Abstract
Computer vision-based automation has become popular in detecting and monitoring plants’ nutrient deficiencies in recent times. The predictive model developed by various researchers were so designed that it can be used in an embedded system, keeping in mind the availability of computational resources. Nevertheless, the enormous popularity of smart phone technology has opened the door of opportunity to common farmers to have access to high computing resources. To facilitate smart phone users, this study proposes a framework of hosting high end systems in the cloud where processing can be done, and farmers can interact with the cloud-based system. With the availability of high computational power, many studies have been focused on applying convolutional Neural Networks-based Deep Learning (CNN-based DL) architectures, including Transfer learning (TL) models on agricultural research. Ensembling of various TL architectures has the potential to improve the performance of predictive models by a great extent. In this work, six TL architectures viz. InceptionV3, ResNet152V2, Xception, DenseNet201, InceptionResNetV2, and VGG19 are considered, and their various ensemble models are used to carry out the task of deficiency diagnosis in rice plants. Two publicly available datasets from Mendeley and Kaggle are used in this study. The ensemble-based architecture enhanced the highest classification accuracy to 100% from 99.17% in the Mendeley dataset, while for the Kaggle dataset; it was enhanced to 92% from 90%.
1. Introduction
Agriculture is a major component of the global economy and food supply, which is under strain as a result of the enormous rise in population. Digital agriculture is a new scientific field that strengthens the agricultural production [1]. One of the many factors that influences agricultural yield is the absence of soil nutrients. Micronutrient acquisition by crops must be understood in order to develop appropriate strategies to prevent a deficiency in crops. It is found that 30–40%, 30%, 59%, and 30% of global agricultural soil is deficient in nutrients such as Phosphorous (P), Iron (Fe), Nitrogen (N), and Zinc (Zn), respectively, which impacts the agricultural yield severely [2]. Rice is one of the most commonly consumed grains worldwide. According to 2021 data of Statista [3], China stands first in rice consumption, followed by India and Bangladesh. The country-wise consumption of rice is shown in Figure 1. Rice production in the world totals 510.6 million tonnes [4]. Nitrogen (N), phosphorus (P), potassium (K), magnesium (Mg), calcium (Ca), sulphur (S), zinc (Zn), iron (Fe), manganese (Mn), copper (Cu), boron (B), molybdenum (Mo), and chlorine (Cl) are the major nutrients required to be present in the soil for the proper growth of rice plants. Nitrogen is responsible for the onset of panicles, grain development, and booting stages. By promoting early flowering and maturity, phosphorus aids in the maintenance of the root growth process as well as disease resistance and drought tolerance. Potassium is essential for plant metabolism as well as stress tolerance [5,6]. Magnesium and molybdenum are required for enzyme phosphate transfer and nitrate-to-nitrite reduction. Cu and Fe are important photosynthesis components. Boron aids in flower formation, pollen germination, and the development of new plant cells. Rice diseases are controlled by Mn and Cl, and the latter intensifies disease resistance. S is in charge of amino acid and lipid synthesis, whereas Ca is in charge of cell division in plants [7,8]. The task of determining the amount of nutrients lacking in soil is known as plant nutrient deficiency identification. The detection of nutrient deficiencies in crops is essential so that future courses of action can be taken to boost the yield as well as to maintain the growth in crops. This has a series of economic and environmental impacts, such as preventing damage and yield loss, less financial loss, reduced food, etc. Agricultural experts struggle to diagnose the nutrient deficiencies because agricultural production sites are dispersed and many nutrient deficiencies are spread across a large region [9,10].
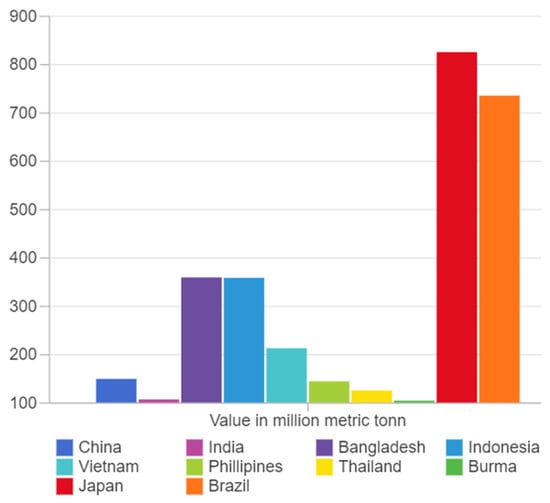
Figure 1.
Country-wise (top ten) consumption of rice.
