Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
2.2. Serum Metabolite Profiles Cluster According to NAFL, Early-NASH and Advanced-NASH
2.3. Individual Metabolites were Associated with Histological Severity in NAFLD
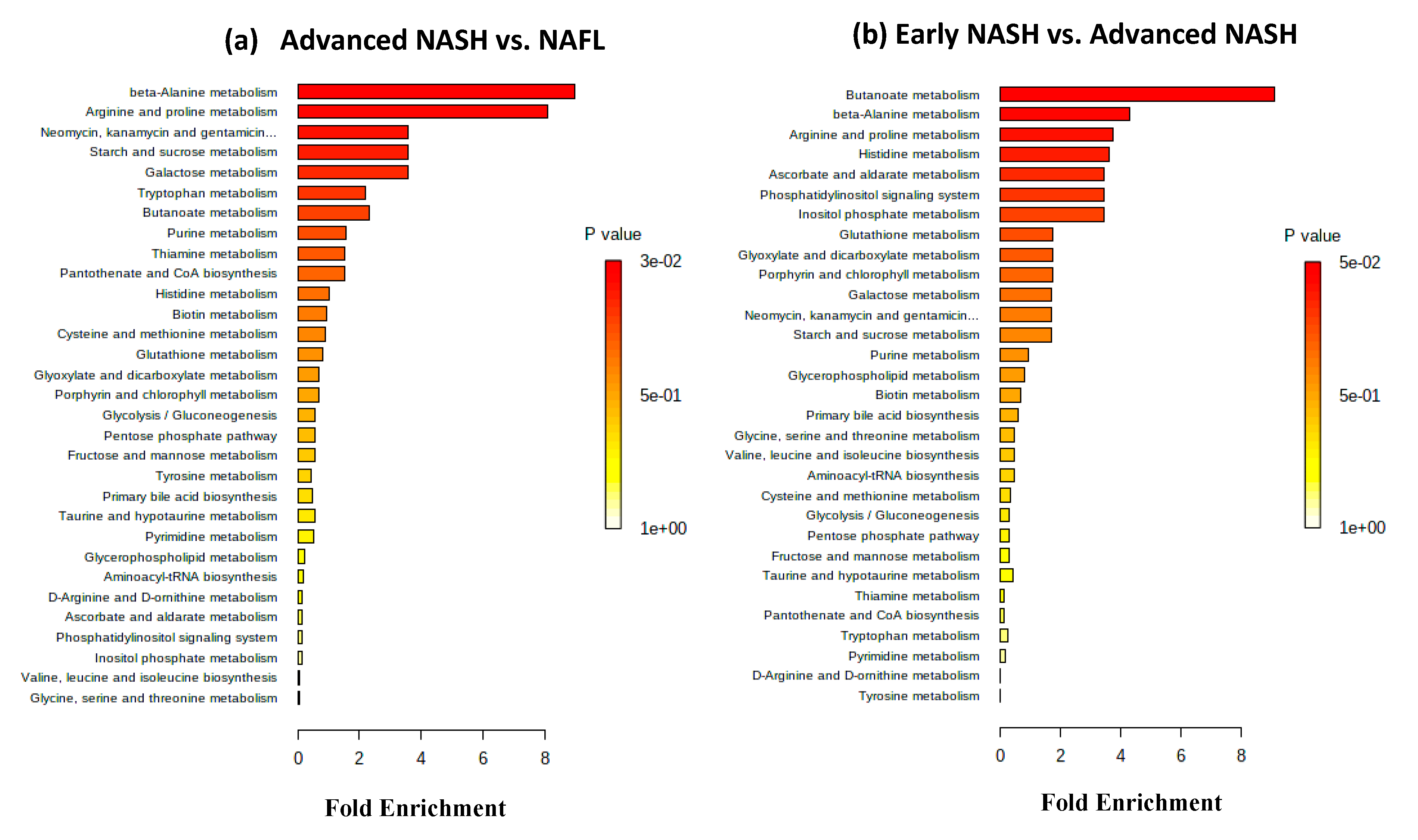
2.4. Metabolic Pathways and Metabolite Enrichment Distinguished NAFL, Early-NASH, Advanced-NASH
2.5. Association of the GSG-Index with Histological Severity
2.6. Evaluation of the Confounding Effects of Gender and Diabetes on the Biomarkers of NASH/NAFL
3. Discussion
4. Materials and Methods
4.1. Study Population
- NAFL (n = 12): Steatosis grade 1–3 with no or minimal inflammation (grade 0–1), no ballooning degeneration (grade 0) and no fibrosis (stage F0).
