A Review of Automatic Phenotyping Approaches using Electronic Health Records
Abstract
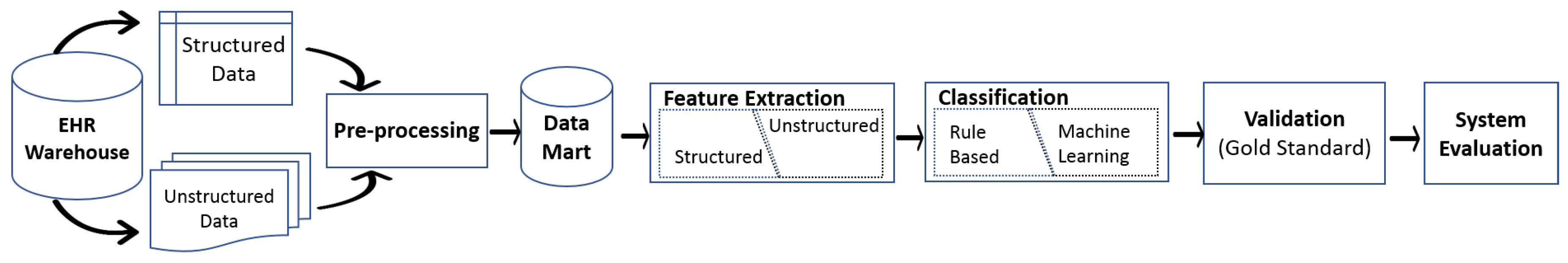
1. Introduction
2. EHR Warehouse
- A large number of clinical studies have developed phenotyping algorithms for specific institutions and it is difficult to generalize these algorithms across other institutions.
- Building an EHR-based phenotyping algorithm requires participation from clinical experts to review subsets within a patient’s record to label it as a case (healthy) or a control (affected) and to validate the algorithm output; these efforts take a considerable amount of time [21].
- EHRs contain noisy data due to missing or incomplete information. Moreover, temporal information may be conflicting or incompatible [22].
- EHR data are highly relational and multi-modal. The information in EHRs is stored in a complex relational structure, and the information of a single record is spread across multiple tables. Restructuring an EHR database to a more simple structure by performing flattening techniques, such as a “join” operation, may result to loss of information [23].
3. Pre-Processing
4. Feature Extraction
4.1. Structured Feature Extraction
4.2. Unstructured Feature Extraction
4.2.1. Bag of Words (BoW)
4.2.2. Keywords Search
4.2.3. Concept Extraction
4.2.4. Feature Selection
5. Classification
5.1. Rule-Based
5.1.1. Rules Based on Clinical Judgment
5.1.2. Rules Based on Healthcare Guidelines
5.2. Machine Learning
5.2.1. Supervised Learning
5.2.2. Unsupervised Learning
5.2.3. Combined Approaches
6. Validation and Evaluation
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Häyrinen, K.; Saranto, K.; Nykänen, P. Definition, structure, content, use and impacts of electronic health records: A review of the research literature. Int. J. Med. Inform. 2008, 77, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Hersh, W.R.; Weiner, M.G.; Embi, P.J.; Logan, J.R.; Payne, P.R.; Bernstam, E.V.; Lehmann, H.P.; Hripcsak, G.; Hartzog, T.H.; Cimino, J.J.; et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med. Care 2013, 51, S30. [Google Scholar] [CrossRef]
- Botsis, T.; Hartvigsen, G.; Chen, F.; Weng, C. Secondary use of EHR: Data quality issues and informatics opportunities. Summit Transl. Bioinform. 2010, 2010, 1. [Google Scholar]
- Richesson, R.; Smerek, M.; Electronic Health Records-Based Phenotyping. Rethinking Clinical Trials: A Living Textbook of Pragmatic Clinical Trials. Available online: http://sites.duke.edu/rethinkingclinicaltrials/informed-consent-in-pragmatic-clinical-trials/ (accessed on 22 October 2019).
- Shivade, C.; Raghavan, P.; Fosler-Lussier, E.; Embi, P.J.; Elhadad, N.; Johnson, S.B.; Lai, A.M. A review of approaches to identifying patient phenotype cohorts using electronic health records. J. Am. Med. Inform. Assoc. 2014, 21, 221–230. [Google Scholar] [CrossRef]
- Cahill, K.N.; Johns, C.B.; Cui, J.; Wickner, P.; Bates, D.W.; Laidlaw, T.M.; Beeler, P.E. Automated identification of an aspirin-exacerbated respiratory disease cohort. J. Allergy Clin. Immunol. 2017, 139, 819–825. [Google Scholar] [CrossRef]
- NIH. Suggestions for Identifying Phenotype Definitions Used in Published Research @ONLINE. Available online: https://www.nihcollaboratory.org/Products/Phenotype_lit_search_suggestions_02-18-2014.pdf (accessed on 10 October 2017).