These deficiencies are often visible to the human eye only when the plant has already been damaged. Hence, technological advancement is vital to assist farmers and experts in identifying these deficiencies at an early stage. Significant innovative progress has been made to solve this problem. The traditional way of identifying a nutrient deficiency in plants involves the use of chemicals and mashing the plants. However, this task requires a lot of effort from agricultural experts and use of machineries. To overcome these limitations, non-invasive techniques, such as sensors, image processing, spectral imaging, and computer vision, have been developed to manage and understand the deficiencies in plants [11,12]. Several techniques have been used to acquire images of nutrient deficiencies in plants from digital cameras, satellites, and unmanned aerial vehicles (UAV). Satellites do not provide supportive information for individual plant analysis and are costly. The UAVs’ dependency on weather, requirement of specific flight directions, and crashes lead to degraded UAV images. Internet of Things (IoT)-based systems work well in monitoring soil temperature, moisture level, and nutrient contents of plants. Image processing techniques involving image acquisition, preprocessing, segmentation, feature extraction, and classification have also been applied effectively for the same cause. Optical sensors and spectral imaging require an expensive experimental setup, which is non-feasible to farmers [13,14,15]. In the past few years, techniques such as computer vision, image processing, and machine learning have been found to be popular in detecting and monitoring plants’ nutrient deficiencies. The classification algorithms such as K-Nearest Neighbor (KNN), Multilayer Perceptron Neural Network, regression, Naïve Bayes, Radial Basis Function, Random Forests, Self-Organizing Maps, fuzzy c-means, and Support Vector Machine (SVM) have gained exceptional attention in smart agriculture. Some of the studies have used feature selection and reduction techniques in combination with classical machine learning techniques to reduce the computational complexity, which is highly required if the predictive model is used in an embedded system. Nevertheless, the performance of the deficiency diagnosis system is more important than the speed with which the results are obtained. At the same time, the enormous popularity of smart phone technology has opened the door of opportunity to common farmers to have access to high computing resources. To address the issue of resource constraint, the study proposes a framework of hosting high-end systems in the cloud where processing can be done and the farmer can interact with the system hosted in the cloud. Figure 2 gives a cloud-based nutrient deficiency diagnosis framework that can be deployed for the benefit of farmers.
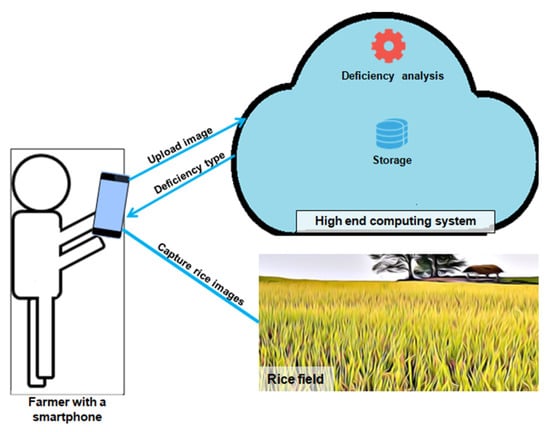
Figure 2.
Cloud-based nutrient deficiency diagnosis framework in agriculture.
With the availability of high computational power, it is feasible to enhance the performance of a predictive model by use of advanced Deep Learning (DL) techniques. A few of the studies have reported using CNN, CNN-based DL techniques, and transfer learning (TL) techniques for a deficiency diagnosis in various crops. However, ensemble of TL architectures in this domain is yet to be explored. The ensemble TL model can play a significant role in improving the performance of nutritional deficiency identification. This motivated us to apply an ensemble averaging technique on various TL models viz. InceptionResNetV2, Xception, DenseNet201, and VGG19. All possible combinations of binary, ternary, and quaternary ensemble classifiers are designed from these base TL models, and the experiment is carried out on two publicly available rice deficiency image datasets.
2. Related Work
In the domain of agricultural research, Artificial Intelligence (AI) is widely used for plant identification and pathology, with the prime focus on designing DL/ML (Machine Learning) architectures to handle various challenges in this field. Supervised ML techniques are widely used in crop identification, yield prediction, disease detection, deficiency identification, etc. Abbaspour-Gilandeh et al. performed research on 13 Iranian rice types and categorized them as white rice, brown rice, and paddy using Discriminant Analysis (DA) and Artificial Neural Networks (ANN) [16]. An Expert Learning Model (ELM) for wheat yield forecasting incorporated Support-vector Machine (SVM), Least Square-SVM, and Proximal SVM classifiers, where ELM beats Support Vector Regression (SVR) and other approaches [17]. K-means and SVM, when applied in the classification of five types of rice deficiencies, achieved accuracy in the range of 85.06% to 93% [18]. However, deep learning shows promising results in identifying macronutrient deficiencies in maize when compared to other machine learning techniques, such as ANN, SVM, and K-Nearest Neighbor (KNN) [19].
For identification of disease and deficiency from image data, CNN based DL models are most popularly used. Pre-trained convolutional encoders performed better than recurrent attention neural network (RAN)-CNN in detection of iron deficiency of Soybean [20]. TL model ResNet-50 showed an accuracy of 65.44% in detecting seven nutrient deficiencies in a dataset of 4088 images of black gram [21]. In another experiment, ensemble of Inception-ResNet and auto-encoder was designed to classify three nutrient deficiencies in 571 tomato images which achieved 91% accuracy [22]. Fine tuning the layers of the pre-trained CNN models increases the accuracy in plant’s nutrient deficiency identification systems [23]. TL models perform better than the conventional CNN. Pre-trained CNN models like InceptionV3, ResNet50, NasNet-large and DenseNet121 were used to detect 11 types of nutrient deficiencies in rice plant leaves, with highest accuracy shown by DenseNet121 (97.44%) followed by NasNet-large (96.25%) [24]. With the increase in number of training steps, the accuracy of pre-trained InceptionV3 was observed to increase while detecting NPK deficiency in maize plant [25]. During prediction of four levels of N deficiency in rice and NPK deficiency in oilseed rape, pre-trained CNN models were combined with SVM or time series model producing best results in the range of 99.84% and 95% [26,27]. Although CNNs have shown their satisfactory performance in the field of nutrient deficiency identification in plants, region based CNN lacked detailed classification when applied in Chilli plants [28]. The pre-trained CNN model (MobileNetV2) can be adjusted to show a statistically significant increase in cassava leaf disease identification accuracy on lower-quality testing images [29]. Deep Residual Convolutional Neural Networks (DRCNN) can be employed in conjunction with unique block processing to identify and classify Kaggle cassava mosaic illness dataset, with DRCNN obtaining a maximum success rate of 96.75% [30]. When RGB, HSV color, and textural Local Binary Pattern descriptors were utilized, the Bagged tree ensemble classifier and the Complex tree classifier outperformed other classifiers in a system for recognizing and categorizing diseases in guava plants [31]. The pre-trained and fine-tuned custom-made convolution classifier used in a combined framework of Internet of Things (IoT) and DL was proved to be low-cost in the prediction of Pearl Millet disease [32]. InceptionV3 model was found to be more suitable to classify the different degrees of ginkgo leaf disease in different field conditions [33]. Many research studies related to crop management found that different variants of deep learning models (InceptionV3, InceptionResNetV2, EfficientNet, MobileNet, DenseNet and others) performed better in terms of accuracy and training time [34]. Furthermore, studies showed that when the proper parameters were employed, the MobileNetV2 architecture was found to be compatible with mobile devices [35]. Table 1 summarizes the state of art research in the domain of nutrient deficiency identification in plants using various CNN based architectures.