- Early-NASH (n = 31): No or mild fibrosis (stage F0-F1) and NAFLD activity score (NAS) 3–4, including steatosis grade ≥1, inflammation ≥1, and ballooning degeneration score ≥1. The NAS score is the sum of steatosis (0–3) plus inflammation (0–3) plus ballooning degeneration (0–2) grades and takes values ranging from 1–8 [43].
- Advanced-NASH (n = 14): Fibrosis score F1-F4 and NAS score 5–8, including ballooning degeneration score = 2.
4.2. Serum Metabolite Profiling: LC-MS
4.3. NMR Spectroscopy
4.4. Statistical, Enrichment, and Pathway Analyses
4.5. Evaluation of the GSG-Index
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marchesini, G.; Brizi, M.; Bianchi, G.; Tomassetti, S.; Bugianesi, E.; Lenzi, M.; McCullough, A.J.; Natale, S.; Forlani, G.; Melchionda, N. Nonalcoholic fatty liver disease: A feature of the metabolic syndrome. Diabetes 2001, 50, 1844–1850. [Google Scholar] [CrossRef] [PubMed]
- Pagano, G.; Pacini, G.; Musso, G.; Gambino, R.; Mecca, F.; Depetris, N.; Cassader, M.; David, E.; Cavallo-Perin, P.; Rizzetto, M. Nonalcoholic steatohepatitis, insulin resistance, and metabolic syndrome: Further evidence for an etiologic association. Hepatology 2002, 35, 367–372. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [PubMed]
- Matteoni, C.A.; Younossi, Z.M.; Gramlich, T.; Boparai, N.; Liu, Y.C.; McCullough, A.J. Nonalcoholic fatty liver disease: A spectrum of clinical and pathological severity. Gastroenterology 1999, 116, 1413–1419. [Google Scholar] [CrossRef]
- Bugianesi, E.; Leone, N.; Vanni, E.; Marchesini, G.; Brunello, F.; Carucci, P.; Musso, A.; De Paolis, P.; Capussotti, L.; Salizzoni, M.; et al. Expanding the natural history of nonalcoholic steatohepatitis: From cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology 2002, 123, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Angulo, P.; Kleiner, D.E.; Dam-Larsen, S.; Adams, L.A.; Bjornsson, E.S.; Charatcharoenwitthaya, P.; Mills, P.R.; Keach, J.C.; Lafferty, H.D.; Stahler, A.; et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2015, 149, 389–397. [Google Scholar] [CrossRef]
- Ekstedt, M.; Hagström, H.; Nasr, P.; Fredrikson, M.; Stål, P.; Kechagias, S.; Hultcrantz, R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 2015, 61, 1547–1554. [Google Scholar] [CrossRef]
- Lindon, J.C.; Nicholson, J.K. The emergent role of metabolic phenotyping in dynamic patient stratification. Expert Opin. Drug Metab. Toxicol. 2014, 10, 915–919. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Biomarker Discovery and Translation in Metabolomics. Curr. Metab. 2013, 1, 227–240. [Google Scholar] [CrossRef]
- Dang, N.H.; Singla, A.K.; Mackay, E.M.; Jirik, F.R.; Weljie, A.M. Targeted cancer therapeutics: Biosynthetic and energetic pathways characterized by metabolomics and the interplay with key cancer regulatory factors. Curr. Pharm. Des. 2014, 20, 2637–2647. [Google Scholar] [CrossRef]
- Raftery, D. Mass Spectrometry in Metabolomics-Methods and Protocols. Methods in Molecular Biology; Walker, J., Ed.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Nagana Gowda, G.A.; Raftery, D. Recent Advances in NMR-Based Metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Suryani, Y.; Nagana Gowda, G.A.; Skill, N.; Maluccio, M.; Raftery, D. Differentiating hepatocellular carcinoma from hepatitis C using metabolite profiling. Metabolites 2012, 2, 701–716. [Google Scholar] [CrossRef] [PubMed]