- Ford, E.; Carroll, J.A.; Smith, H.E.; Scott, D.; Cassell, J.A. Extracting information from the text of electronic medical records to improve case detection: A systematic review. J. Med. Inform. 2016, 23, 1007–1015. [Google Scholar] [CrossRef]
- Xu, J.; Rasmussen, L.V.; Shaw, P.L.; Jiang, G.; Kiefer, R.C.; Mo, H.; Pacheco, J.A.; Speltz, P.; Zhu, Q.; Denny, J.C.; et al. Review and evaluation of electronic health records-driven phenotype algorithm authoring tools for clinical and translational research. Int. J. Med. Inform. 2015, 22, 1251–1260. [Google Scholar] [CrossRef]
- Hripcsak, G.; Albers, D.J. Next-generation phenotyping of electronic health records. Int. J. Med. Inform. 2013, 20, 117–121. [Google Scholar] [CrossRef]
- Ford, E.; Nicholson, A.; Koeling, R.; Tate, A.R.; Carroll, J.; Axelrod, L.; Smith, H.E.; Rait, G.; Davies, K.A.; Petersen, I.; et al. Optimising the use of electronic health records to estimate the incidence of rheumatoid arthritis in primary care: What information is hidden in free text? BMC Med. Res. Methodol. 2013, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Barnado, A.; Casey, C.; Carroll, R.J.; Wheless, L.; Denny, J.C.; Crofford, L.J. Developing Electronic Health Record Algorithms That Accurately Identify Patients With Systemic Lupus Erythematosus. Arthritis Care Res. 2017, 69, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Meystre, S.M.; Savova, G.K.; Kipper-Schuler, K.C.; Hurdle, J.F. Extracting information from textual documents in the electronic health record: A review of recent research. Yearb. Med. Inf. 2008, 35, 44. [Google Scholar]
- Liao, K.P.; Ananthakrishnan, A.N.; Kumar, V.; Xia, Z.; Cagan, A.; Gainer, V.S.; Goryachev, S.; Chen, P.; Savova, G.K.; Agniel, D.; et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS ONE 2015, 10, e0136651. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Cai, T.; Savova, G.; Cheng, S.C.; Chen, P.; Perez, R.G.; Gainer, V.S.; Murphy, S.N.; Szolovits, P.; Xia, Z.; et al. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: A novel informatics approach. Inflamm. Bowel Dis. 2013, 19, 1411. [Google Scholar] [CrossRef] [PubMed]
- Abhyankar, S.; Demner-Fushman, D.; Callaghan, F.M.; McDonald, C.J. Combining structured and unstructured data to identify a cohort of ICU patients who received dialysis. J. Am. Med. Inform. Assoc. 2014, 21, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.Q.; Teixeira, P.L.; Mo, H.; Cronin, R.M.; Warner, J.L.; Denny, J.C. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. 2015, 23, e20–e27. [Google Scholar] [CrossRef] [PubMed]
- Morley, K.I.; Wallace, J.; Denaxas, S.C.; Hunter, R.J.; Patel, R.S.; Perel, P.; Shah, A.D.; Timmis, A.D.; Schilling, R.J.; Hemingway, H. Defining disease phenotypes using national linked electronic health records: A case study of atrial fibrillation. PLoS ONE 2014, 9, e110900. [Google Scholar] [CrossRef] [PubMed]
- Glock, J.; Herold, R.; Pommerening, K. Personal identifiers in medical research networks: Evaluation of the personal identifier generator in the Competence Network Paediatric Oncology and Haematology. GMS Medizinische Informatik Biometrie und Epidemiologie 2006, 2, 6. [Google Scholar]
- Feldman, H.; Reti, S.; Kaldany, E.; Safran, C. Deployment of a highly secure clinical data repository in an insecure international environment. Stud. Health Technol. Inform. 2010, 160, 869–873. [Google Scholar]
- Pathak, J.; Kho, A.N.; Denny, J.C. Electronic health records-driven phenotyping: Challenges, recent advances, and perspectives. J. Am. Med. Inform. Assoc. 2013, 20, e206–e211. [Google Scholar] [CrossRef]
- Peissig, P.L.; Costa, V.S.; Caldwell, M.D.; Rottscheit, C.; Berg, R.L.; Mendonca, E.A.; Page, D. Relational machine learning for electronic health record-driven phenotyping. J. Biomed. Inform. 2014, 52, 260–270. [Google Scholar] [CrossRef]
- Koller, D.; Friedman, N.; Džeroski, S.; Sutton, C.; McCallum, A.; Pfeffer, A.; Abbeel, P.; Wong, M.F.; Heckerman, D.; Meek, C.; et al. Introduction to Statistical Relational Learning; MIT Press: Cambridge, UK, 2007. [Google Scholar]
- McCarty, C.A.; Chisholm, R.L.; Chute, C.G.; Kullo, I.J.; Jarvik, G.P.; Larson, E.B.; Li, R.; Masys, D.R.; Ritchie, M.D.; Roden, D.M.; et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genom. 2011, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Chute, C.G.; Pathak, J.; Savova, G.K.; Bailey, K.R.; Schor, M.I.; Hart, L.A.; Beebe, C.E.; Huff, S.M. The SHARPn project on secondary use of Electronic Medical Record data: Progress, plans, and possibilities. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: Bethesda, MD, USA, 2011; pp. 248–256. [Google Scholar]
- Collins, F.S.; Hudson, K.L.; Briggs, J.P.; Lauer, M.S. PCORnet: Turning a dream into reality. J. Am. Med. Inform. Assoc. 2014, 21, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.M.; Peissig, P.L.; Kho, A.N.; Bielinski, S.J.; Berg, R.L.; Choudhary, V.; Basford, M.; Chute, C.G.; Kullo, I.J.; Li, R.; et al. Validation of electronic medical record-based phenotyping algorithms: Results and lessons learned from the eMERGE network. J. Am. Med. Inform. Assoc. 2013, 20, e147–e154. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Secor, E.; Chibnik, L.B.; Bove, R.M.; Cheng, S.; Chitnis, T.; Cagan, A.; Gainer, V.S.; Chen, P.J.; Liao, K.P.; et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS ONE 2013, 8, e78927. [Google Scholar] [CrossRef] [PubMed]
