activated receptor α in bile acid homeostasis. J. Lipid Res. 2012, 53, 1625–1635. [Google Scholar] [CrossRef]
- Cho, J.-Y.; Kang, D.W.; Ma, X.; Ahn, S.-H.; Krausz, K.W.; Luecke, H.; Idle, J.R.; Gonzalez, F.J. Metabolomics reveals a novel vitamin E metabolite and attenuated vitamin E metabolism upon PXR activation. J. Lipid Res. 2009, 50, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Montagner, A.; Korecka, A.; Polizzi, A.; Lippi, Y.; Blum, Y.; Canlet, C.; Tremblay-Franco, M.; Gautier-Stein, A.; Burcelin, R.; Yen, Y.-C.; et al. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci. Rep. 2016, 6, 20127. [Google Scholar] [CrossRef]
- Lukowicz, C.; Ellero-Simatos, S.; Régnier, M.; Oliviero, F.; Lasserre, F.; Polizzi, A.; Montagner, A.; Smati, S.; Boudou, F.; Lenfant, F.; et al. Dimorphic metabolic and endocrine disorders in mice lacking the constitutive androstane receptor. Sci. Rep. 2019, 9, 20169. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.-P.; Le Net, J.-L.; Baker, D.; Walley, R.J.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Niu, M.; Wei, A.-W.; Tang, J.-F.; Tu, C.; Bai, Z.-F.; Zou, Z.-S.; Xiao, X.-H.; Liu, Y.-P.; Wang, J.-B. Risk profiling using metabolomic characteristics for susceptible individuals of drug-induced liver injury caused by Polygonum multiflorum. Arch. Toxicol. 2019, 295, 113. [Google Scholar] [CrossRef]
- Yip, L.Y.; Aw, C.C.; Lee, S.H.; Hong, Y.S.; Ku, H.C.; Xu, W.H.; Chan, J.M.X.; Cheong, E.J.Y.; Chng, K.R.; Ng, A.H.Q.; et al. The liver-gut microbiota axis modulates hepatotoxicity of tacrine in the rat. Hepatology 2018, 67, 282–295. [Google Scholar] [CrossRef]
- Cabaton, N.J.; Poupin, N.; Canlet, C.; Tremblay-Franco, M.; Audebert, M.; Cravedi, J.-P.; Riu, A.; Jourdan, F.; Zalko, D. An Untargeted Metabolomics Approach to Investigate the Metabolic Modulations of HepG2 Cells Exposed to Low Doses of Bisphenol A and 17β-Estradiol. Front. Endocrinol. 2018, 9, 571. [Google Scholar] [CrossRef]
- Lukowicz, C.; Ellero-Simatos, S.; Régnier, M.; Polizzi, A.; Lasserre, F.; Montagner, A.; Lippi, Y.; Jamin, E.L.; Martin, J.-F.; Naylies, C.; et al. Metabolic Effects of a Chronic Dietary Exposure to a Low-Dose Pesticide Cocktail in Mice: Sexual Dimorphism and Role of the Constitutive Androstane Receptor. Environ. Health Perspect. 2018, 126, 067007. [Google Scholar] [CrossRef]
- Weiss, J.M.; Davies, L.C.; Karwan, M.; Ileva, L.; Ozaki, M.K.; Cheng, R.Y.; Ridnour, L.A.; Annunziata, C.M.; Wink, D.A.; McVicar, D.W. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 2018, 128, 3794–3805. [Google Scholar] [CrossRef]
- Su, L.; Mao, J.; Hao, M.; Lu, T.; Mao, C.; Ji, D.; Tong, H.; Fei, C. Integrated Plasma and Bile Metabolomics Based on an UHPLC-Q/TOF-MS and Network Pharmacology Approach to Explore the Potential Mechanism of Schisandra chinensis-Protection From Acute Alcoholic Liver Injury. Front. Pharmacol. 2019, 10, 1543. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Swann, J.R. Nutrimetabonomics: Applications for nutritional sciences, with specific reference to gut microbial interactions. Annu. Rev. Food Sci. Technol. 2013, 4, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.L.; Rimm, E.B.; Kraft, P.; Clish, C.B.; Cassidy, A.; Hodgson, J.; Croft, K.; Wolpin, B.; Liang, L. Identifying the metabolomic fingerprint of high and low flavonoid consumers. J. Nutr. Sci. 2017, 6, e34. [Google Scholar] [CrossRef]