- Baniasadi, H.; Nagana Gowda, G.A.; Gu, H.; Zeng, A.; Zhuang, S.; Skill, N.; Maluccio, M.; Raftery, D. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis 2013, 34, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.; Hughes, E.; Skill, N.; Maluccio, M.; Raftery, D. Detection of hepatocellular carcinoma in hepatitis C patients: Biomarker discovery by LC-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014, 966, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hong, Z.; Tan, G.; Dong, X.; Yang, G.; Zhao, L.; Chen, X.; Zhu, Z.; Lou, Z.; Qian, B.; et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int. J. Cancer 2014, 135, 658–668. [Google Scholar] [CrossRef]
- Huang, Q.; Tan, Y.; Yin, P.; Ye, G.; Gao, P.; Lu, X.; Wang, H.; Xu, G. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013, 73, 4992–5002. [Google Scholar] [CrossRef]
- Tan, Y.; Yin, P.; Tang, L.; Xing, W.; Huang, Q.; Cao, D.; Zhao, X.; Wang, W.; Lu, X.; Xu, Z.; et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: Potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol. Cell Proteom. 2012, 11, M111 010694. [Google Scholar] [CrossRef]
- Patterson, A.D.; Maurhofer, O.; Beyoglu, D.; Lanz, C.; Krausz, K.W.; Pabst, T.; Gonzalez, F.J.; Dufour, J.F.; Idle, J.R. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011, 71, 6590–6600. [Google Scholar] [CrossRef]
- Safaei, A.; Arefi Oskouie, A.; Mohebbi, S.R.; Rezaei-Tavirani, M.; Mahboubi, M.; Peyvandi, M.; Okhovatian, F.; Zamanian-Azodi, M. Metabolomic analysis of human cirrhosis, hepatocellular carcinoma, non-alcoholic fatty liver disease and non-alcoholic steatohepatitis diseases. Gastroenterol. Hepatol. Bed. Bench. 2016, 9, 158–173. [Google Scholar]
- Di Poto, C.; Ferrarini, A.; Zhao, Y.; Varghese, R.S.; Tu, C.; Zuo, Y.; Wang, M.; Nezami Ranjbar, M.R.; Luo, Y.; Zhang, C.; et al. Metabolomic Characterization of Hepatocellular Carcinoma in Patients with Liver Cirrhosis for Biomarker Discovery. Cancer Epidemiol. Biomark. Prev. 2017, 26, 675–683. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Li, W.; Zou, J.; Jiang, X.; Xu, G.; Huang, H.; Liu, L. Spermidine Prolongs Lifespan and Prevents Liver Fibrosis and Hepatocellular Carcinoma by Activating MAP1S-Mediated Autophagy. Cancer Res. 2017, 77, 2938–2951. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Ryu, S.H.; Kim, S.; Lee, H.W.; Lim, M.S.; Seong, S.J.; Kim, S.; Yoon, Y.R.; Kim, K.B. Pattern recognition analysis for hepatotoxicity induced by acetaminophen using plasma and urinary 1H NMR-based metabolomics in humans. Anal. Chem. 2013, 85, 11326–11334. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Patterson, A.D.; Yang, Q.; Krausz, K.W.; Idle, J.R.; Fornace, A.J.; Gonzalez, F.J. UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J. Proteome Res. 2011, 10, 4120–4133. [Google Scholar] [CrossRef]
- Bradford, B.U.; O’Connell, T.M.; Han, J.; Kosyk, O.; Shymonyak, S.; Ross, P.K.; Winnike, J.; Kono, H.; Rusyn, I. Metabolomic profiling of a modified alcohol liquid diet model for liver injury in the mouse uncovers new markers of disease. Toxicol. Appl. Pharmacol. 2008, 232, 236–243. [Google Scholar] [CrossRef]
- van Waeg, G.; Lööf, L.; Groth, T.; Niklasson, F. Allopurinol kinetics in humans as a means to assess liver function: Evaluation of an allopurinol loading test. Scand. J. Clin. Lab Invest. 1988, 48, 45–57. [Google Scholar] [CrossRef]