- Bellows, B.K.; LaFleur, J.; Kamauu, A.W.; Ginter, T.; Forbush, T.B.; Agbor, S.; Supina, D.; Hodgkins, P.; DuVall, S.L. Automated identification of patients with a diagnosis of binge eating disorder from narrative electronic health records. J. Am. Med. Inform. Assoc. 2014, 21, e163–e168. [Google Scholar] [CrossRef] [PubMed]
- Afzal, Z.; Schuemie, M.J.; van Blijderveen, J.C.; Sen, E.F.; Sturkenboom, M.C.; Kors, J.A. Improving sensitivity of machine learning methods for automated case identification from free-text electronic medical records. BMC Med. Inform. Decis. Mak. 2013, 13, 30. [Google Scholar] [CrossRef]
- Afzal, N.; Sohn, S.; Abram, S.; Liu, H.; Kullo, I.J.; Arruda-Olson, A.M. Identifying peripheral arterial disease cases using natural language processing of clinical notes. In Proceedings of the 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), Las Vegas, NV, USA, 24–27 February 2016; pp. 126–131. [Google Scholar]
- Restrepo, N.A.; Farber-Eger, E.; Crawford, D.C. Searching in the Dark: Phenotyping Diabetic Retinopathy in a De-Identified Electronic Medical Record Sample of African Americans. AMIA Summits Transl. Sci. Proc. 2016, 2016, 221. [Google Scholar]
- Li, D.; Simon, G.; Chute, C.G.; Pathak, J. Using association rule mining for phenotype extraction from electronic health records. AMIA Summits Transl. Sci. Proc. 2013, 2013, 142. [Google Scholar]
- Doss, J.; Mo, H.; Carroll, R.J.; Crofford, L.J.; Denny, J.C. Phenome-wide association study of rheumatoid arthritis subgroups identifies association between seronegative disease and fibromyalgia. Arthritis Rheumatol. 2017, 69, 291–300. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Oake, J.; Godwin, M.; Aubrey-Bassler, K.; Duke, P.; Mahdavian, M.; Asghari, S. Identification of Dyslipidemic Patients Attending Primary Care Clinics Using Electronic Medical Record (EMR) Data from the Canadian Primary Care Sentinel Surveillance Network (CPCSSN) Database. J. Med. Syst. 2017, 41, 45. [Google Scholar] [CrossRef]
- Bobo, W.V.; Pathak, J.; Kremers, H.M.; Yawn, B.P.; Brue, S.M.; Stoppel, C.J.; Croarkin, P.E.; St Sauver, J.; Frye, M.A.; Rocca, W.A. An electronic health record driven algorithm to identify incident antidepressant medication users. J. Am. Med. Inform. Assoc. 2014, 21, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liao, K.P.; Shaw, S.Y.; Gainer, V.S.; Churchill, S.E.; Szolovits, P.; Murphy, S.N.; Kohane, I.S.; Cai, T. Toward high-throughput phenotyping: Unbiased automated feature extraction and selection from knowledge sources. J. Am. Med. Inform. Assoc. 2015, 22, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Michalik, D.E.; Taylor, B.W.; Panepinto, J.A. Identification and validation of a sickle cell disease cohort within electronic health records. Acad. Pediatr. 2017, 17, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Connolly, B.; Miller, T.; Ni, Y.; Cohen, K.B.; Savova, G.; Dexheimer, J.W.; Pestian, J. Natural Language Processing–Overview and History. In Pediatric Biomedical Informatics; Springer: Berlin, Germany, 2016; pp. 203–230. [Google Scholar]
- Nicholson, A.; Tate, A.R.; Koeling, R.; Cassell, J.A. What does validation of cases in electronic record databases mean? The potential contribution of free text. Arthritis Rheumatol. 2011, 20, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Rizzoli, P.; Loder, E.; Joshi, S. Validity of cluster headache diagnoses in an electronic health record data repository. Headache J. Head Face Pain 2016, 56, 1132–1136. [Google Scholar] [CrossRef]
- Garg, R.; Dong, S.; Shah, S.; Jonnalagadda, S.R. A Bootstrap Machine Learning Approach to Identify Rare Disease Patients from Electronic Health Records. arXiv 2016, arXiv:1609.01586. [Google Scholar]
- Gundlapalli, A.V.; Redd, A.; Carter, M.; Divita, G.; Shen, S.; Palmer, M.; Samore, M.H. Validating a strategy for psychosocial phenotyping using a large corpus of clinical text. J. Am. Med. Inform. Assoc. 2013, 20, e355–e364. [Google Scholar] [CrossRef]
- Spyns, P. Natural language processing. Methods Inf. Med. 1996, 35, 285–301. [Google Scholar]
- Walsh, S.H. The clinician’s perspective on electronic health records and how they can affect patient care. BMJ 2004, 328, 1184–1187. [Google Scholar] [CrossRef]
- Earl, M.F. Information retrieval in biomedicine: Natural language processing for knowledge integration. J. Med. Libr. Assoc. JMLA 2010, 98, 190. [Google Scholar] [CrossRef]
- Byrd, R.J.; Steinhubl, S.R.; Sun, J.; Ebadollahi, S.; Stewart, W.F. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int. J. Med. Inform. 2014, 83, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K. The promise of electronic records: Around the corner or down the road? JAMA 2011, 306, 880–881. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.; McCoy, A.B.; Henkin, S.; Kale, A.; Sittig, D.F. Use of a support vector machine for categorizing free-text notes: Assessment of accuracy across two institutions. Int. J. Med. Inform. 