Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis
Abstract
1. Introduction
1.1. The Family of SWItch/Sucrose Non-Fermentable (SWI/SNF) Chromatin Remodeling Enzymes
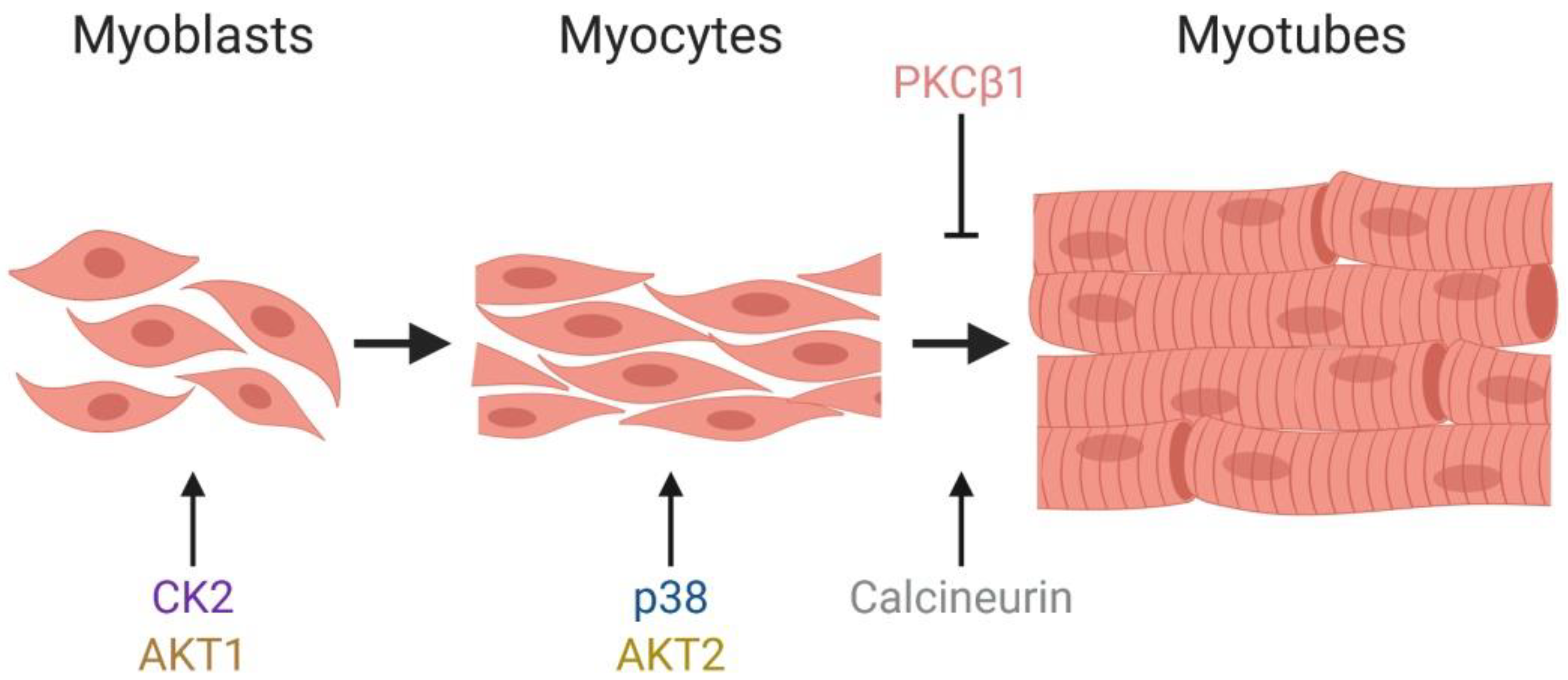
1.2. Myogenesis
2. Cell Signaling Pathways Modulate Chromatin Remodeling Enzyme Function during Skeletal Muscle Myogenesis
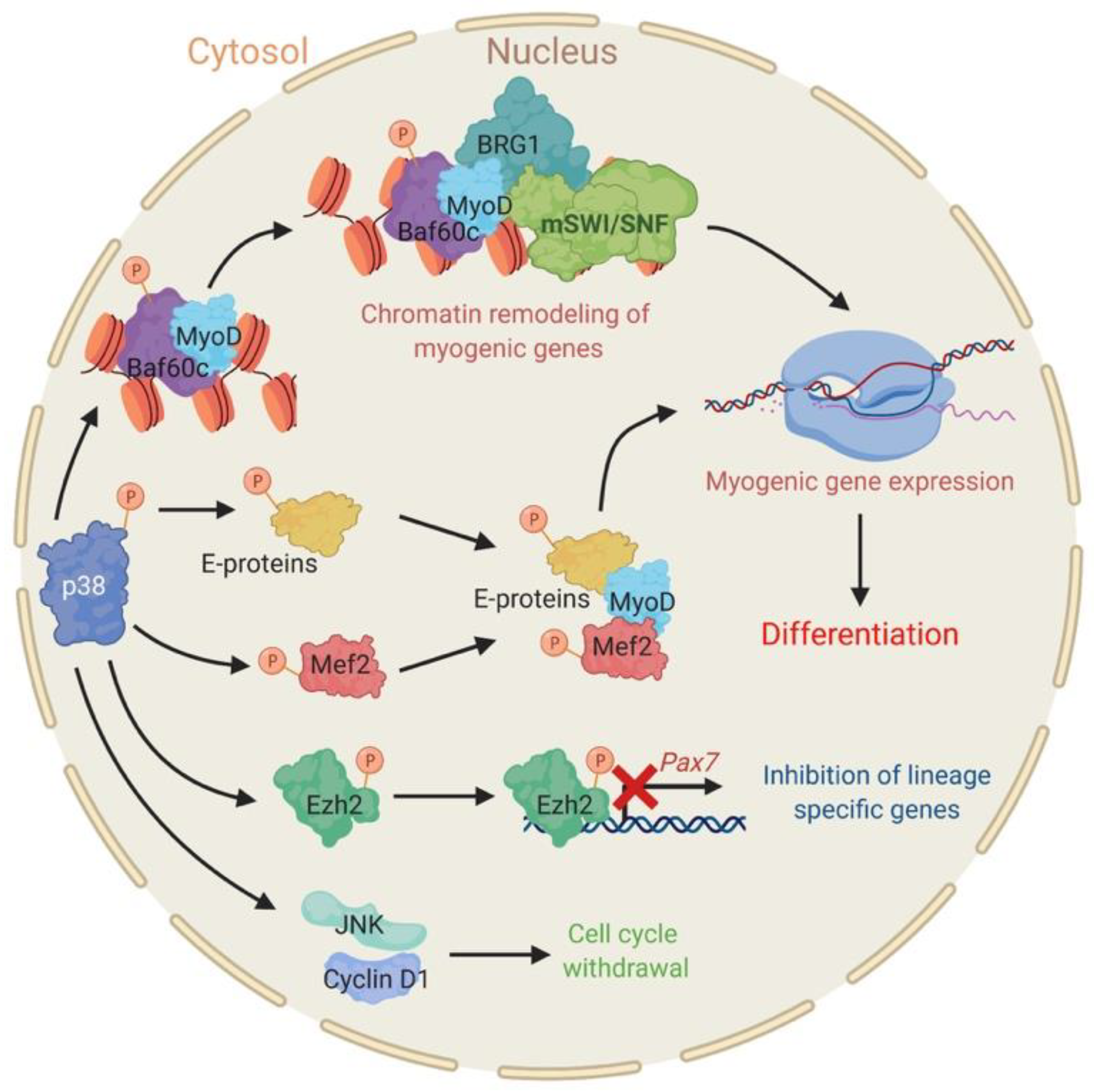
2.1. The Stress Responsive P38 Kinase Is a Regulator of Chromatin Remodeling Enzymes during Myogenesis
2.2. The Pleiotropic Network of AKT Converges on BAF60c to Promote Myogenesis
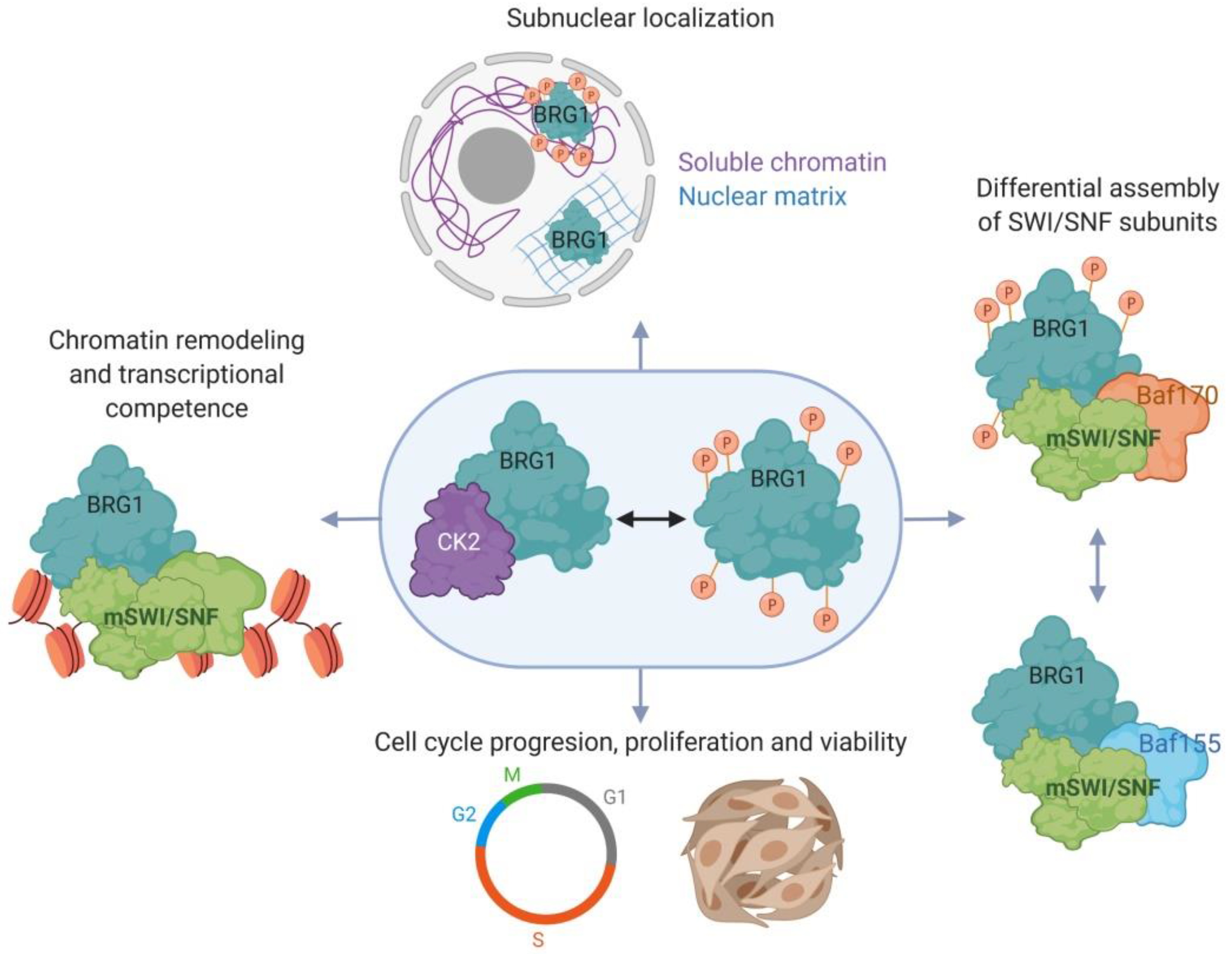
2.3. Myoblast Cell Cycle Progression and Viability Are Regulated by Casein Kinase 2 (CK2)-Mediated Phosphorylation of BRG1
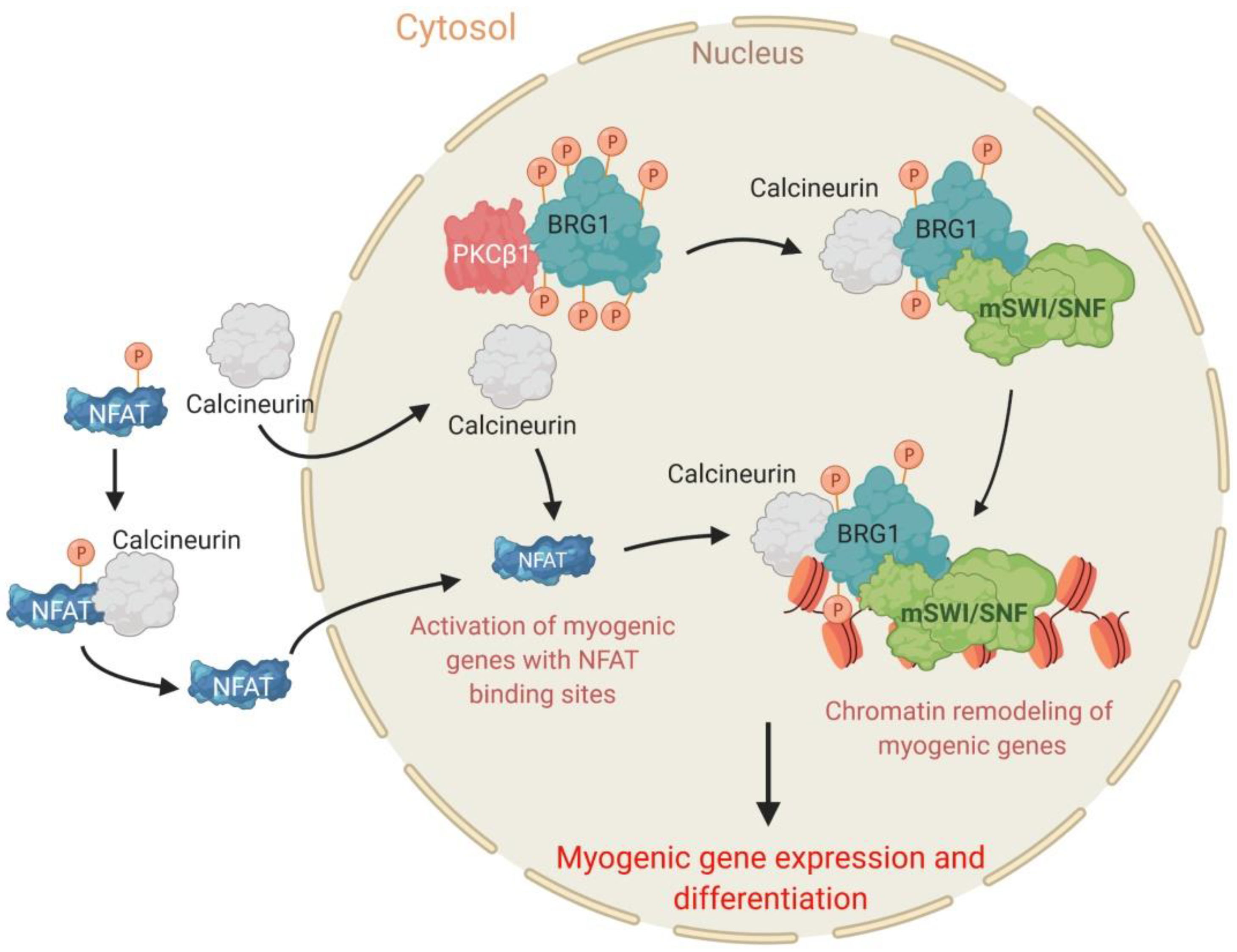
2.4. The PKCβ1 Kinase and the Calcineurin (Cn) Phosphatase Act in Opposition to Regulate BRG1 Phosphorylation and Myogenic Differentiation
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mills, A.A. The chromodomain helicase DNA-binding chromatin remodelers: Family traits that protect from and promote cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026450. [Google Scholar] [CrossRef] [PubMed]
- Marfella, C.G.; Imbalzano, A.N. The chd family of chromatin remodelers. Mutat. Res. 2007, 618, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Poli, J.; Gasser, S.M.; Papamichos-Chronakis, M. The ino80 remodeller in transcription, replication and repair. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160290. [Google Scholar] [CrossRef] [PubMed]
- Gerhold, C.B.; Hauer, M.H.; Gasser, S.M. Ino80-c and swr-c: Guardians of the genome. J. Mol. Biol. 2015, 427, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, L.R.; Picketts, D.J. The role of iswi chromatin remodeling complexes in brain development and neurodevelopmental disorders. Mol. Cell. Neurosci. 2018, 87, 55–64. [Google Scholar] [CrossRef]
- Kadoch, C.; Copeland, R.A.; Keilhack, H. Prc2 and swi/snf chromatin remodeling complexes in health and disease. Biochemistry 2016, 55, 1600–1614. [Google Scholar] [CrossRef]
- Euskirchen, G.; Auerbach, R.K.; Snyder, M. Swi/snf chromatin-remodeling factors: Multiscale analyses and diverse functions. J. Biol. Chem. 2012, 287, 30897–30905. [Google Scholar] [CrossRef]
- Neigeborn, L.; Carlson, M. Genes affecting the regulation of suc2 gene expression by glucose repression in saccharomyces cerevisiae. Genetics 1984, 108, 845–858. [Google Scholar]
- Peterson, C.L.; Herskowitz, I. Characterization of the yeast swi1, swi2, and swi3 genes, which encode a global activator of transcription. Cell 1992, 68, 573–583. [Google Scholar] [CrossRef]
- Stern, M.; Jensen, R.; Herskowitz, I. Five swi genes are required for expression of the ho gene in yeast. J. Mol. Biol. 1984, 178, 853–868. [Google Scholar] [CrossRef]
- Cote, J.; Quinn, J.; Workman, J.L.; Peterson, C.L. Stimulation of gal4 derivative binding to nucleosomal DNA by the yeast swi/snf complex. Science 1994, 265, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Cairns, B.R.; Kim, Y.J.; Sayre, M.H.; Laurent, B.C.; Kornberg, R.D. A multisubunit complex containing the swi1/adr6, swi2/snf2, swi3, snf5, and snf6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 1994, 91, 1950–1954. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Imbalzano, A.N.; Khavari, P.A.; Kingston, R.E.; Green, M.R. Nucleosome disruption and enhancement of activator binding by a human sw1/snf complex. Nature 1994, 370, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Imbalzano, A.N.; Kwon, H.; Green, M.R.; Kingston, R.E. Facilitated binding of tata-binding protein to nucleosomal DNA. Nature 1994, 370, 481–485. [Google Scholar] [CrossRef]
- Wang, W.; Cote, J.; Xue, Y.; Zhou, S.; Khavari, P.A.; Biggar, S.R.; Muchardt, C.; Kalpana, G.V.; Goff, S.P.; Yaniv, M.; et al. Purification and biochemical heterogeneity of the mammalian swi-snf complex. EMBO J. 1996, 15, 5370–5382. [Google Scholar] [CrossRef]
- Logie, C.; Peterson, C.L. Catalytic activity of the yeast swi/snf complex on reconstituted nucleosome arrays. EMBO J. 1997, 16, 6772–6782. [Google Scholar] [CrossRef]
- Papoulas, O.; Beek, S.J.; Moseley, S.L.; McCallum, C.M.; Sarte, M.; Shearn, A.; Tamkun, J.W. The drosophila trithorax group proteins brm, ash1 and ash2 are subunits of distinct protein complexes. Development 1998, 125, 3955–3966. [Google Scholar]
- Dingwall, A.K.; Beek, S.J.; McCallum, C.M.; Tamkun, J.W.; Kalpana, G.V.; Goff, S.P.; Scott, M.P. The drosophila snr1 and brm proteins are related to yeast swi/snf proteins and are components of a large protein complex. Mol. Biol. Cell 1995, 6, 777–791. [Google Scholar] [CrossRef]
- Elfring, L.K.; Daniel, C.; Papoulas, O.; Deuring, R.; Sarte, M.; Moseley, S.; Beek, S.J.; Waldrip, W.R.; Daubresse, G.; DePace, A.; et al. Genetic analysis of brahma: The drosophila homolog of the yeast chromatin remodeling factor swi2/snf2. Genetics 1998, 148, 251–265. [Google Scholar]