Table 1.
CNN studies on rice deficiency classification.
Various classification models employed in the literature can be categorized into four different groups. Firstly, supervised ML models were used in the earlier literature. Secondly, with the availability of image data, CNN gained popularity. Thirdly, researchers tried to work with hybrid models that take advantage of both CNN and conventional ML techniques. Fourthly, pre-trained CNN models have recently become popular. Figure 3 shows the different techniques of nutrient deficiency identification in crops articulated in the literature.
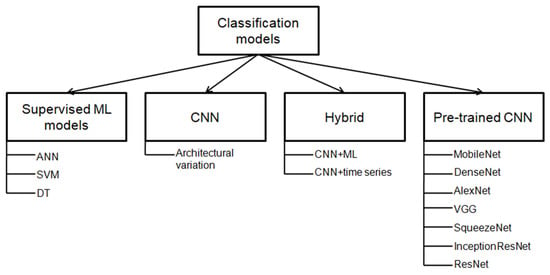
Figure 3.
Types of techniques used for nutrient deficiency classification in crops (in previous studies).
3. Materials and Method
This paper proposes an ensemble of six TL models for diagnosing deficiencies in rice plants. Two publicly available datasets were used to carry out the experiment. The following section explains the experimental setup in detail.
3.1. Dataset Details
This study worked on two rice deficiency datasets available in Kaggle and Mendeley. The Kaggle rice NPK deficiency dataset has three types of deficiencies: N, P, and K, each with 440, 333, and 383 images, respectively [36]. The images in each of the three classes are of varying sizes. The Mendeley rice N deficiency dataset, having a resolution of 100×100 pixels, takes into account four swaps ranging from minor to severe deficiency [37]. This dataset already has 400 test images (100 for each swap) and 5390 training images (1407, 1203, 1400, and 1380 images for each swap). Figure 4 depicts the class distribution of rice deficiency in both datasets.
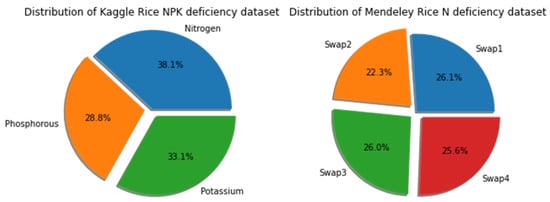
Figure 4.
Class distribution of rice deficiency in the Kaggle dataset and Mendeley dataset.
3.2. Data Augmentation
The rice deficiency datasets were worked with augmentation techniques using Python’s ImageDataGenerator module [38]. The resolution of all images in the Kaggle rice NPK deficiency dataset was scaled to 300 × 300 pixels. Augmentation techniques, such as horizontal flip, rotation without automatic cropping, skew, and a zooming effect, were applied to images from each class in the datasets. Table 2 shows the augmentation parameters. The expanded Mendeley dataset and the Kaggle dataset each have 5790 and 3356 number of images, respectively. These expanded datasets were divided into three sets for assessing the performance of the rice deficiency diagnosis system: training, testing, and validation. Table 3 contains a summary of the datasets’ details.

Table 2.
Parameters of the augmentation techniques applied in the current study.

Table 3.
Details of the rice deficiency datasets used in the current study.
3.3. Proposed Rice Deficiency Identification System
This study presents an ensemble of TL models for diagnosing a deficiency in rice plants as shown in Figure 5. The rice deficiency datasets from Mendeley and Kaggle were gathered and augmented using techniques discussed in the previous section. The extended Mendeley and Kaggle datasets were divided into three sets: training, testing, and validation. Then, six TL models were trained individually on these datasets. The Adam optimizer and categorical cross-entropy loss function were utilized to precisely train the networks. Out of them, the best performing four TL models were ensembled to generate 11 ensemble classifiers. All the classifiers were assessed using performance measures such as a confusion matrix, precision, recall, F1-score, and accuracy to choose the optimal model for a rice deficiency diagnosis [39,40,41].
