Transcriptome Assembly and Comparative Analysis of the Superoxide Dismutase (SOD) Gene Family in Three Hyotissa Species
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
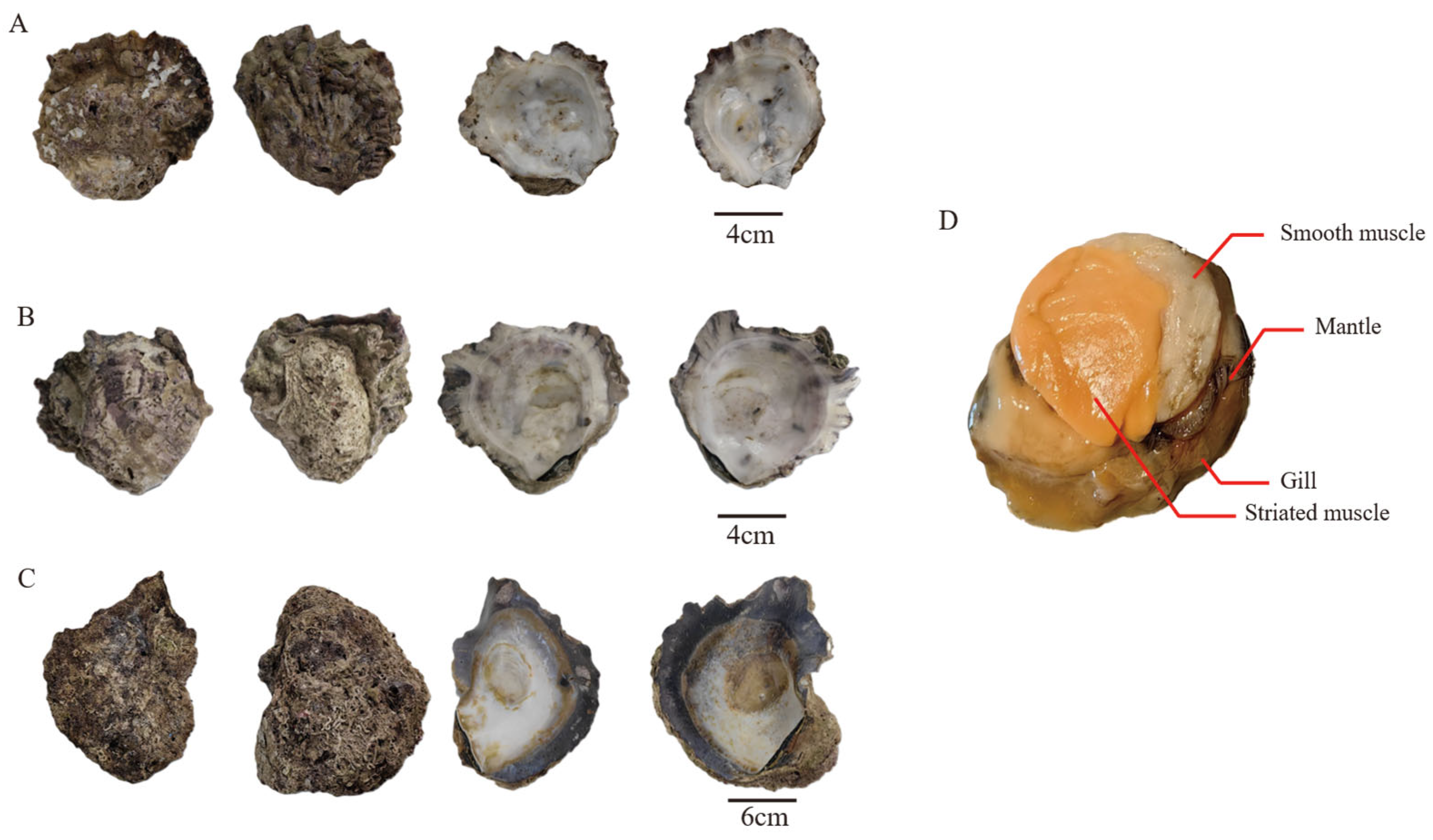
2.1. Sample Collection
2.2. Total RNA Extraction and Quality Control
2.3. Library Construction and Sequencing
2.4. RNA-Seq Data Processing and Quality Control
2.5. Transcriptome Assembly
2.6. Transcriptome Assembly and SOD Gene Identification
2.7. Molecular Characterization of SOD Family Members
2.8. Sequence Alignment and Phylogenetic Analysis
3. Results
3.1. Technical Validation of Transcriptome Sequencing
3.2. Identification of the SOD Gene Family
3.3. Phylogenetic Analysis of Hyotissa SOD
3.4. Sequence Analysis of the ecSOD Homolog Dominin in Hyotissa
3.5. Sequence and Domain Comparison of Hyotissa CSRPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouchet, P.; Rocroi, J.-P.; Bieler, R.; Carter, J.G.; Coan, E.V. Nomenclator of bivalve families with a classification of bivalve families. Malacologia 2010, 52, 1–184. [Google Scholar] [CrossRef]
- Guo, X.; Li, C.; Wang, H.; Xu, Z. Diversity and evolution of living oysters. J. Shellfish. Res. 2018, 37, 755–771. [Google Scholar] [CrossRef]
- Li, F.; Liu, H.; Heng, X.; Zhang, Y.; Fan, M.; Wang, S.; Liu, C.; Gu, Z.; Wang, A.; Yang, Y. The complete mitochondrial genome of Hyotissa sinensis (Bivalvia, Ostreoidea) indicates the genetic diversity within Gryphaeidae. Biodivers. Data J. 2023, 11, e101333. [Google Scholar] [CrossRef]
- Coan, E.V.; Valentich-Scott, P. Bivalve Seashells of Tropical West America: Marine Bivalve Mollusks from Baja California to Northern Peru; Santa Barbara Museum of Natural History: Santa Barbara, CA, USA, 2012; Volume 6. [Google Scholar]
- Harry, H.W. Synopsis of the supraspecific classification of living oysters (Bivalvia: Gryphaeidae and Ostreidae). Veliger 1985, 28, 121–158. [Google Scholar]
- Checa, A.G.; Linares, F.; Maldonado-Valderrama, J.; Harper, E.M. Foamy oysters: Vesicular microstructure production in the Gryphaeidae via emulsification. J. R. Soc. Interface 2020, 17, 20200505. [Google Scholar] [CrossRef]
- Sancho Vaquer, A.; Griesshaber, E.; Salas, C.; Harper, E.M.; Checa, A.G.; Schmahl, W.W. The diversity of crystals, microstructures and texture that form Ostreoidea shells. Crystals 2025, 15, 286. [Google Scholar] [CrossRef]
- Zuschin, M.; Baal, C. Large gryphaeid oysters as habitats for numerous sclerobionts: A case study from the northern Red Sea. Facies 2007, 53, 319–327. [Google Scholar] [CrossRef]
- Duprat-Bertazzi, G.; Garcia-Dominguez, F. Reproductive cycle of the rock oyster Hyotissa hyotis (Linné, 1758) (Gryphaeidae) at the la Ballena Island, Gulf of California, México. J. Shellfish. Res. 2005, 24, 987–993. [Google Scholar]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, J.M.; Hekimi, S. Superoxide dismutase is dispensable for normal animal lifespan. Proc. Natl. Acad. Sci. USA 2012, 109, 5785–5790. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Perry, J.; Shin, D.; Getzoff, E.; Tainer, J. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wei, M.; Zhu, Q.; Zheng, X.; Chen, X. Whole-genome identification of SOD gene family revealed the expansion of SOD in Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 56, 101618. [Google Scholar] [CrossRef]