- Tamkun, J.W.; Deuring, R.; Scott, M.P.; Kissinger, M.; Pattatucci, A.M.; Kaufman, T.C.; Kennison, J.A. Brahma: A regulator of drosophila homeotic genes structurally related to the yeast transcriptional activator snf2/swi2. Cell 1992, 68, 561–572. [Google Scholar] [CrossRef]
- Treisman, J.E.; Luk, A.; Rubin, G.M.; Heberlein, U. Eyelid antagonizes wingless signaling during drosophila development and has homology to the bright family of DNA-binding proteins. Genes Dev. 1997, 11, 1949–1962. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Malakar, S.; Krebs, J.E. How many remodelers does it take to make a brain? Diverse and cooperative roles of atp-dependent chromatin-remodeling complexes in development. Biochem. Cell Biol. 2007, 85, 444–462. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef] [PubMed]
- De la Serna, I.L.; Ohkawa, Y.; Imbalzano, A.N. Chromatin remodelling in mammalian differentiation: Lessons from atp-dependent remodellers. Nat. Rev. Genet. 2006, 7, 461–473. [Google Scholar] [CrossRef]
- Khavari, P.A.; Peterson, C.L.; Tamkun, J.W.; Mendel, D.B.; Crabtree, G.R. Brg1 contains a conserved domain of the swi2/snf2 family necessary for normal mitotic growth and transcription. Nature 1993, 366, 170–174. [Google Scholar] [CrossRef]
- Muchardt, C.; Yaniv, M. A human homologue of saccharomyces cerevisiae snf2/swi2 and drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993, 12, 4279–4290. [Google Scholar] [CrossRef]
- Chiba, H.; Muramatsu, M.; Nomoto, A.; Kato, H. Two human homologues of saccharomyces cerevisiae swi2/snf2 and drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994, 22, 1815–1820. [Google Scholar] [CrossRef]
- Fryer, C.J.; Archer, T.K. Chromatin remodelling by the glucocorticoid receptor requires the brg1 complex. Nature 1998, 393, 88–91. [Google Scholar] [CrossRef]
- De la Serna, I.L.; Carlson, K.A.; Imbalzano, A.N. Mammalian swi/snf complexes promote myod-mediated muscle differentiation. Nat. Genet. 2001, 27, 187–190. [Google Scholar] [CrossRef]
- Lee, D.; Sohn, H.; Kalpana, G.V.; Choe, J. Interaction of e1 and hsnf5 proteins stimulates replication of human papillomavirus DNA. Nature 1999, 399, 487–491. [Google Scholar] [CrossRef]
- Murphy, D.J.; Hardy, S.; Engel, D.A. Human swi-snf component brg1 represses transcription of the c-fos gene. Mol. Cell. Biol. 1999, 19, 2724–2733. [Google Scholar] [CrossRef] [PubMed]
- Patenge, N.; Elkin, S.K.; Oettinger, M.A. Atp-dependent remodeling by swi/snf and iswi proteins stimulates v(d)j cleavage of 5 s arrays. J. Biol. Chem. 2004, 279, 35360–35367. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Lee, C.C.; Kohwi-Shigematsu, T. Satb1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006, 38, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Morshead, K.B.; Ciccone, D.N.; Taverna, S.D.; Allis, C.D.; Oettinger, M.A. Antigen receptor loci poised for v(d)j rearrangement are broadly associated with brg1 and flanked by peaks of histone h3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA 2003, 100, 11577–11582. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, Q.E.; Ray, A.; Wani, G.; Han, C.; Milum, K.; Wani, A.A. Modulation of nucleotide excision repair by mammalian swi/snf chromatin-remodeling complex. J. Biol. Chem. 2009, 284, 30424–30432. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Q.; Jones, K.; Patel, M.; Gong, F. The chromatin remodeling factor brg1 stimulates nucleotide excision repair by facilitating recruitment of xpc to sites of DNA damage. Cell Cycle 2009, 8, 3953–3959. [Google Scholar] [CrossRef]
- Wang, X.; Wang, S.; Troisi, E.C.; Howard, T.P.; Haswell, J.R.; Wolf, B.K.; Hawk, W.H.; Ramos, P.; Oberlick, E.M.; Tzvetkov, E.P.; et al. Brd9 defines a swi/snf sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat. Commun. 2019, 10, 1881. [Google Scholar] [CrossRef]
- Alpsoy, A.; Dykhuizen, E.C. Glioma tumor suppressor candidate region gene 1 (gltscr1) and its paralog gltscr1-like form swi/snf chromatin remodeling subcomplexes. J. Biol. Chem. 2018, 293, 3892–3903. [Google Scholar] [CrossRef]
- Michel, B.C.; D’Avino, A.R.; Cassel, S.H.; Mashtalir, N.; McKenzie, Z.M.; McBride, M.J.; Valencia, A.M.; Zhou, Q.; Bocker, M.; Soares, L.M.M.; et al. A non-canonical swi/snf complex is a synthetic lethal target in cancers driven by baf complex perturbation. Nat. Cell Biol. 2018, 20, 1410–1420. [Google Scholar] [CrossRef]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular organization and assembly of swi/snf family chromatin remodeling complexes. Cell 2018, 175, 1272–1288.e1220. [Google Scholar] [CrossRef]
- Wang, W.; Xue, Y.; Zhou, S.; Kuo, A.; Cairns, B.R.; Crabtree, G.R. Diversity and specialization of mammalian swi/snf complexes. Genes Dev. 1996, 10, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Muchardt, C.; Sardet, C.; Bourachot, B.; Onufryk, C.; Yaniv, M. A human protein with homology to saccharomyces cerevisiae snf5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995, 23, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Kalpana, G.V.; Marmon, S.; Wang, W.; Crabtree, G.R.; Goff, S.P. Binding and stimulation of hiv-1 integrase by a human homolog of yeast transcription factor snf5. Science 1994, 266, 2002–2006. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.I. Diverse functions of atp-dependent chromatin remodeling complexes in development and cancer. Acta Biochim. Biophys. Sin. 2012, 44, 54–69. [Google Scholar] [CrossRef]