2013, 20, 887–890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afzal, Z.; Engelkes, M.; Verhamme, K.; Janssens, H.M.; Sturkenboom, M.C.; Kors, J.A.; Schuemie, M.J. Automatic generation of case-detection algorithms to identify children with asthma from large electronic health record databases. Pharmacoepidemiol. Drug Saf. 2013, 22, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Karlson, E.W.; Canhao, H.; Miller, T.A.; Dligach, D.; Chen, P.J.; Perez, R.N.G.; Shen, Y.; Weinblatt, M.E.; Shadick, N.A.; et al. Automatic prediction of rheumatoid arthritis disease activity from the electronic medical records. PLoS ONE 2013, 8, e69932. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.B.; Glass, B.; Greiner, H.M.; Holland-Bouley, K.; Standridge, S.; Arya, R.; Faist, R.; Morita, D.; Mangano, F.; Connolly, B.; et al. Methodological Issues in Predicting Pediatric Epilepsy Surgery Candidates Through Natural Language Processing and Machine Learning. Biomed. Inform. Insights 2016, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Kimia, A.A.; Savova, G.; Landschaft, A.; Harper, M.B. An introduction to natural language processing: How you can get more from those electronic notes you are generating. Pediatric Emerg. Care 2015, 31, 536–541. [Google Scholar] [CrossRef]
- Nelson, R.E.; Butler, J.; LaFleur, J.; Knippenberg, K.C.; Kamauu, A.W.; DuVall, S.L. Determining Multiple Sclerosis Phenotype from Electronic Medical Records. J. Manag. Care Spec. Pharm. 2016, 22, 1377–1382. [Google Scholar] [CrossRef]
- Castro, V.M.; Minnier, J.; Murphy, S.N.; Kohane, I.; Churchill, S.E.; Gainer, V.; Cai, T.; Hoffnagle, A.G.; Dai, Y.; Block, S.; et al. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am. J. Psychiatry 2015, 172, 363–372. [Google Scholar] [CrossRef]
- Zeng, Q.T.; Goryachev, S.; Weiss, S.; Sordo, M.; Murphy, S.N.; Lazarus, R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: Evaluation of a natural language processing system. BMC Med. Inform. Decis. Mak. 2006, 6, 30. [Google Scholar] [CrossRef]
- Harkema, H.; Dowling, J.N.; Thornblade, T.; Chapman, W.W. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports. J. Biomed. Inform. 2009, 42, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Pathak, J.; Murphy, S.; Durski, M.; Kirsch, P.S.; Chute, C.G.; Ryu, E.; Murray, J.A. Use of computerized algorithm to identify individuals in need of testing for celiac disease. J. Am. Med. Inform. Assoc. 2013, 20, e306–e310. [Google Scholar] [CrossRef] [PubMed]
- Gundlapalli, A.V.; Divita, G.; Redd, A.; Carter, M.E.; Ko, D.; Rubin, M.; Samore, M.; Strymish, J.; Krein, S.; Gupta, K.; et al. Detecting the presence of an indwelling urinary catheter and urinary symptoms in hospitalized patients using natural language processing. J. Biomed. Inform. 2017, 71, S39–S45. [Google Scholar] [CrossRef] [PubMed]
- Hanauer, D.A.; Gardner, M.; Sandberg, D.E. Unbiased identification of patients with disorders of sex development. PLoS ONE 2014, 9, e108702. [Google Scholar] [CrossRef]
- Chary, M.; Parikh, S.; Manini, A.F.; Boyer, E.W.; Radeos, M. A Review of Natural Language Processing in Medical Education. Western J. Emergency Med. 2019, 20, 78. [Google Scholar] [CrossRef]
- Snomed, C. International Health Terminology Standards Development Organisation Web site, London, UK. 2014. Available online: http://www.snomed.org/ (accessed on 16 September 2017).
- Fact, S.U.; Metathesaurus® National Library of Medicine. Metathesaurus [en línea]. Available online: http://www. nlm. nih. gov/pubs/factsheets/umlsmeta. html (accessed on 8 May 2012).
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef]
- Aronson, A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. J. Am. Med. Inform. Assoc. 2001, 2001, 17–21. [Google Scholar]
- Hristovski, D.; Friedman, C.; Rindflesch, T.C.; Peterlin, B. Exploiting semantic relations for literature-based discovery. AMIA annual symposium proceedings. J. Am. Med. Inform. Assoc. 2006, 2006, 349. [Google Scholar]
- Denny, J.C.; Smithers, J.D.; Miller, R.A.; Spickard, A. “Understanding” medical school curriculum content using KnowledgeMap. J. Am. Med. Inform. Assoc. 2003, 10, 351–362. [Google Scholar] [CrossRef]
- Lin, C.; Karlson, E.W.; Dligach, D.; Ramirez, M.P.; Miller, T.A.; Mo, H.; Braggs, N.S.; Cagan, A.; Gainer, V.; Denny, J.C.; et al. Automatic identification of methotrexate-induced liver toxicity in patients with rheumatoid arthritis from the electronic medical record. J. Am. Med. Inform. Assoc. 2014, 22, e151–e161. [Google Scholar] [CrossRef]
- Lingren, T.; Chen, P.; Bochenek, J.; Doshi-Velez, F.; Manning-Courtney, P.; Bickel, J.; Welchons, L.W.; Reinhold, J.; Bing, N.; Ni, Y.; et al. Electronic Health Record Based Algorithm to Identify Patients with Autism Spectrum Disorder. PLoS ONE 2016, 11, e0159621. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.L.; Wei, W.Q.; Cronin, R.M.; Mo, H.; VanHouten, J.P.; Carroll, R.J.; LaRose, E.; Bastarache, L.A.; Rosenbloom, S.T.; Edwards, T.L.; et al. Evaluating electronic health record data sources and algorithmic approaches to identify hypertensive individuals. J. Am. Med. Inform. Assoc. 2016, 24, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Cagan, A.; Cai, T.; Gainer, V.S.; Shaw, S.Y.; Savova, G.; Churchill, S.; Karlson, E.W.; Murphy, S.N.; Liao, K.P.; et al. Identification of nonresponse to treatment using narrative data in an electronic health record inflammatory bowel disease cohort. Inflammatory Bowel Dis. 2016, 22, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Tsui, F.; Wagner, M.; Espino, J.U.; Li, Q. Influenza detection from emergency department reports using natural language processing and Bayesian network classifiers. J. Am. Med. Inform. Assoc. 2014, 21, 815–823. [Google Scholar] [CrossRef]