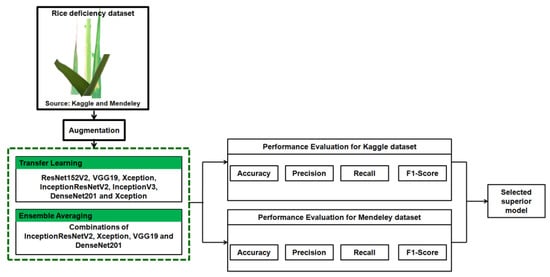
Figure 5.
The proposed rice deficiency identification system.
3.3.1. Transfer Learning Models
Transfer learning architectures are being used in the agricultural domain to assist farmers with innovative solutions. In this work, various TL models viz. InceptionResNetV2, InceptionV3, ResNet152V2, VGG19, DenseNet201 and Xception were fine-tuned with rice nutrition deficiency datasets [42,43,44,45,46,47]. The classification layer of each CNN pre-trained model was replaced with a pooling layer, a dense layer, and a softmax layer with the number of classes, which are 3 for the Kaggle dataset and 4 for the Mendeley dataset in this study. Training the models involved 50 epochs, and an early stopping technique was used to obtain the best weights. A dropout value of 0.5 was used to handle over-fitting. All models were tested and validated using an Adam optimizer, categorical cross-entropy, and batch size of 32. The total parameters used for each of the TL architectures are shown in Table 4.

Table 4.
TL architectures applied in the study.
3.3.2. Ensemble TL Models
The approach of ensemble averaging combines the estimated probability of several base models [48]. Ensemble learning seeks to integrate several algorithms and merge the outcomes with various voting processes in order to outperform any one algorithm [49,50]. The base models used in this study are InceptionResNetV2, Xception, DenseNet201, and VGG19. These four baseline learners are used to form binary, ternary, or quaternary ensemble classifiers. Each of the 11 ensemble classifiers averages the prediction probability produced from the two datasets. The ensemble averaging approach utilized in this investigation is visualized through Figure 6 and Algorithm 1.
| Algorithm 1: Ensemble averaging | ||||||||
| Input:Test_set S: Models Mk (k = 1 to n) where k is the number of models | ||||||||
| Output: Ix Ensemble_model E = [M1,M2,…Mk] | ||||||||
| For i = 1 to k do | ||||||||
| Predict, P = generate(S) A = add (P, along y axis) Ix = index_max (A, along x axis) | ||||||||
| Confusion_matrix (Ix, S) Classification_matrices (Ix, S) | ||||||||
| End | ||||||||
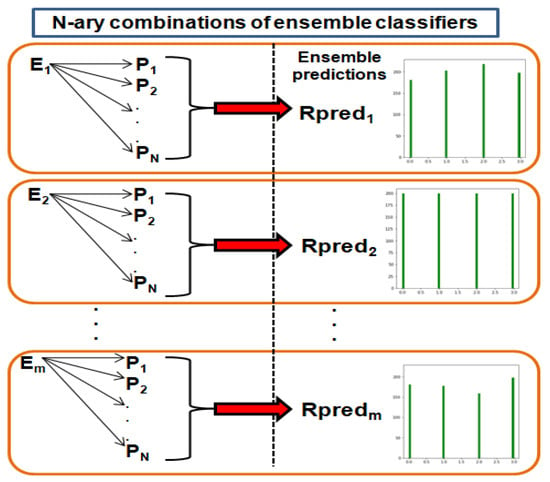
Figure 6.
Ensemble averaging technique used in this study.
4. Results
The training and the validation of various learning models were carried out using GPU available in the Google Colab platform. Table 5 depicts the performance metrics of transfer learning models and the proposed ensemble models considered in this study.

Table 5.
Performance metrics for transfer learning models and the proposed ensemble models used in this work.
4.1. Results of Transfer Learning Models
The nutrient deficient images from both the datasets were worked out with six TL models, i.e., InceptionResNetV2, InceptionV3, ResNet152V2, VGG 19, DenseNet201, and Xception. The accuracy scores of all these models for the datasets were in the range of (51.17–87.17%) and (86.67–99.16%) for Kaggle and Mendeley, respectively. The performance of InceptionResNetV2 and Xceptionwere the highest as compared to other TL models in the Kaggle rice NPK deficiency dataset and Mendeley rice N deficiency dataset, respectively. The variation of validation accuracy across the epochs is shown in Figure 7.
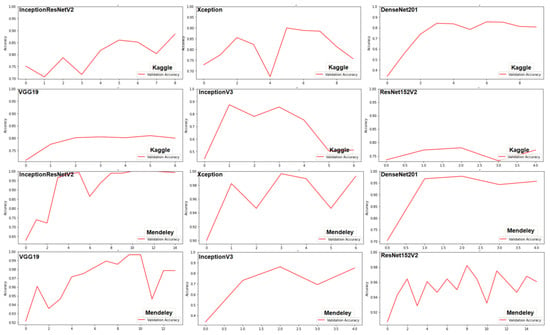
Figure 7.
Validation accuracy versus number of epochs.
Figure 8 presents the confusion matrices of the TL models for both the datasets, which shows both the classification and misclassification information. Using the confusion matrices, it has been noticed that in the Kaggle rice NPK deficiency dataset, K deficiency and N deficiency showed the least and highest diagnostic accuracies, respectively. P and K deficiencies were misclassified in most of the cases by DenseNet201 and Xception. Out of the four swap categories of the Mendeley rice N deficiency dataset, swap1 tended to be misdiagnosed as the other three swaps (swap1, swap2, and swap3); hence, it showed the lowest diagnostic accuracy. Swap2 was correctly classified by all the TL models. Although swap4 was accurately classified by InceptionResNetV2, Xception, and DenseNet201, in some cases, it was misclassified by VGG19, InceptionV3, and ResNet152V2.