- Hargreaves, D.C.; Crabtree, G.R. Atp-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Lyon, D.; Young, C.; Jensen, L.J.; Vertegaal, A.C.; Nielsen, M.L. Site-specific mapping of the human sumo proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017, 24, 325–336. [Google Scholar] [CrossRef]
- Guo, A.; Gu, H.; Zhou, J.; Mulhern, D.; Wang, Y.; Lee, K.A.; Yang, V.; Aguiar, M.; Kornhauser, J.; Jia, X.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteom. 2014, 13, 372–387. [Google Scholar] [CrossRef]
- Nasipak, B.T.; Padilla-Benavides, T.; Green, K.M.; Leszyk, J.D.; Mao, W.; Konda, S.; Sif, S.; Shaffer, S.A.; Ohkawa, Y.; Imbalzano, A.N. Opposing calcium-dependent signalling pathways control skeletal muscle differentiation by regulating a chromatin remodelling enzyme. Nat. Commun. 2015, 6, 7441. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Nasipak, B.T.; Paskavitz, A.L.; Haokip, D.T.; Schnabl, J.M.; Nickerson, J.A.; Imbalzano, A.N. Casein kinase 2-mediated phosphorylation of brahma-related gene 1 controls myoblast proliferation and contributes to swi/snf complex composition. J. Biol. Chem. 2017, 292, 18592–18607. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Haokip, D.T.; Yoon, Y.; Reyes-Gutierrez, P.; Rivera-Perez, J.A.; Imbalzano, A.N. Ck2-dependent phosphorylation of the brg1 chromatin remodeling enzyme occurs during mitosis. Int. J. Mol. Sci. 2020, 21, 923. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wong, R.H.; Tang, T.; Hudak, C.S.; Yang, D.; Duncan, R.E.; Sul, H.S. Phosphorylation and recruitment of baf60c in chromatin remodeling for lipogenesis in response to insulin. Mol. Cell 2013, 49, 283–297. [Google Scholar] [CrossRef]
- Puri, P.L.; Mercola, M. Baf60 a, b, and cs of muscle determination and renewal. Genes Dev. 2012, 26, 2673–2683. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Roberts, C.W. Carma: Carm1 methylation of swi/snf in breast cancer. Cancer Cell 2014, 25, 3–4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Basson, M.A. Signaling in cell differentiation and morphogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008151. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Elowitz, M.B. Communication codes in developmental signaling pathways. Development 2019, 146. [Google Scholar] [CrossRef]
- Perrimon, N.; Pitsouli, C.; Shilo, B.Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 2012, 4, a005975. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Caenepeel, S.; Charydczak, G.; Sudarsanam, S.; Hunter, T.; Manning, G. The mouse kinome: Discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. USA 2004, 101, 11707–11712. [Google Scholar] [CrossRef]
- Wilson, L.J.; Linley, A.; Hammond, D.E.; Hood, F.E.; Coulson, J.M.; MacEwan, D.J.; Ross, S.J.; Slupsky, J.R.; Smith, P.D.; Eyers, P.A.; et al. New perspectives, opportunities, and challenges in exploring the human protein kinome. Cancer Res. 2018, 78, 15–29. [Google Scholar] [CrossRef]
- Fleuren, E.D.; Zhang, L.; Wu, J.; Daly, R.J. The kinome ‘at large’ in cancer. Nat. Rev. Cancer 2016, 16, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, S.; Buckingham, M. The birth of muscle progenitor cells in the mouse: Spatiotemporal considerations. Curr. Top. Dev. Biol. 2000, 48, 225–268. [Google Scholar] [PubMed]
- Bryson-Richardson, R.J.; Currie, P.D. The genetics of vertebrate myogenesis. Nat. Rev. Genet. 2008, 9, 632–646. [Google Scholar] [CrossRef] [PubMed]
- Borycki, A.G.; Emerson, C.P., Jr. Multiple tissue interactions and signal transduction pathways control somite myogenesis. Curr. Top. Dev. Biol. 2000, 48, 165–224. [Google Scholar]
- Sambasivan, R.; Tajbakhsh, S. Skeletal muscle stem cell birth and properties. Semin. Cell Dev. Biol. 2007, 18, 870–882. [Google Scholar] [CrossRef]
- Chal, J.; Pourquie, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef]
- Yabe, T.; Takada, S. Molecular mechanism for cyclic generation of somites: Lessons from mice and zebrafish. Dev. Growth Differ. 2016, 58, 31–42. [Google Scholar] [CrossRef]
- Mallo, M. Revisiting the involvement of signaling gradients in somitogenesis. FEBS J. 2016, 283, 1430–1437. [Google Scholar] [CrossRef]
- Shi, X.; Garry, D.J. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006, 20, 1692–1708. [Google Scholar] [CrossRef]
- Zammit, P.S.; Relaix, F.; Nagata, Y.; Ruiz, A.P.; Collins, C.A.; Partridge, T.A.; Beauchamp, J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Biressi, S.; Tagliafico, E.; Lamorte, G.; Monteverde, S.; Tenedini, E.; Roncaglia, E.; Ferrari, S.; Cusella-De Angelis, M.G.; Tajbakhsh, S.; Cossu, G. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev. Biol. 2007, 304, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.H.; Seale, P.; Rudnicki, M.A. Looking back to the embryo: Defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003, 4, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Biressi, S.; Molinaro, M.; Cossu, G. Cellular heterogeneity during vertebrate skeletal muscle development. Dev. Biol. 2007, 308, 281–293. [Google Scholar] [CrossRef]
