Structural Insights into the Evolutionarily Conserved BAF Chromatin Remodeling Complex
Abstract
1. Introduction
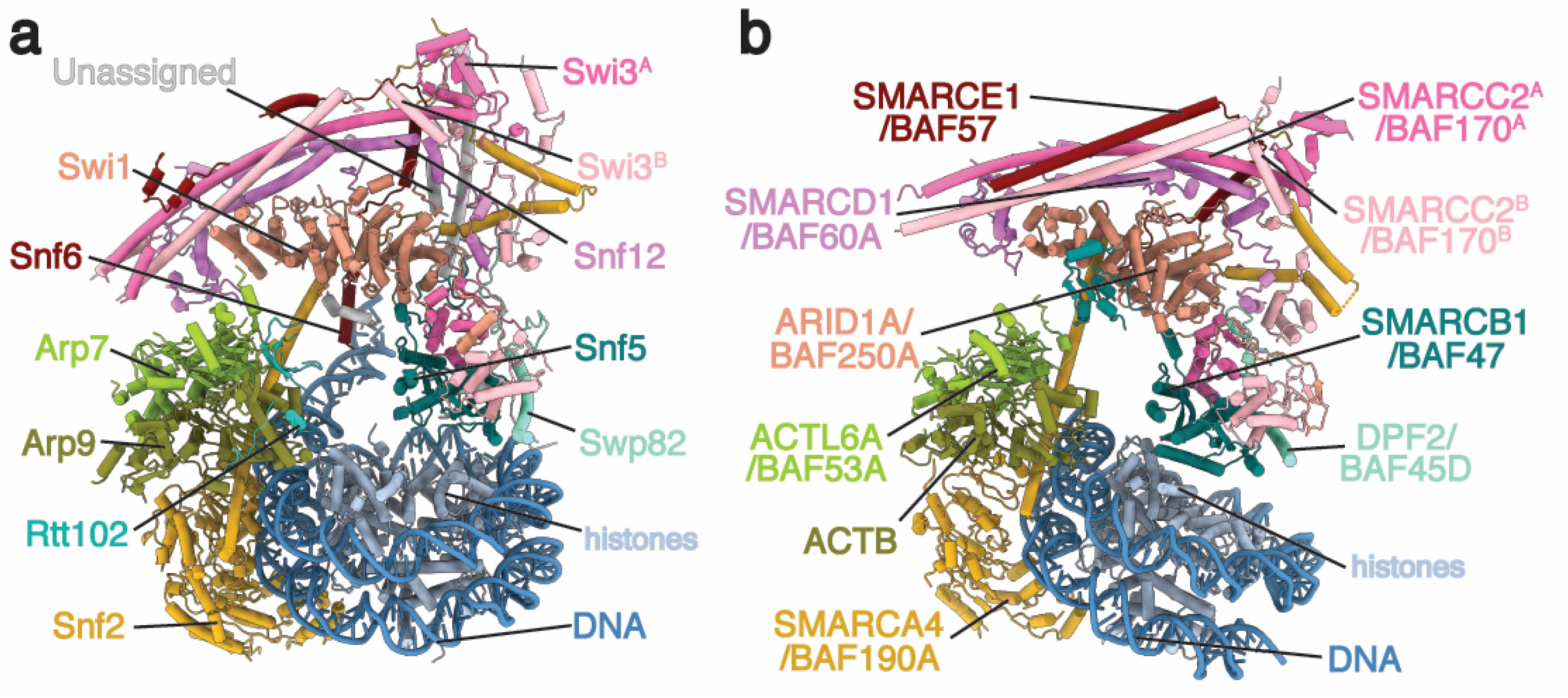
2. Architecture of the BAF Family Remodeler
2.1. ATPase Module
2.2. Arp Module
2.3. Body Module
3. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, C.Y.; Johnson, S.L.; Gamarra, N.I.; Narlikar, G.J. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 2016, 45, 153–181. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Shain, A.H.; Pollack, J.R. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 2013, 8, e55119. [Google Scholar] [CrossRef]
- Arnaud, O.; Le Loarer, F.; Tirode, F. BAFfling pathologies: Alterations of BAF complexes in cancer. Cancer Lett. 2018, 419, 266–279. [Google Scholar] [CrossRef]
- St Pierre, R.; Kadoch, C. Mammalian SWI/SNF complexes in cancer: Emerging therapeutic opportunities. Curr. Opin. Genet. Dev. 2017, 42, 56–67. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- Han, Y.; Reyes, A.A.; Malik, S.; He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 2020, 579, 452–455. [Google Scholar] [CrossRef]
- He, S.; Wu, Z.; Tian, Y.; Yu, Z.; Yu, J.; Wang, X.; Li, J.; Liu, B.; Xu, Y. Structure of nucleosome-bound human BAF complex. Science 2020, 367, 875–881. [Google Scholar] [CrossRef]
- Ye, Y.; Wu, H.; Chen, K.; Clapier, C.R.; Verma, N.; Zhang, W.; Deng, H.; Cairns, B.R.; Gao, N.; Chen, Z. Structure of the RSC complex bound to the nucleosome. Science 2019, 366, 838–843. [Google Scholar] [CrossRef]
- Patel, A.B.; Moore, C.M.; Greber, B.J.; Luo, J.; Zukin, S.A.; Ranish, J.; Nogales, E. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. Elife 2019, 8, e54449. [Google Scholar] [CrossRef]
- Zofall, M.; Persinger, J.; Kassabov, S.R.; Bartholomew, B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006, 13, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xia, X.; Tian, Y.; Jia, Q.; Liu, X.; Lu, Y.; Li, M.; Li, X.; Chen, Z. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 2019, 567, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, M.; Xia, X.; Li, X.; Chen, Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 2017, 544, 440–445. [Google Scholar] [CrossRef]
- Hassan, A.H.; Awad, S.; Prochasson, P. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J. Biol. Chem. 2006, 281, 18126–18134. [Google Scholar] [CrossRef]
- Hassan, A.H.; Prochasson, P.; Neely, K.E.; Galasinski, S.C.; Chandy, M.; Carrozza, M.J.; Workman, J.L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 2002, 111, 369–379. [Google Scholar] [CrossRef]
- Shen, W.; Xu, C.; Huang, W.; Zhang, J.; Carlson, J.E.; Tu, X.; Wu, J.; Shi, Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 2007, 46, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Vivas, P.; Dechassa, M.L.; Mooney, A.M.; Poirier, M.G.; Bartholomew, B. The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol. Cell. Biol. 2013, 33, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.A.; Sanchez, J.C.; Ronan, J.L.; Farrell, D.P.; Varzavand, K.; Johnson, J.K.; Gu, B.X.; Crabtree, G.R.; Musselman, C.A. DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat. Commun. 2017, 8, 16080. [Google Scholar] [CrossRef]