- Luo, Y.; Sohani, A.R.; Hochberg, E.P.; Szolovits, P. Automatic lymphoma classification with sentence subgraph mining from pathology reports. J. Am. Med. Inform. Assoc. 2014, 21, 824–832. [Google Scholar] [CrossRef]
- Hinz, E.R.M.; Bastarache, L.; Denny, J.C. A natural language processing algorithm to define a venous thromboembolism phenotype. J. Am. Med. Inform. Assoc. 2013, 2013, 975. [Google Scholar]
- Yadav, K.; Sarioglu, E.; Smith, M.; Choi, H.A. Automated outcome classification of emergency department computed tomography imaging reports. Acad. Emerg. Med. 2013, 20, 848–854. [Google Scholar] [CrossRef]
- Liao, K.P.; Cai, T.; Savova, G.K.; Murphy, S.N.; Karlson, E.W.; Ananthakrishnan, A.N.; Gainer, V.S.; Shaw, S.Y.; Xia, Z.; Szolovits, P.; et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015, 350, h1885. [Google Scholar] [CrossRef]
- Pineda, A.L.; Ye, Y.; Visweswaran, S.; Cooper, G.F.; Wagner, M.M.; Tsui, F.R. Comparison of machine learning classifiers for influenza detection from emergency department free-text reports. J Biomed. Inf. 2015, 58, 60–69. [Google Scholar] [CrossRef]
- Chapman, W.W.; Bridewell, W.; Hanbury, P.; Cooper, G.F.; Buchanan, B.G. A simple algorithm for identifying negated findings and diseases in discharge summaries. J. Biomed. Inf. 2001, 34, 301–310. [Google Scholar] [CrossRef]
- Chu, D. Clinical Feature Extraction from Emergency Department Reports for Biosurveillance. Master’s Thesis, University of Pittsburgh, Pittsburgh, PA, USA, 2007. [Google Scholar]
- Yu, S.; Cai, T. A short introduction to NILE. arXiv 2013, arXiv:1311.6063. [Google Scholar]
- Wagholikar, K.; Torii, M.; Jonnalagadda, S.; Liu, H. Feasibility of pooling annotated corpora for clinical concept extraction. AMIA Summits Transl. Sci. Proc. 2012, 2012, 38. [Google Scholar] [PubMed]
- Xu, H.; Stenner, S.P.; Doan, S.; Johnson, K.B.; Waitman, L.R.; Denny, J.C. MedEx: A medication information extraction system for clinical narratives. J. Am. Med. Inform. Assoc. 2010, 17, 19–24. [Google Scholar] [CrossRef] [PubMed]
- de Quirós, F.G.B.; Otero, C.; Luna, D. Terminology Services: Standard Terminologies to Control Health Vocabulary. Yearbook Med. Inf. 2018, 27, 227–233. [Google Scholar]
- Ma, S.; Huang, J. Penalized feature selection and classification in bioinformatics. Brief. Bioinform. 2008, 9, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Morstatter, F.; Sharma, S.; Alelyani, S.; Anand, A.; Liu, H. Advancing feature selection research. ASU Feature Sel. Repos. 2010, 1–28. [Google Scholar]
- Garla, V.N.; Brandt, C. Ontology-guided feature engineering for clinical text classification. J. Biomed. Inf. 2012, 45, 992–998. [Google Scholar] [CrossRef]
- Bejan, C.A.; Xia, F.; Vanderwende, L.; Wurfel, M.M.; Yetisgen-Yildiz, M. Pneumonia identification using statistical feature selection. J. Am. Med. Inform. Assoc. 2012, 19, 817–823. [Google Scholar] [CrossRef]
- Alzubi, R.; Ramzan, N.; Alzoubi, H.; Amira, A. A hybrid feature selection method for complex diseases SNPs. IEEE Access 2017, 6, 1292–1301. [Google Scholar] [CrossRef]
- Greenwood, P.E.; Nikulin, M.S. A Guide to Chi-Squared Testing; John Wiley & Sons: Hoboken, NJ, USA, 1996. [Google Scholar]
- Zhong, V.W.; Obeid, J.S.; Craig, J.B.; Pfaff, E.R.; Thomas, J.; Jaacks, L.M.; Beavers, D.P.; Carey, T.S.; Lawrence, J.M.; Dabelea, D.; et al. An efficient approach for surveillance of childhood diabetes by type derived from electronic health record data: The SEARCH for Diabetes in Youth Study. J. Am. Med. Inform. Assoc. 2016, 23, 1060–1067. [Google Scholar] [CrossRef]
- Fan, J.; Arruda-Olson, A.M.; Leibson, C.L.; Smith, C.; Liu, G.; Bailey, K.R.; Kullo, I.J. Billing code algorithms to identify cases of peripheral artery disease from administrative data. J. Am. Med. Inform. Assoc. 2013, 20, e349–e354. [Google Scholar] [CrossRef] [PubMed]
- Oake, J.; Aref-Eshghi, E.; Godwin, M.; Collins, K.; Aubrey-Bassler, K.; Duke, P.; Mahdavian, M.; Asghari, S. Using electronic medical record to identify patients with dyslipidemia in primary care settings: International classification of disease code matters from one region to a national database. Biomed. Inform. Insights 2017, 9. [Google Scholar] [CrossRef]
- Kagawa, R.; Kawazoe, Y.; Ida, Y.; Shinohara, E.; Tanaka, K.; Imai, T.; Ohe, K. Development of Type 2 Diabetes Mellitus Phenotyping Framework Using Expert Knowledge and Machine Learning Approach. J. Diabetes Sci. Technol. 2017, 11, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Wing, K.; Bhaskaran, K.; Smeeth, L.; van Staa, T.P.; Klungel, O.H.; Reynolds, R.F.; Douglas, I. Optimising case detection within UK electronic health records: Use of multiple linked databases for detecting liver injury. BMJ Open 2016, 6, e012102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.M.; Fernandez-Gutierrez, F.; Kennedy, J.; Cooksey, R.; Atkinson, M.; Denaxas, S.; Siebert, S.; Dixon, W.G.; O’Neill, T.W.; Choy, E.; et al. Defining disease phenotypes in primary care electronic health records by a machine learning approach: A case study in identifying rheumatoid arthritis. PLoS ONE 2016, 11, e0154515. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.E.; Kerr, W.T.; Thames, A.; Li, T.; Xiao, J.; Cohen, M.S. Electronic health record phenotyping improves detection and screening of type 2 diabetes in the general United States population: A cross-sectional, unselected, retrospective study. J. Biomed. Inform. 2016, 60, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.T.; Sohn, S.; Ravikumar, K.; Wagholikar, K.; Jonnalagadda, S.R.; Liu, H.; Juhn, Y.J. Automated chart review for asthma cohort identification using natural language processing: An exploratory study. Ann. Allergy Asthma Immunol. 2013, 111, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Mowery, D.L.; Chapman, B.E.; Conway, M.; South, B.R.; Madden, E.; Keyhani, S.; Chapman, W.W. Extracting a stroke phenotype risk factor from Veteran Health Administration clinical reports: An information content analysis. J. Biomed. Semant. 2016, 7, 26. [Google Scholar] [CrossRef]
- DeLisle, S.; Kim, B.; Deepak, J.; Siddiqui, T.; Gundlapalli, A.; Samore, M.; D’Avolio, L. Using the electronic medical record to identify community-acquired pneumonia: Toward a replicable automated strategy. PLoS ONE 2013, 8, e70944. [Google Scholar] [CrossRef]
- Valkhoff, V.E.; Coloma, P.M.; Masclee, G.M.; Gini, R.; Innocenti, F.; Lapi, F.; Molokhia, M.; Mosseveld, M.; Nielsson, M.S.; Schuemie, M.; et al. Validation study in four health-care databases: Upper gastrointestinal bleeding misclassification affects precision but not magnitude of drug-related upper gastrointestinal bleeding risk. J. Clin. Epidemiol. 2014, 67, 921–931. [Google Scholar] [CrossRef]
- Liu, H.; Bielinski, S.J.; Sohn, S.; Murphy, S.; Wagholikar, K.B.; Jonnalagadda, S.R.; Ravikumar, K.; Wu, S.T.; Kullo, I.J.; Chute, C.G. An information extraction framework for cohort identification using electronic health records. AMIA Summits Trans. Sci. Proc. 2013, 2013, 149. [Google Scholar]
- Mo, H.; Thompson, W.K.; Rasmussen, L.V.; Pacheco, J.A.; Jiang, G.; Kiefer, R.; Zhu, Q.; Xu, J.; Montague, E.; Carrell, D.S.; et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J. Am. Med. Inform. Assoc. 2015, 22, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Xi, N.; Wallace, R.; Agarwal, G.; Chan, D.; Gershon, A.; Gupta, S. Identifying patients with asthma in primary care electronic medical record systems. Can. Fam. Physician 2015, 61, e474–e483. [Google Scholar]
- Roch, A.M.; Mehrabi, S.; Krishnan, A.; Schmidt, H.E.; Kesterson, J.; Beesley, C.; Dexter, P.R.; Palakal, M.; Schmidt, C.M. Automated pancreatic cyst screening using natural language processing: A new tool in the early detection of pancreatic cancer. HPB 2015, 17, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Zheng, C.; Jung, H.; Chang, A.; Kim, B.; Gelfond, J.; Slezak, J.; Porter, K.; Jacobsen, S.J.; Chien, G.W. Extracting data from electronic medical records: Validation of a natural language processing program to assess prostate biopsy results. World J. Urol. 2014, 32, 99–103. [Google Scholar] [CrossRef]
- Jackson, K.L.; Mbagwu, M.; Pacheco, J.A.; Baldridge, A.S.; Viox, D.J.; Linneman, J.G.; Shukla, S.K.; Peissig, P.L.; Borthwick, K.M.; Carrell, D.A.; et al. Performance of an electronic health record-based phenotype algorithm to identify community associated methicillin-resistant Staphylococcus aureus cases and controls for genetic association studies. BMC Infect. Dis. 2016, 16, 684. [Google Scholar] [CrossRef]
- Safarova, M.S.; Liu, H.; Kullo, I.J. Rapid identification of familial hypercholesterolemia from electronic health records: The SEARCH study. J. Clin. Lipidol. 2016, 10, 1230–1239. [Google Scholar] [CrossRef]
- Chartrand, S.; Swigris, J.J.; Stanchev, L.; Lee, J.S.; Brown, K.K.; Fischer, A. Clinical features and natural history of interstitial pneumonia with autoimmune features: A single center experience. Respir. Med. 2016, 119, 150–154. [Google Scholar] [CrossRef]
- Alpaydin, E. Introduction to Machine Learning; MIT Press: Cambridge, UK, 2014. [Google Scholar]
- Henriksson, A. Semantic Spaces of Clinical Text: Leveraging Distributional Semantics for Natural Language Processing of Electronic Health Records. Ph.D. Thesis, Department of Computer and Systems Sciences, Stockholm University, Stockholm, Sweden, 2013. [Google Scholar]
- Alzoubi, H.; Ramzan, N.; Alzubi, R.; Mesbahi, E. An Automated System for Identifying Alcohol Use Status from Clinical Text. In Proceedings of the 2018 IEEE International Conference on Computing, Southend, UK, 16–17 Auguest 2018; pp. 41–46. [Google Scholar]
- Huda, S.; Abawajy, J.; Alazab, M.; Abdollalihian, M.; Islam, R.; Yearwood, J. Hybrids of support vector machine wrapper and filter based framework for malware detection. Future Gener. Comp. Syst. 2016, 55, 376–390. [Google Scholar] [CrossRef]
- Lasko, T.A.; Denny, J.C.; Levy, M.A. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PLoS ONE 2013, 8, e66341. [Google Scholar] [CrossRef]
- Lipton, Z.C.; Kale, D.C.; Elkan, C.; Wetzel, R. Learning to diagnose with LSTM recurrent neural networks. arXiv 2015, arXiv:1511.03677. [Google Scholar]
- Gehrmann, S.; Dernoncourt, F.; Li, Y.; Carlson, E.T.; Wu, J.T.; Welt, J.; Foote, J.J.; Moseley, E.T.; Grant, D.W.; Tyler, P.D.; et al. Comparing Rule-Based and Deep Learning Models for Patient Phenotyping. arXiv 2017, arXiv:1703.08705. [Google Scholar]