- Messina, G.; Cossu, G. The origin of embryonic and fetal myoblasts: A role of pax3 and pax7. Genes Dev. 2009, 23, 902–905. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef]
- Dhawan, J.; Rando, T.A. Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005, 15, 666–673. [Google Scholar] [CrossRef]
- Relaix, F. Skeletal muscle progenitor cells: From embryo to adult. Cell. Mol. Life Sci. CMLS 2006, 63, 1221–1225. [Google Scholar] [CrossRef]
- Zammit, P.S. All muscle satellite cells are equal, but are some more equal than others? J. Cell Sci. 2008, 121, 2975–2982. [Google Scholar] [CrossRef]
- Motohashi, N.; Asakura, A. Muscle satellite cell heterogeneity and self-renewal. Front Cell Dev. Biol. 2014, 2, 1. [Google Scholar] [CrossRef]
- Chang, N.C.; Rudnicki, M.A. Satellite cells: The architects of skeletal muscle. Curr. Top. Dev. Biol. 2014, 107, 161–181. [Google Scholar]
- Sambasivan, R.; Tajbakhsh, S. Adult skeletal muscle stem cells. Results Probl. Cell Differ. 2015, 56, 191–213. [Google Scholar] [PubMed]
- Rudnicki, M.A.; Le Grand, F.; McKinnell, I.; Kuang, S. The molecular regulation of muscle stem cell function. Cold Spring Harbor Symp. Quant. Biol. 2008, 73, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef] [PubMed]
- Montarras, D.; L’Honore, A.; Buckingham, M. Lying low but ready for action: The quiescent muscle satellite cell. FEBS J. 2013, 280, 4036–4050. [Google Scholar] [CrossRef] [PubMed]
- Brack, A.S.; Rando, T.A. Tissue-specific stem cells: Lessons from the skeletal muscle satellite cell. Cell Stem Cell 2012, 10, 504–514. [Google Scholar] [CrossRef]
- Mansouri, A.; Stoykova, A.; Torres, M.; Gruss, P. Dysgenesis of cephalic neural crest derivatives in pax7-/-mutant mice. Development 1996, 122, 831–838. [Google Scholar]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef]
- Oustanina, S.; Hause, G.; Braun, T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004, 23, 3430–3439. [Google Scholar] [CrossRef]
- Relaix, F.; Montarras, D.; Zaffran, S.; Gayraud-Morel, B.; Rocancourt, D.; Tajbakhsh, S.; Mansouri, A.; Cumano, A.; Buckingham, M. Pax3 and pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006, 172, 91–102. [Google Scholar] [CrossRef]
- Kuang, S.; Gillespie, M.A.; Rudnicki, M.A. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell 2008, 2, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; von Maltzahn, J.; Rudnicki, M.A. Extrinsic regulation of satellite cell specification. Stem Cell Res. Ther. 2010, 1, 27. [Google Scholar] [CrossRef] [PubMed]
- Muchardt, C.; Reyes, J.C.; Bourachot, B.; Leguoy, E.; Yaniv, M. The hbrm and brg-1 proteins, components of the human snf/swi complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996, 15, 3394–3402. [Google Scholar] [CrossRef] [PubMed]
- Sif, S.; Stukenberg, P.T.; Kirschner, M.W.; Kingston, R.E. Mitotic inactivation of a human swi/snf chromatin remodeling complex. Genes Dev. 1998, 12, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Arakawa, N.; Hirano, H. Mass spectrometric analysis of the phosphorylation levels of the swi/snf chromatin remodeling/tumor suppressor proteins arid1a and brg1 in ovarian clear cell adenocarcinoma cell lines. J. Proteome Res. 2014, 13, 4959–4969. [Google Scholar] [CrossRef] [PubMed]
- Dallas, P.B.; Yaciuk, P.; Moran, E. Characterization of monoclonal antibodies raised against p300: Both p300 and cbp are present in intracellular tbp complexes. J. Virol. 1997, 71, 1726–1731. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Nasipak, B.T.; Imbalzano, A.N. Brg1 controls the expression of pax7 to promote viability and proliferation of mouse primary myoblasts. J. Cell. Physiol. 2015, 230, 2990–2997. [Google Scholar] [CrossRef]
- Albini, S.; Coutinho Toto, P.; Dall’Agnese, A.; Malecova, B.; Cenciarelli, C.; Felsani, A.; Caruso, M.; Bultman, S.J.; Puri, P.L. Brahma is required for cell cycle arrest and late muscle gene expression during skeletal myogenesis. EMBO Rep. 2015, 16, 1037–1050. [Google Scholar] [CrossRef]
- Lickert, H.; Takeuchi, J.K.; Von Both, I.; Walls, J.R.; McAuliffe, F.; Adamson, S.L.; Henkelman, R.M.; Wrana, J.L.; Rossant, J.; Bruneau, B.G. Baf60c is essential for function of baf chromatin remodelling complexes in heart development. Nature 2004, 432, 107–112. [Google Scholar] [CrossRef]
- Phelan, M.L.; Sif, S.; Narlikar, G.J.; Kingston, R.E. Reconstitution of a core chromatin remodeling complex from swi/snf subunits. Mol. Cell 1999, 3, 247–253. [Google Scholar] [CrossRef]