- Phelan, M.L.; Sif, S.; Narlikar, G.J.; Kingston, R.E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 1999, 3, 247–253. [Google Scholar] [CrossRef]
- Wang, W.; Cote, J.; Xue, Y.; Zhou, S.; Khavari, P.A.; Biggar, S.R.; Muchardt, C.; Kalpana, G.V.; Goff, S.P.; Yaniv, M.; et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996, 15, 5370–5382. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; McKenzie, Z.M.; D’Avino, A.R.; Mashtalir, N.; Lareau, C.A.; St Pierre, R.; Wang, L.; Shilatifard, A.; Kadoch, C. The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat. Genet. 2019, 51, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.Z.; Hodges, C.; Calarco, J.P.; Braun, S.M.; Ku, W.L.; Kadoch, C.; Zhao, K.; Crabtree, G.R. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 2017, 49, 282–288. [Google Scholar] [CrossRef]
- Strobeck, M.W.; Knudsen, K.E.; Fribourg, A.F.; DeCristofaro, M.F.; Weissman, B.E.; Imbalzano, A.N.; Knudsen, E.S. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 2000, 97, 7748–7753. [Google Scholar] [CrossRef]
- Dunaief, J.L.; Strober, B.E.; Guha, S.; Khavari, P.A.; Alin, K.; Luban, J.; Begemann, M.; Crabtree, G.R.; Goff, S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994, 79, 119–130. [Google Scholar] [CrossRef]
- Reisman, D.N.; Strobeck, M.W.; Betz, B.L.; Sciariotta, J.; Funkhouser, W.; Jr Murchardt, C.; Yaniv, M.; Sherman, L.S.; Knudsen, E.S.; Weissman, B.E. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: Differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene 2002, 21, 1196–1207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bultman, S.; Gebuhr, T.; Yee, D.; La Mantia, C.; Nicholson, J.; Gilliam, A.; Randazzo, F.; Metzger, D.; Chambon, P.; Crabtree, G.; et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 2000, 6, 1287–1295. [Google Scholar] [CrossRef]
- Bultman, S.J.; Herschkowitz, J.I.; Godfrey, V.; Gebuhr, T.C.; Yaniv, M.; Perou, C.M.; Magnuson, T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 2008, 27, 460–468. [Google Scholar] [CrossRef]
- Karnezis, A.N.; Wang, Y.; Ramos, P.; Hendricks, W.P.; Oliva, E.; D’Angelo, E.; Prat, J.; Nucci, M.R.; Nielsen, T.O.; Chow, C.; et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J. Pathol. 2016, 238, 389–400. [Google Scholar] [CrossRef]
- Witkowski, L.; Carrot-Zhang, J.; Albrecht, S.; Fahiminiya, S.; Hamel, N.; Tomiak, E.; Grynspan, D.; Saloustros, E.; Nadaf, J.; Rivera, B.; et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 2014, 46, 438–443. [Google Scholar] [CrossRef]
- Jelinic, P.; Mueller, J.J.; Olvera, N.; Dao, F.; Scott, S.N.; Shah, R.; Gao, J.; Schultz, N.; Gonen, M.; Soslow, R.A.; et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 2014, 46, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Le Loarer, F.; Watson, S.; Pierron, G.; de Montpreville, V.T.; Ballet, S.; Firmin, N.; Auguste, A.; Pissaloux, D.; Boyault, S.; Paindavoine, S.; et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat. Genet. 2015, 47, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Carretero, J.; Fraga, M.F.; Esteller, M.; Sidransky, D.; Sanchez-Cespedes, M. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRG1, in lung tumors. Genes Chromosomes Cancer 2004, 41, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Tawfik, O.; Gayed, B.; Thrasher, J.B.; Hoestje, S.; Li, C.; Li, B. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate 2007, 67, 203–213. [Google Scholar] [CrossRef]