- Robertson, D.G.; Ruepp, S.U.; Stryker, S.A.; Hnatyshyn, S.Y.; Shipkova, P.A.; Aranibar, N.; Mcnaney, C.A.; Fiehn, O.; Reily, M.D. Metabolomic and transcriptomic changes induced by overnight (16 h) fasting in male and female Sprague-Dawley rats. Chem. Res. Toxicol. 2011, 24, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Abbondante, S.; Eckel-Mahan, K.L.; Ceglia, N.J.; Baldi, P.; Sassone-Corsi, P. Comparative Circadian Metabolomics Reveal Differential Effects of Nutritional Challenge in the Serum and Liver. J. Biol. Chem. 2016, 291, 2812–2828. [Google Scholar] [CrossRef] [PubMed]
- Zierer, J.; Jackson, M.A.; Kastenmüller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Beaumont, M.; Portune, K.J.; Steuer, N.; Lan, A.; Cerrudo, V.; Audebert, M.; Dumont, F.; Mancano, G.; Khodorova, N.; Andriamihaja, M.; et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: A randomized, parallel, double-blind trial in overweight humans. Am. J. Clin. Nutr. 2017, 106, 1005–1019. [Google Scholar] [CrossRef]
- Zhang, X.; Grosfeld, A.; Williams, E.; Vasiliauskas, D.; Barretto, S.; Smith, L.; Mariadassou, M.; Philippe, C.; Devime, F.; Melchior, C.; et al. Fructose malabsorption induces cholecystokinin expression in the ileum and cecum by changing microbiota composition and metabolism. FASEB J. 2019, 33, 7126–7142. [Google Scholar] [CrossRef]
- Choi, W.; Namkung, J.; Hwang, I.; Kim, H.; Lim, A.; Park, H.J.; Lee, H.W.; Han, K.-H.; Park, S.; Jeong, J.-S.; et al. Serotonin signals through a gut-liver axis to regulate hepatic steatosis. Nat. Commun. 2018, 9, 4824. [Google Scholar] [CrossRef]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961. [Google Scholar] [CrossRef]
- Huang, W.; Ramsey, K.M.; Marcheva, B.; Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Invest. 2011, 121, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.-K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.E.; DiTacchio, L.; Hayes, K.R.; Vollmers, C.; Pulivarthy, S.; Baggs, J.E.; Panda, S.; Hogenesch, J.B. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009, 5, e1000442. [Google Scholar] [CrossRef] [PubMed]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The human circadian metabolome. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef]
- Deprez, S.; Sweatman, B.C.; Connor, S.C.; Haselden, J.N.; Waterfield, C.J. Optimisation of collection, storage and preparation of rat plasma for 1H NMR spectroscopic analysis in toxicology studies to determine inherent variation in biochemical profiles. J. Pharm. Biomed. Anal. 2002, 30, 1297–1310. [Google Scholar] [CrossRef]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Rankin, K.N.; Wagner, J.; Fu, H.; Chang, D.; Weljie, A.M.; Saude, E.J.; Lix, B.; Adamko, D.J.; Shah, S.; et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal. Chem. 2007, 79, 6995–7004. [Google Scholar] [CrossRef]
- Stevens, V.L.; Hoover, E.; Wang, Y.; Zanetti, K.A. Pre-Analytical Factors that Affect Metabolite Stability in Human Urine, Plasma, and Serum: A Review. Metabolites 2019, 9, 156. [Google Scholar] [CrossRef]
- Eriksson, E.; Royo, F.; Lyberg, K.; Carlsson, H.-E.; Hau, J. Effect of metabolic cage housing on immunoglobulin A and corticosterone excretion in faeces and urine of young male rats. Exp. Physiol. 2004, 89, 427–433. [Google Scholar] [CrossRef]
- Hoffman, J.F.; Fan, A.X.; Neuendorf, E.H.; Vergara, V.B.; Kalinich, J.F. Hydrophobic Sand Versus Metabolic Cages: A Comparison of Urine Collection Methods for Rats (Rattus norvegicus). J. Am. Assoc. Lab. Anim. Sci. 2018, 57, 51–57. [Google Scholar]