- Adi, S.; Bin-Abbas, B.; Wu, N.Y.; Rosenthal, S.M. Early stimulation and late inhibition of extracellular signal-regulated kinase 1/2 phosphorylation by igf-i: A potential mechanism mediating the switch in igf-i action on skeletal muscle cell differentiation. Endocrinology 2002, 143, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Coolican, S.A.; Samuel, D.S.; Ewton, D.Z.; McWade, F.J.; Florini, J.R. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J. Biol. Chem. 1997, 272, 6653–6662. [Google Scholar] [CrossRef] [PubMed]
- Gredinger, E.; Gerber, A.N.; Tamir, Y.; Tapscott, S.J.; Bengal, E. Mitogen-activated protein kinase pathway is involved in the differentiation of muscle cells. J. Biol. Chem. 1998, 273, 10436–10444. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.A.; Ouellette, S.E.; Liu, X.; Allen, R.E.; Johnson, S.E. E2f5 and lek1 translocation to the nucleus is an early event demarcating myoblast quiescence. J. Cell. Biochem. 2007, 101, 1394–1408. [Google Scholar] [CrossRef]
- Cuenda, A.; Rousseau, S. P38 map-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 2007, 1773, 1358–1375. [Google Scholar] [CrossRef]
- Lee, J.K.; Kim, N.J. Recent advances in the inhibition of p38 mapk as a potential strategy for the treatment of alzheimer’s disease. Molecules 2017, 22, 1287. [Google Scholar] [CrossRef]
- Risco, A.; Cuenda, A. New insights into the p38gamma and p38delta mapk pathways. J. Signal Transduct. 2012, 2012, 520289. [Google Scholar] [CrossRef] [PubMed]
- Escos, A.; Risco, A.; Alsina-Beauchamp, D.; Cuenda, A. P38gamma and p38delta mitogen activated protein kinases (mapks), new stars in the mapk galaxy. Front Cell Dev. Biol. 2016, 4, 31. [Google Scholar] [CrossRef]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 mapk signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef]
- Kumar, S.; Boehm, J.; Lee, J.C. P38 map kinases: Key signalling molecules as therapeutic targets for inflammatory diseases. Nat. Rev. Drug Discov. 2003, 2, 717–726. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Protein kinase cascades activated by stress and inflammatory cytokines. BioEssays News Rev. Mol. Cell. Dev. Biol. 1996, 18, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Choi, E.J. Compromised mapk signaling in human diseases: An update. Arch. Toxicol. 2015, 89, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Yu, L.; Liu, L.; Cheung, C.F.; Li, X.; Yee, S.P.; Yang, X.J.; Wu, Z. P38 mitogen-activated protein kinase-, calcium-calmodulin-dependent protein kinase-, and calcineurin-mediated signaling pathways transcriptionally regulate myogenin expression. Mol. Biol. Cell 2002, 13, 1940–1952. [Google Scholar] [CrossRef] [PubMed]
- Simone, C.; Forcales, S.V.; Hill, D.A.; Imbalzano, A.N.; Latella, L.; Puri, P.L. P38 pathway targets swi-snf chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004, 36, 738–743. [Google Scholar] [CrossRef]
- Lluis, F.; Ballestar, E.; Suelves, N.; Esteller, M.; Munoz-Canoves, P. E47 phosphorylation by p38 mapk promotes myod/e47 association and muscle-specific gene transcription. EMBO J. 2005, 24, 974–984. [Google Scholar] [CrossRef]
- Lluis, F.; Perdiguero, E.; Nebreda, A.R.; Munoz-Canoves, P. Regulation of skeletal muscle gene expression by p38 map kinases. Trends Cell. Biol. 2006, 16, 36–44. [Google Scholar] [CrossRef]
- Keren, A.; Tamir, Y.; Bengal, E. The p38 mapk signaling pathway: A major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Rampalli, S.; Li, L.; Mak, E.; Ge, K.; Brand, M.; Tapscott, S.J.; Dilworth, F.J. P38 mapk signaling regulates recruitment of ash2l-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 2007, 14, 1150–1156. [Google Scholar] [CrossRef]
- Perdiguero, E.; Ruiz-Bonilla, V.; Serrano, A.L.; Munoz-Canoves, P. Genetic deficiency of p38alpha reveals its critical role in myoblast cell cycle exit: The p38alpha-jnk connection. Cell Cycle 2007, 6, 1298–1303. [Google Scholar] [CrossRef]
- Perdiguero, E.; Ruiz-Bonilla, V.; Gresh, L.; Hui, L.; Ballestar, E.; Sousa-Victor, P.; Baeza-Raja, B.; Jardi, M.; Bosch-Comas, A.; Esteller, M.; et al. Genetic analysis of p38 map kinases in myogenesis: Fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 2007, 26, 1245–1256. [Google Scholar] [CrossRef]
- Ruiz-Bonilla, V.; Perdiguero, E.; Gresh, L.; Serrano, A.L.; Zamora, M.; Sousa-Victor, P.; Jardi, M.; Wagner, E.F.; Munoz-Canoves, P. Efficient adult skeletal muscle regeneration in mice deficient in p38beta, p38gamma and p38delta map kinases. Cell Cycle 2008, 7, 2208–2214. [Google Scholar] [CrossRef] [PubMed]
- Penn, B.H.; Bergstrom, D.A.; Dilworth, F.J.; Bengal, E.; Tapscott, S.J. A myod-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 2004, 18, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, D.A.; Penn, B.H.; Strand, A.; Perry, R.L.; Rudnicki, M.A.; Tapscott, S.J. Promoter-specific regulation of myod binding and signal transduction cooperate to pattern gene expression. Mol. Cell 2002, 9, 587–600. [Google Scholar] [CrossRef]
- Gillespie, M.A.; Le Grand, F.; Scime, A.; Kuang, S.; von Maltzahn, J.; Seale, V.; Cuenda, A.; Ranish, J.A.; Rudnicki, M.A. P38-[190]-dependent gene silencing restricts entry into the myogenic differentiation program. J. Cell Biol. 2009, 187, 991–1005. [Google Scholar] [CrossRef] [PubMed]