- Hodges, H.C.; Stanton, B.Z.; Cermakova, K.; Chang, C.Y.; Miller, E.L.; Kirkland, J.G.; Ku, W.L.; Veverka, V.; Zhao, K.; Crabtree, G.R. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 2018, 25, 61–72. [Google Scholar] [CrossRef]
- Bartlett, C.; Orvis, T.J.; Rosson, G.S.; Weissman, B.E. BRG1 mutations found in human cancer cell lines inactivate Rb-mediated cell-cycle arrest. J. Cell. Physiol. 2011, 226, 1989–1997. [Google Scholar] [CrossRef]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288. [Google Scholar] [CrossRef]
- Trotter, K.W.; Fan, H.Y.; Ivey, M.L.; Kingston, R.E.; Archer, T.K. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell. Biol. 2008, 28, 1413–1426. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, W.; Rando, O.J.; Xue, Y.; Swiderek, K.; Kuo, A.; Crabtree, G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 1998, 95, 625–636. [Google Scholar] [CrossRef]
- Shen, X.; Ranallo, R.; Choi, E.; Wu, C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 2003, 12, 147–155. [Google Scholar] [CrossRef]
- Son, E.Y.; Crabtree, G.R. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 333–349. [Google Scholar] [CrossRef]
- Goutham, A. In silico screening of cancer-associated mutations in the HSA domain of BRG1 and its role in affecting the Arp-HSA sub-complex of SWI/SNF. Comput. Biol. Chem. 2018, 77, 109–115. [Google Scholar]
- Jones, S.; Li, M.; Parsons, D.W.; Zhang, X.; Wesseling, J.; Kristel, P.; Schmidt, M.K.; Markowitz, S.; Yan, H.; Bigner, D.; et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum. Mutat. 2012, 33, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Cajuso, T.; Hanninen, U.A.; Kondelin, J.; Gylfe, A.E.; Tanskanen, T.; Katainen, R.; Pitkanen, E.; Ristolainen, H.; Kaasinen, E.; Taipale, M.; et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int. J. Cancer 2014, 135, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.N.; Roberts, C.W. ARID1A mutations in cancer: Another epigenetic tumor suppressor? Cancer Discov. 2013, 3, 35–43. [Google Scholar] [CrossRef]
- Guan, B.; Gao, M.; Wu, C.H.; Wang, T.L.; Shih Ie, M. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia 2012, 14, 986–993. [Google Scholar] [CrossRef]
- Zang, Z.J.; Cutcutache, I.; Poon, S.L.; Zhang, S.L.; McPherson, J.R.; Tao, J.; Rajasegaran, V.; Heng, H.L.; Deng, N.; Gan, A.; et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 2012, 44, 570–574. [Google Scholar] [CrossRef]
- Mamo, A.; Cavallone, L.; Tuzmen, S.; Chabot, C.; Ferrario, C.; Hassan, S.; Edgren, H.; Kallioniemi, O.; Aleynikova, O.; Przybytkowski, E.; et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene 2012, 31, 2090–2100. [Google Scholar] [CrossRef]