- Barr, J.; Caballeria, J.; Martinez-Arranz, I.; Domínguez-DíezM, A.; Alonso, C.; Muntané, J.; Pérez-Cormenzana, M.; García-Monzón, C.; Mayo, R.; Martín-Duce, A.; et al. Obesitydependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J. Proteome Res. 2012, 11, 2521–2532. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Ijare, O.B.; Somashekar, B.S.; Sharma, A.; Kapoor, V.K.; Khetrapal, C.L. Single-step analysis of individual conjugated bile acids in human bile using 1H NMR spectroscopy. Lipids 2006, 41, 591–603. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 2011, 60, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G.; Marchesini, G.; Brunetti, N.; Manicardi, E.; Montuschi, F.; Chianese, R.; Zoli, M. Impaired insulin-mediated amino acid plasma disappearance in non-alcoholic fatty liver disease: A feature of insulin resistance. Dig. Liver Dis. 2003, 35, 722–727. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Agren, R.; Kampf, C.; Asplund, A.; Uhlen, M.; Nielsen, J. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 3083. [Google Scholar] [CrossRef] [PubMed]
- Lake, A.D.; Novak, P.; Shipkova, P.; Aranibar, N.; Robertson, D.G.; Reily, M.D.; Lehman-McKeeman, L.D.; Vaillancourt, R.R.; Cherrington, N.J. Branched chain amino acid metabolism profiles in progressive human nonalcoholic fatty liver disease. Amino Acids 2015, 47, 603–615. [Google Scholar] [CrossRef]
- Hyotylainen, T.; Jerby, L.; Petaja, E.M.; Mattila, I.; Jantti, S.; Auvinen, P.; Gastaldelli, A.; Yki-Järvinen, H.; Ruppin, E.; Orešič, M. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat. Commun. 2016, 7, 8994. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Bjornson, E.; Zhang, C.; Klevstig, M.; Soderlund, S.; Stahlman, M.; Adiels, M.; Hakkarainen, A.; Lundbom, N.; Kilicarslan, M.; et al. Personal model-assisted identification of NAD+ and glutathione metabolism as intervention target in NAFLD. Mol. Syst. Biol. 2017, 13, 916. [Google Scholar] [CrossRef]
- Gaggini, M.; Carli, F.; Rosso, C.; Buzzigoli, E.; Marietti, M.; Della Latta, V.; Ciociaro, D.; Abate, M.L.; Gambino, R.; Cassader, M.; et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 2018, 67, 145–158. [Google Scholar] [CrossRef]
- Koliaki, C.; Szendroedi, J.; Kaul, K.; Jelenik, T.; Nowotny, P.; Jankowiak, F.; Herder, C.; Carstensen, M.; Krausch, M.; Knoefel, W.T.; et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015, 21, 739–746. [Google Scholar] [CrossRef]
- Dong, S.; Zhan, Z.Y.; Cao, H.Y.; Wu, C.; Bian, Y.Q.; Li, J.Y.; Cheng, G.H.; Liu, P.; Sun, M.Y. Urinary metabolomics analysis identifies key biomarkers of different stages of nonalcoholic fatty liver disease. World J. Gastroenterol. 2017, 23, 2771–2784. [Google Scholar] [CrossRef]
- Alonso, C.; Fernández-Ramos, D.; Varela-Rey, M.; Martínez-Arranz, I.; Navasa, N.; Van Liempd, S.M.; Lavín Trueba, J.L.; Mayo, R.; Ilisso, C.P.; de Juan, V.G.; et al. Metabolomic Identification of Subtypes of Nonalcoholic Steatohepatitis. Gastroenterology 2017, 152, 1449–1461. [Google Scholar] [CrossRef]
- Castera, L.; Friedrich-Rust, M.; Loomba, R. Noninvasive Assessment of Liver Disease in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 156, 1264–1281 e4. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver (EASL). European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1388–1402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Gowda, Y.N.; Raftery, D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal. Chem. 2015, 87, 706–715. [Google Scholar] [CrossRef]

| Parameter | Simple Steatosis n = 12 | Early NASH n = 31 | Advanced NASH n = 14 |
|---|---|---|---|
| Age (yrs), mean (SD) | 50.2 (12) | 50.4 (9.5) | 52.3 (9.1) |
| Race: | |||
| White, non-Hispanic (%) | 100% | 73% | 87% |
| Black, non-Hispanic (%) | 0% | 7% | 0% |
| Other (%) | 0% | 10% | 3% |
| Not-declared (%) | 0% | 10% | 10% |
| Male (%) | 92% | 93% | 80% |
| Diabetes (%) | 75% | 71% | 40% |
| BMI (Kg/m2), mean (SD) | 35 (7) | 33 (5) | 34 (6) |
| Serum Laboratory Tests, mean (SD): | |||
| AST (U/L) | 39 (32) | 39 (20) | 66 (39) |
| ALT (U/L) | 50 (30) | 62 (32) | 97 (42) |