- Cuenda, A.; Cohen, P. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for c2c12 myogenesis. J. Biol. Chem. 1999, 274, 4341–4346. [Google Scholar] [CrossRef]
- Wu, Z.; Woodring, P.J.; Bhakta, K.S.; Tamura, K.; Wen, F.; Feramisco, J.R.; Karin, M.; Wang, J.Y.; Puri, P.L. P38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 2000, 20, 3951–3964. [Google Scholar] [CrossRef]
- Zetser, A.; Gredinger, E.; Bengal, E. P38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the mef2c transcription factor. J. Biol. Chem. 1999, 274, 5193–5200. [Google Scholar] [CrossRef]
- Palacios, D.; Mozzetta, C.; Consalvi, S.; Caretti, G.; Saccone, V.; Proserpio, V.; Marquez, V.E.; Valente, S.; Mai, A.; Forcales, S.V.; et al. Tnf/p38alpha/polycomb signaling to pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 2010, 7, 455–469. [Google Scholar] [CrossRef]
- Ornatsky, O.I.; Cox, D.M.; Tangirala, P.; Andreucci, J.J.; Quinn, Z.A.; Wrana, J.L.; Prywes, R.; Yu, Y.T.; McDermott, J.C. Post-translational control of the mef2a transcriptional regulatory protein. Nucleic Acids Res. 1999, 27, 2646–2654. [Google Scholar] [CrossRef]
- Zhao, M.; New, L.; Kravchenko, V.V.; Kato, Y.; Gram, H.; di Padova, F.; Olson, E.N.; Ulevitch, R.J.; Han, J. Regulation of the mef2 family of transcription factors by p38. Mol. Cell. Biol. 1999, 19, 21–30. [Google Scholar] [CrossRef]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef]
- de la Serna, I.L.; Ohkawa, Y.; Berkes, C.A.; Bergstrom, D.A.; Dacwag, C.S.; Tapscott, S.J.; Imbalzano, A.N. Myod targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005, 25, 3997–4009. [Google Scholar] [CrossRef] [PubMed]
- Forcales, S.V.; Albini, S.; Giordani, L.; Malecova, B.; Cignolo, L.; Chernov, A.; Coutinho, P.; Saccone, V.; Consalvi, S.; Williams, R.; et al. Signal-dependent incorporation of myod-baf60c into brg1-based swi/snf chromatin-remodelling complex. EMBO J. 2012, 31, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Toker, A. Akt/pkb signaling: Navigating the network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Gonzalez, I.; Tripathi, G.; Carter, E.J.; Cobb, L.J.; Salih, D.A.; Lovett, F.A.; Holding, C.; Pell, J.M. Akt2, a novel functional link between p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways in myogenesis. Mol. Cell. Biol. 2004, 24, 3607–3622. [Google Scholar] [CrossRef]
- Fujio, Y.; Mitsuuchi, Y.; Testa, J.R.; Walsh, K. Activation of akt2 inhibits anoikis and apoptosis induced by myogenic differentiation. Cell Death Differ. 2001, 8, 1207–1212. [Google Scholar] [CrossRef]
- Vandromme, M.; Rochat, A.; Meier, R.; Carnac, G.; Besser, D.; Hemmings, B.A.; Fernandez, A.; Lamb, N.J. Protein kinase b beta/akt2 plays a specific role in muscle differentiation. J. Biol. Chem. 2001, 276, 8173–8179. [Google Scholar] [CrossRef]
- Heron-Milhavet, L.; Franckhauser, C.; Rana, V.; Berthenet, C.; Fisher, D.; Hemmings, B.A.; Fernandez, A.; Lamb, N.J. Only akt1 is required for proliferation, while akt2 promotes cell cycle exit through p21 binding. Mol. Cell. Biol. 2006, 26, 8267–8280. [Google Scholar] [CrossRef]
- Heron-Milhavet, L.; Mamaeva, D.; Rochat, A.; Lamb, N.J.; Fernandez, A. Akt2 is implicated in skeletal muscle differentiation and specifically binds prohibitin2/rea. J. Cell. Physiol. 2008, 214, 158–165. [Google Scholar] [CrossRef]
- Sumitani, S.; Goya, K.; Testa, J.R.; Kouhara, H.; Kasayama, S. Akt1 and akt2 differently regulate muscle creatine kinase and myogenin gene transcription in insulin-induced differentiation of c2c12 myoblasts. Endocrinology 2002, 143, 820–828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, S.J.; Hwang, J.; Jeong, H.J.; Yoo, M.; Go, G.Y.; Lee, J.R.; Leem, Y.E.; Park, J.W.; Seo, D.W.; Kim, Y.K.; et al. Pkn2 and cdo interact to activate akt and promote myoblast differentiation. Cell Death Dis. 2016, 7, e2431. [Google Scholar] [CrossRef]
- Serra, C.; Palacios, D.; Mozzetta, C.; Forcales, S.V.; Morantte, I.; Ripani, M.; Jones, D.R.; Du, K.; Jhala, U.S.; Simone, C.; et al. Functional interdependence at the chromatin level between the mkk6/p38 and igf1/pi3k/akt pathways during muscle differentiation. Mol. Cell 2007, 28, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase ck2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, D.W. Protein kinase ck2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef]
- Nunez de Villavicencio-Diaz, T.; Rabalski, A.J.; Litchfield, D.W. Protein kinase ck2: Intricate relationships within regulatory cellular networks. Pharmaceuticals 2017, 10, 27. [Google Scholar] [CrossRef]
- Pepperkok, R.; Lorenz, P.; Jakobi, R.; Ansorge, W.; Pyerin, W. Cell growth stimulation by egf: Inhibition through antisense-oligodeoxynucleotides demonstrates important role of casein kinase ii. Exp. Cell Res. 1991, 197, 245–253. [Google Scholar] [CrossRef]
- Lorenz, P.; Pepperkok, R.; Ansorge, W.; Pyerin, W. Cell biological studies with monoclonal and polyclonal antibodies against human casein kinase ii subunit beta demonstrate participation of the kinase in mitogenic signaling. J. Biol. Chem. 1993, 268, 2733–2739. [Google Scholar]