- Guan, B.; Wang, T.L.; Shih Ie, M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011, 71, 6718–6727. [Google Scholar] [CrossRef]
- Wilsker, D.; Patsialou, A.; Zumbrun, S.D.; Kim, S.; Chen, Y.; Dallas, P.B.; Moran, E. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 2004, 32, 1345–1353. [Google Scholar] [CrossRef]
- Nagl, N.G.; Wang, X., Jr.; Patsialou, A.; Van Scoy, M.; Moran, E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007, 26, 752–763. [Google Scholar] [CrossRef]
- Valencia, A.M.; Collings, C.K.; Dao, H.T.; St Pierre, R.; Cheng, Y.C.; Huang, J.; Sun, Z.Y.; Seo, H.S.; Mashtalir, N.; Comstock, D.E.; et al. Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling. Cell 2019, 179, 1342–1356. [Google Scholar] [CrossRef] [PubMed]
- Versteege, I.; Sevenet, N.; Lange, J.; Rousseau-Merck, M.F.; Ambros, P.; Handgretinger, R.; Aurias, A.; Delattre, O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998, 394, 203–206. [Google Scholar] [CrossRef]
- Roberts, C.W.; Galusha, S.A.; McMenamin, M.E.; Fletcher, C.D.; Orkin, S.H. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 2000, 97, 13796–13800. [Google Scholar] [CrossRef] [PubMed]
- Le Loarer, F.; Zhang, L.; Fletcher, C.D.; Ribeiro, A.; Singer, S.; Italiano, A.; Neuville, A.; Houlier, A.; Chibon, F.; Coindre, J.M.; et al. Consistent SMARCB1 homozygous deletions in epithelioid sarcoma and in a subset of myoepithelial carcinomas can be reliably detected by FISH in archival material. Genes Chromosomes Cancer 2014, 53, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.M.; Folpe, A.L.; Pawel, B.R.; Judkins, A.R.; Biegel, J.A. Epithelioid sarcoma is associated with a high percentage of SMARCB1 deletions. Mod. Pathol. 2013, 26, 385–392. [Google Scholar] [CrossRef]
- Calderaro, J.; Moroch, J.; Pierron, G.; Pedeutour, F.; Grison, C.; Maille, P.; Soyeux, P.; de la Taille, A.; Couturier, J.; Vieillefond, A.; et al. SMARCB1/INI1 inactivation in renal medullary carcinoma. Histopathology 2012, 61, 428–435. [Google Scholar] [CrossRef]
- Liu, Q.; Galli, S.; Srinivasan, R.; Linehan, W.M.; Tsokos, M.; Merino, M.J. Renal medullary carcinoma: Molecular, immunohistochemistry, and morphologic correlation. Am. J. Surg. Pathol. 2013, 37, 368–374. [Google Scholar] [CrossRef]
- Kadoch, C.; Crabtree, G.R. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013, 153, 71–85. [Google Scholar] [CrossRef]
- Hulsebos, T.J.; Plomp, A.S.; Wolterman, R.A.; Robanus-Maandag, E.C.; Baas, F.; Wesseling, P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am. J. Hum. Genet. 2007, 80, 805–810. [Google Scholar] [CrossRef]
- Kohashi, K.; Oda, Y.; Yamamoto, H.; Tamiya, S.; Oshiro, Y.; Izumi, T.; Taguchi, T.; Tsuneyoshi, M. SMARCB1/INI1 protein expression in round cell soft tissue sarcomas associated with chromosomal translocations involving EWS: A special reference to SMARCB1/INI1 negative variant extraskeletal myxoid chondrosarcoma. Am. J. Surg. Pathol. 2008, 32, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh-Lotrario, A.D.; Tomlinson, G.E.; Finegold, M.J.; Gore, L.; Feusner, J.H. Small cell undifferentiated variant of hepatoblastoma: Adverse clinical and molecular features similar to rhabdoid tumors. Pediatric Blood Cancer 2009, 52, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Middeljans, E.; Wan, X.; Jansen, P.W.; Sharma, V.; Stunnenberg, H.G.; Logie, C. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS ONE 2012, 7, e33834. [Google Scholar] [CrossRef]