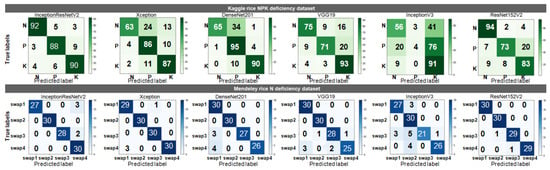
Figure 8.
Confusion matrices of TL models for both datasets.
4.2. Results of Ensemble TL Models
As shown in Table 5, the classification accuracies of binary and ternary ensemble TL classifiers for the Kaggle rice NPK deficiency dataset varied in the range of 82.67–92% and 87–92%, respectively, where EM3.0 and EM2.4 outperformed the rest with an accuracy of 92%. EM3.2 underperformed all other ternary ensemble TL models with an accuracy of 87%. The quaternary ensemble model showed an accuracy of 90.33%. It is noticed that in the Kaggle rice NPK deficiency dataset, the N deficiency and K deficiency showed the highest and lowest diagnostic accuracies, respectively, whereas, in the Mendeley N deficiency dataset, swap3 was classified with a precision of 100%. The confusion matrices of the first eight best ensemble classifiers for the Kaggle rice NPK deficiency dataset is shown in Figure 9. Confusion matrices of the ensemble classifiers having an accuracy between 97.5% and 99.17% for the Mendeley rice N deficiency dataset is shown in Figure 10. The classification accuracies for the Mendeley rice N deficiency dataset of binary ensemble TL classifiers EM2.1 and EM2.2 were 97.5% and 99.17%, respectively. The ternary ensemble TL classifier EM3.0 had a classification accuracy of 99.17%. With an accuracy of 100%, all the remaining ensemble TL classifiers outperformed EM2.1, EM2.2, and EM3.0 in the case of the Mendeley dataset.
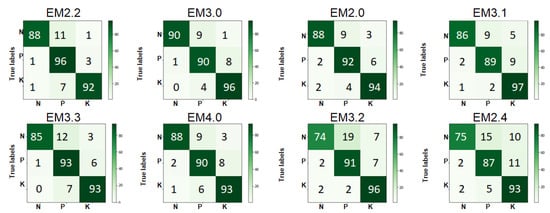
Figure 9.
Confusion matrices of the first eight best ensemble classifiers for the Kaggle rice NPK deficiency dataset.
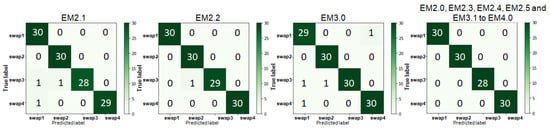
Figure 10.
Confusion matrices of the ensemble classifiers for the Mendeley rice N deficiency dataset.
5. Discussion
This study resolved the issue of resource constraint of an embedded system by proposing a cloud-based framework, where high-end systems responsible for processing will be hosted in the cloud, and a user with internet connectivity can interact with the system. To explore the performance of various TL models, six of them viz. InceptionResNetV2, InceptionV3, ResNet152V2, VGG 19, DenseNet201, and Xception were used for this experiment. Further, model averaging-based ensembling of these TL models were done to generate a range of ensemble classifiers. However, InceptionV3 and ResNet152V2 were removed from the TL model ensembling process because the hybrid of these two models (InceptionResNetV2) performed better than both, as shown in Table 5.
With the increase in the degree of ensemble classifiers, the accuracy was becoming improved as observed from the experiment. In the case of the Kaggle dataset, individual TL, binary, ternary, and quaternary ensemble classifiers hadaverage accuracies of 77.56%, 87.56%, 90%, and 90.33%, respectively, as shown in Figure 11. In the case of the Mendeley dataset, individual TL, binary, ternary, and quaternary ensemble classifiers had average accuracies of 94.39%, 99.45%, 99.79%, and 100%, respectively. The quaternary ensemble model EM4 performed 100% accurately in the Mendeley dataset and also performed well in the Kaggle dataset. Figure 12 shows the plot for the accuracy of various individual TL and ensemble TL classifiers. The mean F1-scores of the ensemble averaging models for the Mendeley and Kaggle rice disease datasets are visualized in Figure 13. In the Kaggle dataset, all ensemble models showed an F1-score approximately equal to 1. Except for EM2.1, EM2.2, and EM3.0, all ensemble models achieved an exact F1-score of 1 for the Mendeley dataset. Figure 12 shows that the average classification accuracy of the ensemble TL models for the entire experiment was in the range of 77.56–100%, whereas individual TL models showed an accuracy of68.50–92.91%, which confirms the superiority of the proposed approach.
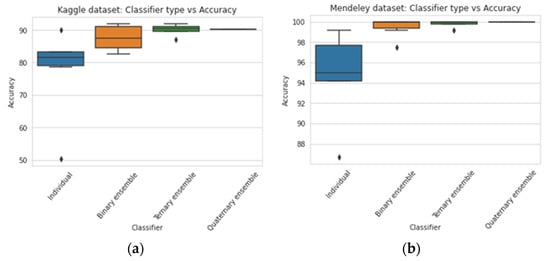
Figure 11.