- Pepperkok, R.; Lorenz, P.; Ansorge, W.; Pyerin, W. Casein kinase ii is required for transition of g0/g1, early g1, and g1/s phases of the cell cycle. J. Biol. Chem. 1994, 269, 6986–6991. [Google Scholar]
- Lorenz, P.; Pepperkok, R.; Pyerin, W. Requirement of casein kinase 2 for entry into and progression through early phases of the cell cycle. Cell. Mol. Biol. Res. 1994, 40, 519–527. [Google Scholar]
- Gotz, C.; Montenarh, M. Protein kinase ck2 in development and differentiation. Biomed. Rep. 2017, 6, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Winter, B.; Kautzner, I.; Issinger, O.G.; Arnold, H.H. Two putative protein kinase ck2 phosphorylation sites are important for myf-5 activity. Biol. Chem. 1997, 378, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.E.; Wang, X.; Hardy, S.; Taparowsky, E.J.; Konieczny, S.F. Casein kinase ii increases the transcriptional activities of mrf4 and myod independently of their direct phosphorylation. Mol. Cell. Biol. 1996, 16, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.N.; Miller, P.J.; Hollenbach, A.D. Phosphorylation of serine 205 by the protein kinase ck2 persists on pax3-foxo1, but not pax3, throughout early myogenic differentiation. Biochemistry 2009, 48, 11786–11795. [Google Scholar] [CrossRef]
- Dietz, K.N.; Miller, P.J.; Iyengar, A.S.; Loupe, J.M.; Hollenbach, A.D. Identification of serines 201 and 209 as sites of pax3 phosphorylation and the altered phosphorylation status of pax3-foxo1 during early myogenic differentiation. Int. J. Biochem. Cell Biol. 2011, 43, 936–945. [Google Scholar] [CrossRef]
- Iyengar, A.S.; Loupe, J.M.; Miller, P.J.; Hollenbach, A.D. Identification of ck2 as the kinase that phosphorylates pax3 at ser209 in early myogenic differentiation. Biochem. Biophys. Res. Commun. 2012, 428, 24–30. [Google Scholar] [CrossRef]
- Dick, S.A.; Chang, N.C.; Dumont, N.A.; Bell, R.A.; Putinski, C.; Kawabe, Y.; Litchfield, D.W.; Rudnicki, M.A.; Megeney, L.A. Caspase 3 cleavage of pax7 inhibits self-renewal of satellite cells. Proc. Natl. Acad. Sci. USA 2015, 112, E5246–E5252. [Google Scholar] [CrossRef]
- Gonzalez, N.; Moresco, J.J.; Cabezas, F.; de la Vega, E.; Bustos, F.; Yates, J.R., 3rd; Olguin, H.C. Ck2-dependent phosphorylation is required to maintain pax7 protein levels in proliferating muscle progenitors. PLoS ONE 2016, 11, e0154919. [Google Scholar] [CrossRef]
- Miller, P.J.; Dietz, K.N.; Hollenbach, A.D. Identification of serine 205 as a site of phosphorylation on pax3 in proliferating but not differentiating primary myoblasts. Protein Sci. 2008, 17, 1979–1986. [Google Scholar] [CrossRef]
- Salizzato, V.; Zanin, S.; Borgo, C.; Lidron, E.; Salvi, M.; Rizzuto, R.; Pallafacchina, G.; Donella-Deana, A. Protein kinase ck2 subunits exert specific and coordinated functions in skeletal muscle differentiation and fusogenic activity. FASEB J. 2019, fj201801833RR. [Google Scholar] [CrossRef]
- Kuang, S.; Charge, S.B.; Seale, P.; Huh, M.; Rudnicki, M.A. Distinct roles for pax7 and pax3 in adult regenerative myogenesis. J. Cell Biol. 2006, 172, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Chin, E.R.; Olson, E.N.; Richardson, J.A.; Yang, Q.; Humphries, C.; Shelton, J.M.; Wu, H.; Zhu, W.; Bassel-Duby, R.; Williams, R.S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998, 12, 2499–2509. [Google Scholar] [CrossRef]
- Narayanan, R.; Pirouz, M.; Kerimoglu, C.; Pham, L.; Wagener, R.J.; Kiszka, K.A.; Rosenbusch, J.; Seong, R.H.; Kessel, M.; Fischer, A.; et al. Loss of baf (mswi/snf) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 2015, 13, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Ronan, J.L.; Wu, J.; Staahl, B.T.; Chen, L.; Kuo, A.; Lessard, J.; Nesvizhskii, A.I.; Ranish, J.; Crabtree, G.R. An embryonic stem cell chromatin remodeling complex, esbaf, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 2009, 106, 5181–5186. [Google Scholar] [CrossRef] [PubMed]
- Gottesfeld, J.M.; Forbes, D.J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997, 22, 197–202. [Google Scholar] [CrossRef]
- Hartl, P.; Gottesfeld, J.; Forbes, D.J. Mitotic repression of transcription in vitro. J. Cell Biol. 1993, 120, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Gottesfeld, J.M.; Wolf, V.J.; Dang, T.; Forbes, D.J.; Hartl, P. Mitotic repression of rna polymerase iii transcription in vitro mediated by phosphorylation of a tfiiib component. Science 1994, 263, 81–84. [Google Scholar] [CrossRef]
- Segil, N.; Guermah, M.; Hoffmann, A.; Roeder, R.G.; Heintz, N. Mitotic regulation of tfiid: Inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996, 10, 2389–2400. [Google Scholar] [CrossRef]
- Martinez-Balbas, M.A.; Dey, A.; Rabindran, S.K.; Ozato, K.; Wu, C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 1995, 83, 29–38. [Google Scholar] [CrossRef]
- Stewart, A.A.; Ingebritsen, T.S.; Manalan, A.; Klee, C.B.; Cohen, P. Discovery of a Ca2+- and calmodulin-dependent protein phosphatase: Probable identity with calcineurin (cam-bp80). FEBS Lett. 1982, 137, 80–84. [Google Scholar] [CrossRef]