| Albumin (g/dL) | 4.4 (0.4) | 4.6 (0.2) | 4.6 (0.2) |
| Bilirubin (g/dL) | 0.7 (0.4) | 0.5 (0.1) | 0.5 (0.3) |
| Liver Histology: | |||
| Steatosis Grade 0/1/2/3 | 0/7/4/1 | 0/19/11/1 | 0/1/9/4 |
| Inflammation Grade 0/1/2/3 | 3/9/0/0 | 0/30/1/0 | 0/4/10/0 |
| Ballooning Degeneration 0/1/2 | 12/0/0 | 0/27/4 | 0/1/13 |
| Fibrosis Stage 0/1/2/3/4 | 12/0/0/0/0 | 10/21/0/0/0 | 0/4/6/3/1 |
| NAS Score (0–8), mean (SD) | 2.4 (0.8) | 3.3 (0.7) | 5.7 (0.6) |
| NASH vs. NAFL | Early NASH vs. NAFL | ||||||
| Metabolite | p Value | Fold * Change | Method | Metabolite | p Value | Fold * Change | Method |
| Acetylglycine | 0.03 | 0.57 | MS | Hydroxyphenylpyruvate | 0.002 | 0.83 | MS |
| Cysteine | 0.04 | 0.88 | MS | Inositol | 0.03 | 0.86 | MS |
| Alanine | 0.02 | 0.96 | NMR | Cysteine | 0.04 | 0.87 | MS |
| Glucose | 0.04 | 1.16 | MS | Acetylcarnitine | 0.04 | 0.90 | MS |
| Erythrose | 0.02 | 1.18 | MS | Phenylalanine | 0.03 | 1.12 | NMR |
| Tyrosine | 0.01 | 1.18 | NMR | Tyrosine | 0.02 | 1.18 | NMR |
| Isovaleric acid | 0.02 | 1.25 | MS | Erythrose | 0.04 | 1.18 | MS |
| Leucic acid | 0.04 | 1.28 | MS | Alanine | 0.03 | 1.18 | NMR |
| Xanthine | 0.02 | 1.49 | MS | Tryptophan | 0.04 | 1.19 | NMR |
| Oxypurinol | 0.01 | 1.54 | MS | ||||
| Glycochenodeoxycholate | 0.04 | 3.13 | MS | ||||
| Advanced NASH vs. Early NASH | Advanced NASH vs. NAFL | ||||||
| Metabolite | p Value | Fold * Change | Method | Metabolite | p Value | Fold * Change | Method |
| Spermidine | 0.005 | 0.49 | MS | Spermidine | 0.005 | 0.33 | MS |
| Oxaloacetate | 0.01 | 0.85 | MS | Acetylglycine | 0.01 | 0.48 | MS |
| Orotate | 0.0009 | 0.85 | MS | Glucose | 0.04 | 1.20 | MS |
| Linoleic acid | 0.01 | 1.32 | MS | Isovaleric acid | 0.04 | 1.30 | MS |
| Linolenic acid | 0.01 | 1.33 | MS | Leucic acid | 0.02 | 1.30 | MS |
| 2-hydroxyglutarate | 0.01 | 1.33 | MS | 2-hydroxyisovaleric acid | 0.03 | 1.49 | MS |
| Xanthine | 0.04 | 2.08 | MS | ||||
| Oxypurinol | 0.04 | 2.17 | MS | ||||
| Glycocholate | 0.02 | 2.22 | MS | ||||
| Glycochenodeoxycholate | 0.01 | 2.38 | MS | ||||
| Fibrosis Stage 2–4 vs. Fibrosis Stage 0–1 | Steatosis Grade 2–3 vs. Steatosis Grade 0–1 | ||||||
|---|---|---|---|---|---|---|---|
| Metabolite | p Value | Fold Change * | Method | Metabolite | p Value | Fold Change * | Method |
| Spermidine | 0.0008 | 0.47 | MS | Erythrose | 0.01 | 1.19 | MS |
| N-acetylglycine | 0.001 | 0.63 | MS | Mannose | 0.002 | 1.33 | NMR |
| Oxaloacetate | 0.004 | 0.83 | MS | Isovaleric acid | 0.01 | 1.33 | MS |
| Orotate | 0.01 | 0.85 | MS | Glucose | 0.02/0.005 | 1.37/1.22 | NMR/MS |
| Adipic acid | 0.03 | 1.20 | MS | ||||
| Sucrose | 0.04 | 1.20 | MS | ||||
| Aconitate | 0.03 | 1.23 | MS | ||||
| Azelaic acid | 0.04 | 1.25 | MS | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ioannou, G.N.; Nagana Gowda, G.A.; Djukovic, D.; Raftery, D. Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach. Metabolites 2020, 10, 168. https://doi.org/10.3390/metabo10040168
Ioannou GN, Nagana Gowda GA, Djukovic D, Raftery D. Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach. Metabolites. 2020; 10(4):168. https://doi.org/10.3390/metabo10040168
Chicago/Turabian StyleIoannou, George N., G. A. Nagana Gowda, Danijel Djukovic, and Daniel Raftery. 2020. "Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach" Metabolites 10, no. 4: 168. https://doi.org/10.3390/metabo10040168
APA StyleIoannou, G. N., Nagana Gowda, G. A., Djukovic, D., & Raftery, D. (2020). Distinguishing NASH Histological Severity Using a Multiplatform Metabolomics Approach. Metabolites, 10(4), 168. https://doi.org/10.3390/metabo10040168