Range of accuracies of various classification models for (a) Kaggle and (b) Mendeley.
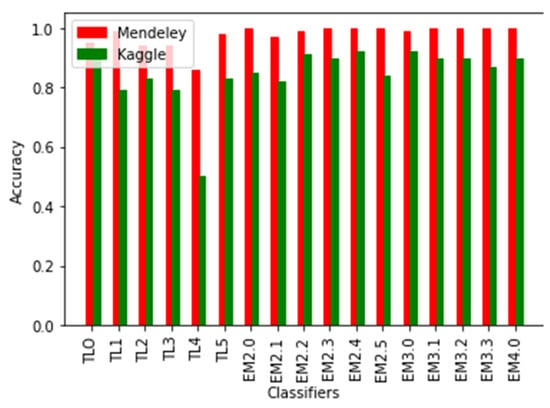
Figure 12.
Average accuracy of various classifiers.
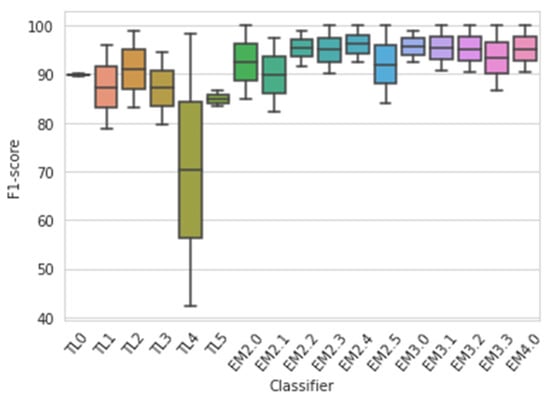
Figure 13.
F1-scores of various models for both datasets taken together.
This research has certain limitations that must be noted. Firstly, no conventional ML classifier is employed in this study, as most of the literature cited the superiority of DL techniques over traditional ML techniques. However, the performance of hybrid machine learning techniques over the present datasets is a matter to explore. Secondly, as the study is focused on DL techniques, pre-processing of the image dataset was ignored, which is an important step while working with ML techniques. Thirdly, only three mineral deficiencies were considered in this experiment. However, for the system to be suitable for real-world applications, all types of deficiencies must be taken into consideration. In addition, this study considered only six major TL models and performed their ensembling. Despite the fact that these TL models effectively satisfied our objective of the experiment, the rest of the TL models can also be incorporated in future studies.
6. Conclusions
The current study is based on an ensemble learning framework that employs transfer learning architectures to diagnose rice plant deficiencies. Six TL architectures viz. Xception, DenseNet201, InceptionResNetV2, InceptionV3, ResNet50V2, and VGG19 were employed to perform this task. The highest performance was achieved with InceptionResNetV2 (90%) and Xception (95.83%) in the Kaggle and Mendeley datasets, respectively. In addition, 11 ensemble averaging models were generated using Xception, DenseNet201, InceptionResNetV2, and VGG19. The ensembled models showed a significant improvement in performance as compared to individual TL models. After applying the ensemble approach to the Kaggle and Mendeley datasets, the average accuracy of the TL models increased from 77.55% to 88.69% and 94.72% to 99.62%, respectively. An accurate determination of nutritional deficiencies in an appropriate time may help farmers to take a future course of action. As a result, severe damage of crops and loss in yield can be avoided. Future works may focus on further improvement of the proposed system, keeping in mind the limitations highlighted. Moreover, researchers are expected to investigate the further extension of state-of-the-art research, from designing a deficiency diagnosis tool to a complete and efficient support system for farmers.
Author Contributions
Conceptualization, R.K.S. and C.J.K.; Investigation, K.N., C.J.K. and A.C.; Methodology, M.S.; Project administration, C.J.K.; Resources, R.K.S.; Visualization, M.S.; Writing—original draft, M.S.; Writing—review & editing, K.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| DL | deep learning |
| CNN | convolutional neural network |
| TL | transfer learning |
| ML | machine learning |
| UAV | unmanned aerial vehicle |
| IoT | internet of things |
| ANN | artificial neural network |
| SVM | support vector machine |
| KNN | k-nearest neighbor |
| RAN | recurrent attention neural network |
| DRCNN | deep residual convolutional neural networks |
References
- Mohapatra, D.; Tripathy, J.; Patra, T.K. Rice disease detection and monitoring using CNN and naive Bayes classification. In Soft Computing Techniques and Applications; Borah, S., Pradhan, R., Dey, N., Gupta, P., Eds.; Springer: Singapore, 2021; Volume 1248. [Google Scholar]
- Mahender, A.; Swamy, B.P.M.; Anandan, A.; Ali, J. Tolerance of Iron-Deficient and -Toxic Soil Conditions in Rice. Plants 2019, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice Consumption by Country 2019 Statista. Available online: https://www.statista.com/statistics/255971/top-countries-based-on-rice-consumption-2012-2013/ (accessed on 19 December 2021).