- Kale, D.C.; Che, Z.; Bahadori, M.T.; Li, W.; Liu, Y.; Wetzel, R. Causal phenotype discovery via deep networks. AMIA Annual Symposium Proceedings. J. Am. Med. Inform. Assoc. 2015, 2015, 677. [Google Scholar]
- Zheng, C.; Rashid, N.; Wu, Y.L.; Koblick, R.; Lin, A.T.; Levy, G.D.; Cheetham, T.C. Using natural language processing and machine learning to identify gout flares from electronic clinical notes. Arthritis Care Res. 2014, 66, 1740–1748. [Google Scholar] [CrossRef]
- Ho, J.C.; Ghosh, J.; Sun, J. Extracting phenotypes from patient claim records using nonnegative tensor factorization. International Conference on Brain Informatics and Health. J. Biomed. Inform. 2014, 52, 199–211. [Google Scholar] [CrossRef]
- Joshi, S.; Gunasekar, S.; Sontag, D.; Joydeep, G. Identifiable phenotyping using constrained non-negative matrix factorization. In Proceedings of the Machine Learning for Healthcare Conference, Los Angeles, CA, USA, 19–20 August 2016; pp. 17–41. [Google Scholar]
- Gunasekar, S.; Ho, J.C.; Ghosh, J.; Kreml, S.; Kho, A.N.; Denny, J.C.; Malin, B.A.; Sun, J. Phenotyping using Structured Collective Matrix Factorization of Multi–source EHR Data. arXiv 2016, arXiv:1609.04466. [Google Scholar]
- Elmasry, W.; Akbulut, A.; Zaim, A.H. Deep learning approaches for predictive masquerade detection. Secur.Commun. Net. 2018, 2018, 1–24. [Google Scholar] [CrossRef]
- Vazquez Guillamet, R.; Ursu, O.; Iwamoto, G.; Moseley, P.L.; Oprea, T. Chronic obstructive pulmonary disease phenotypes using cluster analysis of electronic medical records. Health Inf. J. 2016, 394–409. [Google Scholar] [CrossRef]
- Ho, J.C.; Ghosh, J.; Steinhubl, S.R.; Stewart, W.F.; Denny, J.C.; Malin, B.A.; Sun, J. Limestone: High-throughput candidate phenotype generation via tensor factorization. J. Biomed. Inf. 2014, 52, 199–211. [Google Scholar] [CrossRef]
- Ho, J.C.; Ghosh, J.; Sun, J. Marble: High-throughput phenotyping from electronic health records via sparse nonnegative tensor factorization. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 115–124. [Google Scholar]
- Wang, Y.; Chen, R.; Ghosh, J.; Denny, J.C.; Kho, A.; Chen, Y.; Malin, B.A.; Sun, J. Rubik: Knowledge guided tensor factorization and completion for health data analytics. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, NSW, Australia, 10–13 August 2015; pp. 1265–1274. [Google Scholar]
- Schuler, A.; Liu, V.; Wan, J.; Callahan, A.; Udell, M.; Stark, D.E.; Shah, N.H. Discovering patient phenotypes using generalized low rank models. Biocomputing 2016, 21, 144–155. [Google Scholar]
- Nguyen, D.H.; Patrick, J.D. Supervised machine learning and active learning in classification of radiology reports. J. Am. Med. Inform. Assoc. 2014, 21, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.K.; Aggarwal, C.C. Healthcare Data Analytics; Chapman and Hall/CRC: London, UK, 2015. [Google Scholar]
| Disease | Software | Terminology | Extracted Features | Ref |
|---|---|---|---|---|
| Autism Spectrum Disorder | cTAKES | UMLS (SNOMED-CT and RxNORM) and manual project-specific code | A vector of concept unique identifiers (CUIs) | [69] |
| Hypertensive | KMCI | UMLS (SNOMED-CT) | Count the appearance of the concept in the note | [70] |
| Inflammatory Bowel | cTAKES | UMLS | Customized list of narrative concepts | [71] |
| Rheumatoid Arthritis | NILE | UMLS | Count the appearance of the concept in the note | [37] |
| Influenza | Topaz, MedLEE | UMLS | Assign a single value of ‘present’, ‘absent’, or ‘missing’ (not mentioned) to each influenza-related findings | [72] |
| Rheumatoid Arthritis | cTAKES | UMLS (SNOMED-CT and RxNORM) | A vector of frequency of concept unique identifiers (CUIs) | [68] |
| Lymphoma | UMLS | Relations among a flexible number of medical concepts sentence subgraph features | [73] | |
| Crohn’s Disease and Ulcerative Colitis | cTAKES | SNOMED-CT and RxNORM | The number of times the terms were mentioned in the narrative notes and the number of times the terms were mentioned in the clinical text for each subject | [15] |
| Venous Thromboembolism | KMCI | UMLS | Sentence contains concepts | [74] |
| Multiple Sclerosis | cTAKES | SNOMED-CT and RxNORM | The sum of positive and negative mentioned concepts per patient. | [28] |
| Rheumatoid Arthritis | cTAKES | UMLS | A vector of concept unique identifiers | [51] |
| Blunt facial trauma victims | MedLEE | UMLS | Certainty and temporal status modifiers | [75] |
| Peripheral Arterial Disease | MedTagger | MedLex | Count the appearance of the concept in the note | [31] |
| Crohn’s Disease, Multiple Sclerosis, Rheumatoid Arthritis and Ulcerative Colitis | cTAKES, HITex | UMLS (SNOMED-CT and RxNORM) | Number of mentions of the concept | [76] |
| Influenza | Topaz | UMLS | A vector of concept unique identifiers | [77] |
| Feature Extraction Method | Papers |
|---|---|
| Structured only | [18,35,38,90,91,92,93,94,95,96] |
| BoW only | [30,49] |
| Keyword search only | [29,54,97] |
| Concept extraction only | [31,59,69,72,73,75,98] |
| Structured + BoW | [42,50,52,99] |
| Structured + Keyword search | [6,16,17,32,55,58,60] |
| Structured + Concept extraction | [15,28,36,37,48,68,70,71,74,100] |
| Criteria | Year | Phenotype | Se | Sp | F1 | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|---|
| 2017 [12] | Systemic Lupus Erythematosus | 40 | - | 0.56 | 91 | - | - | |