- Saude, E.J.; Sykes, B.D. Urine stability for metabolomic studies: Effects of preparation and storage. Metabolomics 2007, 3, 19–27. [Google Scholar] [CrossRef]
- Thongboonkerd, V.; Saetun, P. Bacterial overgrowth affects urinary proteome analysis: Recommendation for centrifugation, temperature, duration, and the use of preservatives during sample collection. J. Proteome Res. 2007, 6, 4173–4181. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Peralbo, M.A.; Luque de Castro, M.D. Preparation of urine samples prior to targeted or untargeted metabolomics mass-spectrometry analysis. TrAC Trends Anal. Chem. 2012, 41, 75–85. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Wilson, I.D. Liquid chromatography and ultra-performance liquid chromatography–mass spectrometry fingerprinting of human urine: Sample stability under different handling and storage conditions for metabonomics studies. J. Chromatogr. A 2008, 1189, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, D.K.; Jones, H.; Shah, A.J.; Iles, R.K. Development of Zwitterionic Hydrophilic Liquid Chromatography (ZICⓇHILIC-MS) metabolomics method for Shotgun analysis of human urine. J. Chromatogr. Sep. Tech. 2012, 3. [Google Scholar] [CrossRef]
- Rachita Dhurat, M.S. Principles and Methods of Preparation of Platelet-Rich Plasma: A Review and Author’s Perspective. J. Cutan. Aesthetic Surg. 2014, 7, 189–197. [Google Scholar] [CrossRef]
- Teahan, O.; Gamble, S.; Holmes, E.; Waxman, J.; Nicholson, J.K.; Charlotte Bevan, A.; Keun, H.C. Impact of Analytical Bias in Metabonomic Studies of Human Blood Serum and Plasma. Anal. Chem. 2006, 78, 4307–4318. [Google Scholar] [CrossRef]
- Yin, P.; Lehmann, R.; Xu, G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal. Bioanal. Chem. 2015, 407, 4879–4892. [Google Scholar] [CrossRef]
- Denery, J.R.; Nunes, A.A.K.; Dickerson, T.J. Characterization of differences between blood sample matrices in untargeted metabolomics. Anal. Chem. 2011, 83, 1040–1047. [Google Scholar] [CrossRef]
- Barri, T.; Dragsted, L.O. UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: Effect of experimental artefacts and anticoagulant. Anal. Chim. Acta 2013, 768, 118–128. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Human Serum Metabolome (HUSERMET) Consortium Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Pinto, J.; Domingues, M.R.M.; Galhano, E.; Pita, C.; Almeida, M.D.C.; Carreira, I.M.; Gil, A.M. Human plasma stability during handling and storage: Impact on NMR metabolomics. Analyst 2014, 139, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.L.; Yasui, Y.; Li, C.I.; Fitzpatrick, A.L.; Lampe, P.D. Impact of freeze-thaw cycles and storage time on plasma samples used in mass spectrometry based biomarker discovery projects. Cancer Inform 2005, 1, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Hernandes, V.V.; Barbas, C.; Dudzik, D. A review of blood sample handling and pre-processing for metabolomics studies. Electrophoresis 2017, 38, 2232–2241. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Karu, N.; Deng, L.; Slae, M.; Guo, A.C.; Sajed, T.; Huynh, H.; Wine, E.; Wishart, D.S. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Anal. Chim. Acta 2018, 1030, 1–24. [Google Scholar] [CrossRef]