- FAO. Rice Market Monitor (RMM); FAO: Roma, Italy, 2018; Volume XXI, pp. 1–38. [Google Scholar]
- Wang, Y.; Wu, W.H. Potassium transport and signaling in higher plants. Annu. Rev.Plant. Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieves-Cordones, M.; Ródenas, R.; Lara, A.; Martínez, V.; Rubio, F. The combination of K+ deficiency with other environmental stresses: What is the outcome? Physiol. Plant. 2019, 165, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Deficiencies and Toxicities-IRRI Rice Knowledge Bank. Available online: http://www.knowledgebank.irri.org/step-by-step-production/growth/soil-fertility/deficiencies-and-toxicities (accessed on 19 December 2021).
- Shrestha, J.; Manoj, K.; Subash, S.; Shah, K.K. Role of nutrients in rice (Oryza sativa L.): A review. Agrica 2020, 9, 53–62. [Google Scholar] [CrossRef]
- Cevallos, C.; Ponce, H.; Moya-Albor, E.; Brieva, J. Vision-Based Analysis on Leaves of Tomato Crops for Classifying Nutrient Deficiency using Convolutional Neural Networks. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020. [Google Scholar]
- Patel, A.; Swaminarayan, P.; Patel, M. Identification of Nutrition’s Deficiency in Plant and Prediction of Nutrition Requirement Using Image Processing. In Proceedings of the Second International Conference on Information Management and Machine Intelligence, Jaipur, India, 24–25 July 2020; Springer: Singapore, 2020. [Google Scholar]
- Singh, A.; Budihal, S.V. Non-Invasive Techniques of Nutrient Detection in Plants. In Intelligent Computing and Applications. Advances in Intelligent Systems and Computing; Dash, S.S., Das, S., Panigrahi, B.K., Eds.; Springer: Singapore, 2019; p. 1172. [Google Scholar]
- Lu, T.; Han, B.; Chen, L.; Yu, F.; Xue, C. A generic intelligent tomato classification system for practical applications using DenseNet-201 with transfer learning. Sci. Rep. 2021, 11, 15824. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Detection of nutrition deficiencies in plants using proximal images and machine learning: A review. Comput. Electron. Agric. 2019, 162, 482–492. [Google Scholar] [CrossRef]
- Zha, H.; Miao, Y.; Wang, T.; Li, Y.; Zhang, J.; Sun, W.; Feng, Z.; Kusnierek, K. Improving Unmanned Aerial Vehicle Remote Sensing-Based Rice Nitrogen Nutrition Index Prediction with Machine Learning. Remote Sens. 2020, 12, 215. [Google Scholar] [CrossRef] [Green Version]
- Krishnamoorthy, N.; Prasad, L.V.N.; Kumar, C.S.P.; Subedi, B.; Abraha, H.B.; Sathiskumar, V.E. Rice leaf diseases prediction using deep neural networks with transfer learning. Environ. Res. 2021, 198, 111275. [Google Scholar]
- Abbaspour-Gilandeh, Y.; Molaee, A.; Sabzi, S.; Nabipur, N.; Shamshirband, S.; Mosavi, A. A Combined Method of Image Processing and Artificial Neural Network for the Identification of 13 Iranian Rice Cultivars. Agronomy 2020, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Mostafaeipour, A.; Fakhrzad, M.; Gharaat, S.; Jahangiri, M.; Dhanraj, J.; Band, S.; Issakhov, A.; Mosavi, A. Machine Learning for Prediction of Energy in Wheat Production. Agriculture 2020, 10, 517. [Google Scholar] [CrossRef]
- Sethy, P.K.; Kumari, C.; Barpanda, N.K.; Negi, B.; Behera, S.; Rath, A.K. Identification of Mineral Deficiency in Rice Crop based on SVM in Approach of K-Means & Fuzzy C-Means Clustering. Helix 2017, 7, 1970–1983. [Google Scholar]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- Li, J.; Oswald, C.; George, L.G.; Shi, Y. Improving model robustness for soybean iron deficiency chlorosis rating by unsupervised pre-training on unmanned aircraft system derived images. Comput. Electron. Agric. 2020, 175, 105557. [Google Scholar] [CrossRef]
- Han, K.A.M.; Watchareeruetai, U. Classification of Nutrient Deficiency in Black Gram Using Deep Convolutional Neural Networks. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chonburi, Thailand, 10–12 July 2019; pp. 277–282. [Google Scholar]
- Tran, T.-T.; Choi, J.-W.; Le, T.-T.H.; Kim, J.-W. A Comparative Study of Deep CNN in Forecasting and Classifying the Macronutrient Deficiencies on Development of Tomato Plant. Appl. Sci. 2019, 9, 1601. [Google Scholar] [CrossRef] [Green Version]
- Wulandhari, L.A.; Gunawan, A.A.S.; Qurania, A.; Harsani, P.; Tarawan, T.F.; Hermawan, R.F. Plant nutrient deficiency detection using deep convolutional neural network. ICIC Express Lett. 2019, 13, 971–977. [Google Scholar]
- Xu, Z.; Guo, X.; Zhu, A.; He, X.; Zhao, X.; Han, Y.; Subedi, R. Using Deep Convolutional Neural Networks for Image-Based Diagnosis of Nutrient Deficiencies in Rice. Comput. Intell. Neurosci. 2020, 2020, 7307252. [Google Scholar] [CrossRef]