- Mitchell, P.O.; Pavlath, G.K. Multiple roles of calcineurin in skeletal muscle growth. Clin. Orthop. Relat. Res. 2002, S197–S202. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Role of calcineurin in striated muscle: Development, adaptation, and disease. Biochem. Biophys. Res. Commun. 2003, 311, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Rusnak, F.; Mertz, P. Calcineurin: Form and function. Physiol. Rev. 2000, 80, 1483–1521. [Google Scholar] [CrossRef] [PubMed]
- Manalan, A.S.; Krinks, M.H.; Klee, C.B. Calcineurin: A member of a family of calmodulin-stimulated protein phosphatases. Proc. Soc. Exp. Biol. Med. 1984, 177, 12–16. [Google Scholar] [CrossRef]
- Liu, J.; Farmer, J.D., Jr.; Lane, W.S.; Friedman, J.; Weissman, I.; Schreiber, S.L. Calcineurin is a common target of cyclophilin-cyclosporin a and fkbp-fk506 complexes. Cell 1991, 66, 807–815. [Google Scholar] [CrossRef]
- Shaw, K.T.; Ho, A.M.; Raghavan, A.; Kim, J.; Jain, J.; Park, J.; Sharma, S.; Rao, A.; Hogan, P.G. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor nfat1 in stimulated immune cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11205–11209. [Google Scholar] [CrossRef]
- Flanagan, W.M.; Corthesy, B.; Bram, R.J.; Crabtree, G.R. Nuclear association of a t-cell transcription factor blocked by fk-506 and cyclosporin a. Nature 1991, 352, 803–807. [Google Scholar] [CrossRef]
- Ruff, V.A.; Leach, K.L. Direct demonstration of nfatp dephosphorylation and nuclear localization in activated ht-2 cells using a specific nfatp polyclonal antibody. J. Biol. Chem. 1995, 270, 22602–22607. [Google Scholar] [CrossRef]
- Ravel-Chapuis, A.; Belanger, G.; Cote, J.; Michel, R.N.; Jasmin, B.J. Misregulation of calcium-handling proteins promotes hyperactivation of calcineurin-nfat signaling in skeletal muscle of dm1 mice. Hum. Mol. Genet. 2017, 26, 2192–2206. [Google Scholar] [CrossRef]
- Schiaffino, S.; Sandri, M.; Murgia, M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiol. Bethesda 2007, 22, 269–278. [Google Scholar] [CrossRef]
- Horsley, V.; Friday, B.B.; Matteson, S.; Kegley, K.M.; Gephart, J.; Pavlath, G.K. Regulation of the growth of multinucleated muscle cells by an nfatc2-dependent pathway. J. Cell Biol. 2001, 153, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Dunn, S.E.; Burns, J.L.; Michel, R.N. Calcineurin is required for skeletal muscle hypertrophy. J. Biol. Chem. 1999, 274, 21908–21912. [Google Scholar] [CrossRef] [PubMed]
- Musaro, A.; McCullagh, K.J.; Naya, F.J.; Olson, E.N.; Rosenthal, N. Igf-1 induces skeletal myocyte hypertrophy through calcineurin in association with gata-2 and nf-atc1. Nature 1999, 400, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Semsarian, C.; Wu, M.J.; Ju, Y.K.; Marciniec, T.; Yeoh, T.; Allen, D.G.; Harvey, R.P.; Graham, R.M. Skeletal muscle hypertrophy is mediated by a ca2+-dependent calcineurin signalling pathway. Nature 1999, 400, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Friday, B.B.; Horsley, V.; Pavlath, G.K. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J. Cell Biol. 2000, 149, 657–666. [Google Scholar] [CrossRef]
- Friday, B.B.; Mitchell, P.O.; Kegley, K.M.; Pavlath, G.K. Calcineurin initiates skeletal muscle differentiation by activating mef2 and myod. Differ. Res. Biol. Divers. 2003, 71, 217–227. [Google Scholar] [CrossRef]
- Lai, D.; Wan, M.; Wu, J.; Preston-Hurlburt, P.; Kushwaha, R.; Grundstrom, T.; Imbalzano, A.N.; Chi, T. Induction of tlr4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc. Natl. Acad. Sci. USA 2009, 106, 1169–1174. [Google Scholar] [CrossRef]
- Witwicka, H.; Nogami, J.; Syed, S.A.; Maehara, K.; Padilla-Benavides, T.; Ohkawa, Y.; Imbalzano, A.N. Calcineurin broadly regulates the initiation of skeletal muscle-specific gene expression by binding target promoters and facilitating the interaction of the swi/snf chromatin remodeling enzyme. Mol. Cell. Biol. 2019, 39, e00063-19. [Google Scholar] [CrossRef]
- Kwon, S.J.; Park, J.H.; Park, E.J.; Lee, S.A.; Lee, H.S.; Kang, S.W.; Kwon, J. Atm-mediated phosphorylation of the chromatin remodeling enzyme brg1 modulates DNA double-strand break repair. Oncogene 2015, 34, 303–313. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilla-Benavides, T.; Reyes-Gutierrez, P.; Imbalzano, A.N. Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis. Biology 2020, 9, 152. https://doi.org/10.3390/biology9070152
Padilla-Benavides T, Reyes-Gutierrez P, Imbalzano AN. Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis. Biology. 2020; 9(7):152. https://doi.org/10.3390/biology9070152
Chicago/Turabian StylePadilla-Benavides, Teresita, Pablo Reyes-Gutierrez, and Anthony N. Imbalzano. 2020. "Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis" Biology 9, no. 7: 152. https://doi.org/10.3390/biology9070152
APA StylePadilla-Benavides, T., Reyes-Gutierrez, P., & Imbalzano, A. N. (2020). Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis. Biology, 9(7), 152. https://doi.org/10.3390/biology9070152