- Lee, R.S.; Stewart, C.; Carter, S.L.; Ambrogio, L.; Cibulskis, K.; Sougnez, C.; Lawrence, M.S.; Auclair, D.; Mora, J.; Golub, T.R.; et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J. Clin. Investig. 2012, 122, 2983–2988. [Google Scholar] [CrossRef]
- Crew, A.J.; Clark, J.; Fisher, C.; Gill, S.; Grimer, R.; Chand, A.; Shipley, J.; Gusterson, B.A.; Cooper, C.S. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995, 14, 2333–2340. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Rocques, P.J.; Crew, A.J.; Gill, S.; Shipley, J.; Chan, A.M.; Gusterson, B.A.; Cooper, C.S. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 1994, 7, 502–508. [Google Scholar] [CrossRef]
- Hiraga, H.; Nojima, T.; Abe, S.; Sawa, H.; Yamashiro, K.; Yamawaki, S.; Kaneda, K.; Nagashima, K. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: Analyses of 84 soft tissue and bone tumors. Diagn. Mol. Pathol. 1998, 7, 102–110. [Google Scholar] [CrossRef]
- McBride, M.J.; Pulice, J.L.; Beird, H.C.; Ingram, D.R.; D’Avino, A.R.; Shern, J.F.; Charville, G.W.; Hornick, J.L.; Nakayama, R.T.; Garcia-Rivera, E.M.; et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018, 33, 1128–1141 e1127. [Google Scholar] [CrossRef]
- Kia, S.K.; Gorski, M.M.; Giannakopoulos, S.; Verrijzer, C.P. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 2008, 28, 3457–3464. [Google Scholar] [CrossRef]
- Wilson, B.G.; Wang, X.; Shen, X.; McKenna, E.S.; Lemieux, M.E.; Cho, Y.J.; Koellhoffer, E.C.; Pomeroy, S.L.; Orkin, S.H.; Roberts, C.W. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010, 18, 316–328. [Google Scholar] [CrossRef]
- Nakayama, R.T.; Pulice, J.L.; Valencia, A.M.; McBride, M.J.; McKenzie, Z.M.; Gillespie, M.A.; Ku, W.L.; Teng, M.; Cui, K.; Williams, R.T.; et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet. 2017, 49, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcum, R.D.; Reyes, A.A.; He, Y. Structural Insights into the Evolutionarily Conserved BAF Chromatin Remodeling Complex. Biology 2020, 9, 146. https://doi.org/10.3390/biology9070146
Marcum RD, Reyes AA, He Y. Structural Insights into the Evolutionarily Conserved BAF Chromatin Remodeling Complex. Biology. 2020; 9(7):146. https://doi.org/10.3390/biology9070146
Chicago/Turabian StyleMarcum, Ryan D., Alexis A. Reyes, and Yuan He. 2020. "Structural Insights into the Evolutionarily Conserved BAF Chromatin Remodeling Complex" Biology 9, no. 7: 146. https://doi.org/10.3390/biology9070146
APA StyleMarcum, R. D., Reyes, A. A., & He, Y. (2020). Structural Insights into the Evolutionarily Conserved BAF Chromatin Remodeling Complex. Biology, 9(7), 146. https://doi.org/10.3390/biology9070146