- Probert, C.S.J.; Jones, P.R.H.; Ratcliffe, N.M. A novel method for rapidly diagnosing the causes of diarrhoea. Gut 2004, 53, 58–61. [Google Scholar] [CrossRef]
- Garner, C.E.; Smith, S.; de Lacy Costello, B.; White, P.; Spencer, R.; Probert, C.S.J.; Ratcliffe, N.M. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007, 21, 1675–1688. [Google Scholar] [CrossRef]
- Rose, C.; Parker, A.; Jefferson, B.; Cartmell, E. The Characterization of Feces and Urine: A Review of the Literature to Inform Advanced Treatment Technology. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1827–1879. [Google Scholar] [CrossRef]
- Matysik, S.; Le Roy, C.I.; Liebisch, G.; Claus, S.P. Metabolomics of fecal samples: A practical consideration. Trends Food Sci. Technol. 2016, 57, 244–255. [Google Scholar] [CrossRef]
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484. [Google Scholar] [CrossRef] [PubMed]
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015, 17, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Duparc, T.; Plovier, H.; Marrachelli, V.G.; Van Hul, M.; Essaghir, A.; Ståhlman, M.; Matamoros, S.; Geurts, L.; Pardo-Tendero, M.M.; Druart, C.; et al. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut 2016, 66, 620–632. [Google Scholar] [CrossRef]
- Sberna, A.L.; Assem, M.; Gautier, T.; Grober, J.; Guiu, B.; Jeannin, A.; Pais de Barros, J.-P.; Athias, A.; Lagrost, L.; Masson, D. Constitutive androstane receptor activation stimulates faecal bile acid excretion and reverse cholesterol transport in mice. J. Hepatol. 2011, 55, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.; Sundekilde, U.K.; Blædel, T.; Dalsgaard, T.K.; Larsen, L.H.; Dragsted, L.O.; Astrup, A.; Bertram, H.C. Optimizing sampling strategies for NMR-based metabolomics of human feces: Pooled vs. unpooled analyses. Anal. Methods 2017, 9, 4476–4480. [Google Scholar] [CrossRef]
- Phua, L.C.; Koh, P.K.; Cheah, P.Y.; Ho, H.K.; Chan, E.C.Y. Global gas chromatography/time-of-flight mass spectrometry (GC/TOFMS)-based metabonomic profiling of lyophilized human feces. J. Chromatogr. B 2013, 937, 103–113. [Google Scholar] [CrossRef]
- Lamichhane, S.; Yde, C.C.; Schmedes, M.S.; Jensen, H.M.; Meier, S.; Bertram, H.C. Strategy for Nuclear-Magnetic-Resonance-Based Metabolomics of Human Feces. Anal. Chem. 2015, 87, 5930–5937. [Google Scholar] [CrossRef]
- Le Gall, G.; Noor, S.O.; Ridgway, K.; Scovell, L.; Jamieson, C.; Johnson, I.T.; Colquhoun, I.J.; Kemsley, E.K.; Narbad, A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 2011, 10, 4208–4218. [Google Scholar] [CrossRef]
- Armstrong, C.W.; McGregor, N.R.; Lewis, D.P.; Butt, H.L.; Gooley, P.R. The association of fecal microbiota and fecal, blood serum and urine metabolites in myalgic encephalomyelitis/chronic fatigue syndrome. Metabolomics 2016, 13, 8. [Google Scholar] [CrossRef]
- Bezabeh, T.; Somorjai, R.L.; Smith, I.C.P. MR metabolomics of fecal extracts: Applications in the study of bowel diseases. Magn. Reson. Chem. 2009, 47, S54–S61. [Google Scholar] [CrossRef] [PubMed]
- Gratton, J.; Phetcharaburanin, J.; Mullish, B.H.; Williams, H.R.T.; Thursz, M.; Nicholson, J.K.; Holmes, E.; Marchesi, J.R.; Li, J.V. Optimized Sample Handling Strategy for Metabolic Profiling of Human Feces. Anal. Chem. 2016, 88, 4661–4668. [Google Scholar] [CrossRef] [PubMed]