- Jahagirdar, P.; Budihal, S.V. Framework to Detect NPK Deficiency in Maize Plants Using CNNs. In Progress in Advanced Computing and Intelligent Engineering, Advances in Intelligent Systems and Computing; Panigrahi, C.R., Ed.; Springer: Singapore, 2021; p. 1199. [Google Scholar]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Nitrogen Defciency Prediction of Rice Crop Based on Convolutional Neural Network. J. Ambient. Intell. Humaniz. Comput. 2020, 11, 5703–5711. [Google Scholar] [CrossRef]
- Abdalla, A.; Cen, H.; Wan, L.; Mehmood, K.; He, Y. Nutrient Status Diagnosis of Infield Oilseed Rape via Deep Learning-Enabled Dynamic Model. IEEE Trans. Ind. Inform. 2021, 17, 4379–4389. [Google Scholar] [CrossRef]
- Bahtiar, A.R.; Pranowo; Santoso, A.J.; Juhariah, J. Deep Learning Detected Nutrient Deficiency in Chili Plant, in 8th International conference on Information and Communication Technology. In Proceedings of the 2020 8th International Conference on Information and Communication Technology, Yogyakarta, Indonesia, 24–26 June 2020. [Google Scholar]
- Abayomi-Alli, O.; Damaševičius, R.; Misra, S.; Maskeliūnas, R. Cassava disease recognition from low-quality images using enhanced data augmentation model and deep learning. Expert Syst. 2021, 38, e12746. [Google Scholar] [CrossRef]
- Oyewola, D.O.; Dada, E.G.; Misra, S.; Damaševičius, R. Detecting cassava mosaic disease using a deep residual convolutional neural network with distinct block processing. PeerJ Comput. Sci. 2021, 7, e352. [Google Scholar] [CrossRef]
- Almadhor, A.; Rauf, H.; Lali, M.; Damaševičius, R.; Alouffi, B.; Alharbi, A. AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery. Sensors 2021, 21, 3830. [Google Scholar] [CrossRef] [PubMed]
- Kundu, N.; Rani, G.; Dhaka, V.S.; Gupta, K.; Nayak, S.C.; Verma, S.; Ijaz, M.F.; Woźniak, M. IoT and Interpretable Machine Learning Based Framework for Disease Prediction in Pearl Millet. Sensors 2021, 21, 5386. [Google Scholar] [CrossRef]
- Li, K.; Lin, J.; Liu, J.; Zhao, Y. Using Deep Learning for Image-Based Different Degrees of Ginkgo Leaf Disease Classification. Information 2020, 11, 95. [Google Scholar] [CrossRef] [Green Version]
- Yi, J.; Krusenbaum, L.; Unger, P.; Hüging, H.; Seidel, S.J.; Schaaf, G.; Gall, J. Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors 2020, 20, 5893. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Maji, A.; Jasiński, M.; Leonowicz, Z.; Jasińska, E. Identification of Plant-Leaf Diseases Using CNN and Transfer-Learning Approach. Electronics 2020, 10, 1388. [Google Scholar] [CrossRef]
- Raksarikon, W. Nutrient Deficiency Symptom in Rice, Kaggle V1. 2020. Available online: https://www.kaggle.com/guy007/nutrientdeficiencysymptomsinrice/activity (accessed on 21 October 2021).
- Sethy, P.K. Nitrogen Deficiency of Rice Crop, Mendeley Data, V1. 2020. Available online: https://data.mendeley.com/datasets/gzm5pxntyv/1 (accessed on 21 October 2021).
- Bansal, P.; Kumar, R.; Kumar, S. Disease detection in Apple leaves using deep convolutional neural network. Agriculture 2021, 11, 617. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Ramesh, S.; Vydeki, D. Recognition and classification of paddy leaf diseases using Optimized Deep Neural network with Jaya algorithm. Inf. Processing Agric. 2020, 7, 249–260. [Google Scholar] [CrossRef]
- Bari, S.; Islam, M.N.; Rashid, M.; Hasan, M.J.; Razman, M.A.M.; Musa, R.M.; Ab Nasir, A.F.; Abdul Majeed, A.P.P. A real-time approach of diagnosing rice leaf disease using deep learning-based faster R-CNN framework. PeerJ Comput. Sci. 2021, 7, e432. [Google Scholar] [CrossRef]
- Uğuz, S.; Uysal, N. Classification of olive leaf diseases using deep convolutional neural networks. Neural Comput. Appl. 2021, 33, 4133–4149. [Google Scholar] [CrossRef]
- Sharma, M.; Kumar, C.J.; Deka, A. Early diagnosis of rice plant disease using machine learning techniques. Arch. Phytopathol. Plant. Prot. 2021, 1–25. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erjam, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015. [Google Scholar]
- Peng, J.; Kang, S.; Ning, Z.; Deng, H.; Shen, J.; Xu, Y.; Zhang, J.; Zhao, W.; Li, X.; Gong, W.; et al. Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging. Eur Radiol. 2020, 30, 413–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turkoglu, M.; Yanikoğlu, B.; Hanbay, D. PlantDiseaseNet: Convolutional neural network ensemble for plant disease and pest detection. In Signal Image Video Processing; Springer: London, UK, 2021; pp. 1–9. [Google Scholar]
- Vallabhajosyula, S.; Sistla, V.; Kolli, V.K.K. Transfer learning-based deep ensemble neural network for plant leaf disease detection. J. Plant. Dis. Prot. 2021, 1–14. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, Z.; Ma, Y.; Du, Q.; Williams, P.; Drewry, J.; Luck, B. Alfalfa yield prediction using UAV-based hyperspectral imagery and ensemble learning. Remote Sens. 2020, 12, 2028. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).