| 2016 [6] | Aspirin-exacerbated Respiratory Disease | - | - | - | 81 | - | - | |
| Expert | 2016 [71] | Non-Response to Treatment | - | - | - | - | - | 0.91 |
| 2016 [32] | Diabetic Retinopathy | - | - | - | 75 | 100 | - | |
| 2016 [31] | Peripheral Arterial Disease | 96 | 98 | - | 92 | 99 | - | |
| 2016 [59] | Indwelling Urinary Catheter | 72 | - | - | 98 | - | - | |
| 2016 [98] | Carotid Stenosis | 88 | 84 | - | 70 | 95 | - | |
| 2015 [104] | Pancreatic Cyst | 99 | 98 | - | - | - | - | |
| 2014 [105] | Prostate Biopsy | 99 | 99 | - | 99 | 99 | - | |
| 2014 [36] | Incident Antidepressant Medication | - | - | - | 90 | 98 | - | |
| 2014 [47] | Heart Failure | 93 | - | 0.93 | 92 | - | - | |
| 2014 [29] | Binge Eating Disorder | 96 | - | - | - | - | - | |
| 2014 [16] | Dialysis | 100 | 98 | - | 78 | - | - | |
| 2013 [74] | Venous Thromboembolism | 90 | - | 0.89 | 84 | - | - | |
| 2013 [15] | Crohn’s Disease | 69 | 97 | - | 98 | - | 0.95 | |
| Ulcerative Colitis | 79 | 97 | - | 97 | - | 0.94 | ||
| 2013 [58] | Celiac | 72 | 82 | 0.78 | - | - | - | |
| Healthcare | ||||||||
| Guidelines | 2017 [92] | Dyslipidemia | 94 | - | - | - | 79 | 0.97 |
| 2016 [94] | Liver Injury | 80 | 100 | - | - | - | 0.95 | |
| 2016 [106] | CA-MRSA (Case) | 94-100 | - | - | 68-100 | - | - | |
| CA-MRSA (Control) | 75-100 | - | - | 96-100 | - | - | ||
| 2016 [107] | Familial Hypercholesterolemia | - | - | - | 94 | 97 | - |
| Paper name | Phenotype | ML Method | Se | Sp | F1 | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|---|
| 2016 [42] | Cardiac Amyloidosis | K-NN, SVM, decision tree, Random Forests, AdaBoost, and Naïve Bayes | - | - | 0.98 | - | - | - |
| 2016 [70] | Hypertensive | Random Forests | 90 | - | - | 95 | - | 0.97 |
| 2016 [52] | Epilepsy | SVM, Naive Bayes | - | - | 0.78 | - | - | 0.83 |
| 2016 [95] | Rheumatoid Arthritis | C5.0 decision tree | 93 | 99 | - | 90 | - | - |
| 2016 [96] | Type 2 Diabetes | Multivariate Logistic Regression and a Random-Forests Probabilistic Model | 80 | 74 | - | 40 | 94 | 0.84 |
| Inflammatory Bowel Disease (Crohn’s Disease) | 72 | - | - | 98 | - | - | ||
| 2015 [76] | Inflammatory Bowel Disease (Ulcerative Colitis) | Logistic Regression | 73 | - | - | 97 | - | - |
| Multiple Sclerosis | 78 | - | - | 95 | - | - | ||
| Rheumatoid Arthritis | 63 | - | - | 94 | - | - | ||
| 2015 [37] | Rheumatoid Arthritis | Penalized Logistic Regression | - | - | 0.77 | 70 | - | 0.95 |
| Coronary Artery Disease | - | - | 0.82 | 84 | - | 0.92 | ||
| 2015 [117] | Gout Flares | SVM | 82 | 92 | 0.87 | 77 | 93 | - |
| 2014 [72] | Influenza | Bayesian Network | - | - | - | - | - | 0.73 |
| 2013 [75] | Blunt Facial Trauma Victims | CART Decision Tree | 93 | 97 | 0.97 | 81 | 99 | - |
| 2013 [91] | Peripheral Arterial Disease | Logistic Regression | 68 | 87 | - | 75 | 83 | 0.91 |
| 2013 [49] | Diabetes | SVM | 92 | - | 0.93 | 95 | - | 0.94 |
| 2013 [97] | Asthma | C4.5 Decision Tree | 84 | 96 | 0.86 | 88 | 95 | - |
| 2013 [51] | Rheumatoid Arthritis | Logistic Regression, Naive Bayes, Multilayer perceptron, SVM | - | - | - | - | - | 0.83 |
| 2013 [28] | Multiple Sclerosis | LASSO Penalized Logistic Regression Models with Bayesian Information Criterion | 83 | 95 | - | 92 | 89 | 0.95 |
| 2013 [33] | Type 2 Diabetes Mellitus | Association Rule Mining, DT, Logistic Regression, SVM | 96 | - | 0.91 | 86 | - | - |
| 2013 [30] | Hepatobiliary Disease | C4.5, SVM, RIPPER, MyC | 95 | 56 | - | - | - | - |
| Acute Renal Failure | 86 | 77 | - | - | - | - |
| Performance metrics | Definition | Equation |
|---|---|---|
| Sensitivity (Recall) (Se) | The proportion of all actually positive samples that are correctly detected. | |
| Specificity (Sp) | The proportion of all actually negative samples that are correctly detected. | |
| Positive Predictive Value (precision) (PPV) | The proportion of positively detected samples that are true positive. | |
| Negative Predictive Value (NPV) | The proportion of negatively detected samples that are true negative. | |
| F1-score (F1) | The weighted harmonic mean of precision and recall. | |
| Area under the ROC (AUROC) | ROC is the graph that represent the trade-off between sensitivity and specificity and area under the curve is equal the probability that a predictor will rank a randomly chosen positive sample higher than a randomly chosen negative one |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alzoubi, H.; Alzubi, R.; Ramzan, N.; West, D.; Al-Hadhrami, T.; Alazab, M. A Review of Automatic Phenotyping Approaches using Electronic Health Records. Electronics 2019, 8, 1235. https://doi.org/10.3390/electronics8111235
Alzoubi H, Alzubi R, Ramzan N, West D, Al-Hadhrami T, Alazab M. A Review of Automatic Phenotyping Approaches using Electronic Health Records. Electronics. 2019; 8(11):1235. https://doi.org/10.3390/electronics8111235
Chicago/Turabian StyleAlzoubi, Hadeel, Raid Alzubi, Naeem Ramzan, Daune West, Tawfik Al-Hadhrami, and Mamoun Alazab. 2019. "A Review of Automatic Phenotyping Approaches using Electronic Health Records" Electronics 8, no. 11: 1235. https://doi.org/10.3390/electronics8111235
APA StyleAlzoubi, H., Alzubi, R., Ramzan, N., West, D., Al-Hadhrami, T., & Alazab, M. (2019). A Review of Automatic Phenotyping Approaches using Electronic Health Records. Electronics, 8(11), 1235. https://doi.org/10.3390/electronics8111235