- Yen, S.; McDonald, J.A.K.; Schroeter, K.; Oliphant, K.; Sokolenko, S.; Blondeel, E.J.M.; Allen-Vercoe, E.; Aucoin, M.G. Metabolomic Analysis of Human Fecal Microbiota: A Comparison of Feces-Derived Communities and Defined Mixed Communities. J. Proteome Res. 2015, 14, 1472–1482. [Google Scholar] [CrossRef]
- Couch, R.D.; Navarro, K.; Sikaroodi, M.; Gillevet, P.; Forsyth, C.B.; Mutlu, E.; Engen, P.A.; Keshavarzian, A. The Approach to Sample Acquisition and Its Impact on the Derived Human Fecal Microbiome and VOC Metabolome. PLoS ONE 2013, 8, e81163. [Google Scholar] [CrossRef]
- Loftfield, E.; Vogtmann, E.; Sampson, J.N.; Moore, S.C.; Nelson, H.; Knight, R.; Chia, N.; Sinha, R. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1483–1490. [Google Scholar] [CrossRef]
- Saric, J.; Wang, Y.; Li, J.; Coen, M.; Utzinger, J.; Marchesi, J.R.; Keiser, J.; Veselkov, K.; Lindon, J.C.; Nicholson, J.K.; et al. Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J. Proteome Res. 2008, 7, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Moosmang, S.; Pitscheider, M.; Sturm, S.; Seger, C.; Tilg, H.; Halabalaki, M.; Stuppner, H. Metabolomic analysis-Addressing NMR and LC-MS related problems in human feces sample preparation. Clin. Chim. Acta 2017, 489, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Jungermann, K.; Katz, N. Functional specialization of different hepatocyte populations. Physiol. Rev. 1989, 69, 708–764. [Google Scholar] [CrossRef]
- Braeuning, A.; Ittrich, C.; Köhle, C.; Hailfinger, S.; Bonin, M.; Buchmann, A.; Schwarz, M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006, 273, 5051–5061. [Google Scholar] [CrossRef]
- Isse, K.; Lesniak, A.; Grama, K.; Maier, J.; Specht, S.; Castillo-Rama, M.; Lunz, J.; Roysam, B.; Michalopoulos, G.; Demetris, A.J. Preexisting epithelial diversity in normal human livers: A tissue-tethered cytometric analysis in portal/periportal epithelial cells. Hepatology 2013, 57, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Hu, Z.-P.; Browne, E.R.; Liu, T.; Angel, T.E.; Ho, P.C.; Chan, E.C.Y. Metabonomic profiling of TASTPM transgenic Alzheimer’s disease mouse model. J. Proteome Res. 2012, 11, 5903–5913. [Google Scholar] [CrossRef]
- Garrod, S.; Humpfer, E.; Spraul, M.; Connor, S.C.; Polley, S.; Connelly, J.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. High-resolution magic angle spinning 1H NMR spectroscopic studies on intact rat renal cortex and medulla. Magn. Reson. Med. 1999, 41, 1108–1118. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, W.; Xie, W.; Chen, H.; Yu, W.; Li, H.; Shen, G. Tributyl phosphate impairs the urea cycle and alters liver pathology and metabolism in mice after short-term exposure based on a metabonomics study. Sci. Total Environ. 2017, 603-604, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Abuja, P.M.; Ehrhart, F.; Schoen, U.; Schmidt, T.; Stracke, F.; Dallmann, G.; Friedrich, T.; Zimmermann, H.; Zatloukal, K. Alterations in Human Liver Metabolome during Prolonged Cryostorage. J. Proteome Res. 2015, 14, 2758–2768. [Google Scholar] [CrossRef] [PubMed]
- Torell, F.; Bennett, K.; Cereghini, S.; Rännar, S.; Lundstedt-Enkel, K.; Moritz, T.; Haumaitre, C.; Trygg, J.; Lundstedt, T. Tissue sample stability: Thawing effect on multi-organ samples. Metabolomics 2015, 12, 19. [Google Scholar] [CrossRef]
- Kapoore, R.V.; Coyle, R.; Staton, C.A.; Brown, N.J.; Vaidyanathan, S. Influence of washing and quenching in profiling the metabolome of adherent mammalian cells: A case study with the metastatic breast cancer cell line MDA-MB-231. Analyst 2017, 142, 2038–2049. [Google Scholar] [CrossRef]
- Lorenz, M.A.; Burant, C.F.; Kennedy, R.T. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 2011, 83, 3406–3414. [Google Scholar] [CrossRef]
- Rusz, M.; Rampler, E.; Keppler, B.K.; Jakupec, M.A.; Koellensperger, G. Single Spheroid Metabolomics: Optimizing Sample Preparation of Three-Dimensional Multicellular Tumor Spheroids. Metabolites 2019, 9, 304. [Google Scholar] [CrossRef]
- Shryock, J.C.; Rubio, R.; Berne, R.M. Extraction of adenine nucleotides from cultured endothelial cells. Anal. Biochem. 1986, 159, 73–81. [Google Scholar] [CrossRef]
- Kořínek, M.; Šístek, V.; Mládková, J.; Mikeš, P.; Jiráček, J.; Selicharová, I. Quantification of homocysteine-related metabolites and the role of betaine–homocysteine S-methyltransferase in HepG2 cells. Biomed. Chromatogr. 2013, 27, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, C.; Simó, C.; Valdés, A.; Campone, L.; Piccinelli, A.L.; García-Cañas, V.; Cifuentes, A. Metabolomics of adherent mammalian cells by capillary electrophoresis-mass spectrometry: HT-29 cells as case study. J. Pharm. Biomed. Anal. 2015, 110, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Nürnberger, N.; Kaspar, H.; Gruber, M.A.; Almstetter, M.F.; Oefner, P.J. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: Optimization of harvesting and extraction protocols. Anal. Bioanal. Chem. 2011, 399, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- León, Z.; García-Cañaveras, J.C.; Donato, M.T.; Lahoz, A. Mammalian cell metabolomics: Experimental design and sample preparation. Electrophoresis 2013, 34, 2762–2775. [Google Scholar] [CrossRef]
- Yuan, W.; Anderson, K.W.; Li, S.; Edwards, J.L. Subsecond absolute quantitation of amine metabolites using isobaric tags for discovery of pathway activation in mammalian cells. Anal. Chem. 2012, 84, 2892–2899. [Google Scholar] [CrossRef]
- Poupin, N.; Corlu, A.; Cabaton, N.J.; Dubois-Pot-Schneider, H.; Canlet, C.; Person, E.; Bruel, S.; Frainay, C.; Vinson, F.; Maurier, F.; et al. Large-Scale Modeling Approach Reveals Functional Metabolic Shifts during Hepatic Differentiation. J. Proteome Res. 2019, 18, 204–216. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Højer-Pedersen, J.; Akesson, M.; Smedsgaard, J.; Nielsen, J. Global metabolite analysis of yeast: Evaluation of sample preparation methods. Yeast 2005, 22, 1155–1169. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, L.; Villaret-Cazadamont, J.; Claus, S.P.; Canlet, C.; Guillou, H.; Cabaton, N.J.; Ellero-Simatos, S. Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites 2020, 10, 104. https://doi.org/10.3390/metabo10030104
Smith L, Villaret-Cazadamont J, Claus SP, Canlet C, Guillou H, Cabaton NJ, Ellero-Simatos S. Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites. 2020; 10(3):104. https://doi.org/10.3390/metabo10030104
Chicago/Turabian StyleSmith, Lorraine, Joran Villaret-Cazadamont, Sandrine P. Claus, Cécile Canlet, Hervé Guillou, Nicolas J. Cabaton, and Sandrine Ellero-Simatos. 2020. "Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions" Metabolites 10, no. 3: 104. https://doi.org/10.3390/metabo10030104
APA StyleSmith, L., Villaret-Cazadamont, J., Claus, S. P., Canlet, C., Guillou, H., Cabaton, N. J., & Ellero-Simatos, S. (2020). Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites, 10(3), 104. https://doi.org/10.3390/metabo10030104