- Lian, S.; Zhao, L.; Xun, X.; Lou, J.; Li, M.; Li, X.; Wang, S.; Zhang, L.; Hu, X.; Bao, Z. Genome-wide identification and characterization of SOD s in Zhikong scallop reveals gene expansion and regulation divergence after toxic dinoflagellate exposure. Mar. Drugs 2019, 17, 700. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bao, Z.; Lin, Z.; Xue, Q. Genome-wide identification and characterization of superoxide dismutases in four oyster species reveals functional differentiation in response to biotic and abiotic stress. BMC Genom. 2022, 23, 378. [Google Scholar] [CrossRef]
- Itoh, N.; Xue, Q.-G.; Schey, K.L.; Li, Y.; Cooper, R.K.; La Peyre, J.F. Characterization of the major plasma protein of the eastern oyster, Crassostrea virginica, and a proposed role in host defense. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 158, 9–22. [Google Scholar] [CrossRef]
- Scotti, P.D.; Dearing, S.C.; Greenwood, D.R. Characterisation of cavortin, the major haemolymph protein of the Pacific oyster (Crassostrea gigas). N. Z. J. Mar. Freshw. Res. 2007, 41, 91–101. [Google Scholar] [CrossRef]
- Ruan, Z.; Liu, Y.; Chang, G.; Lin, Z.; Xue, Q. Molecular characterization of two CuZn-SOD family proteins in the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2022, 260, 110736. [Google Scholar] [CrossRef]
- Robinett, N.G.; Peterson, R.L.; Culotta, V.C. Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains. J. Biol. Chem. 2018, 293, 4636–4643. [Google Scholar] [CrossRef]
- Tan, L.; Liu, Y.; Sun, Y.; Liu, S.; Lin, Z.; Xue, Q. Copper only SOD repeat proteins likely act as an extracellular superoxide dismutase in oyster antioxidant defense. Sci. Rep. 2025, 15, 20465. [Google Scholar] [CrossRef]
- Titschack, J.; Zuschin, M.; Spötl, C.; Baal, C. The giant oyster Hyotissa hyotis from the northern Red Sea as a decadal-scale archive for seasonal environmental fluctuations in coral reef habitats. Coral Reefs 2010, 29, 1061–1075. [Google Scholar] [CrossRef]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Caparros, P.; De Filippis, L.; Gul, A.; Hasanuzzaman, M.; Ozturk, M.; Altay, V.; Lao, M.T. Oxidative stress and antioxidant metabolism under adverse environmental conditions: A review. Bot. Rev. 2021, 87, 421–466. [Google Scholar] [CrossRef]
- Sundaray, J.K.; Dixit, S.; Rather, A.; Rasal, K.D.; Sahoo, L. Aquaculture omics: An update on the current status of research and data analysis. Mar. Genom. 2022, 64, 100967. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, Y.; Tan, Y.; Li, Q.; Liu, S. Chromosome-level haplotype-resolved genome assembly of the giant honeycomb oyster, Hyotissa hyotis. Sci. Data 2025, 12, 1327. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Chen, M.; Wang, H.; Ma, P. Diversity and distribution of oysters from Weizhou Island, China. J. Oceanol. Limnol. 2025, 1–22. [Google Scholar] [CrossRef]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Peng, J.; Li, Q.; Xu, L.; Wei, P.; He, P.; Zhang, X.; Zhang, L.; Guan, J.; Zhang, X.; Lin, Y.; et al. Chromosome-level analysis of the Crassostrea hongkongensis genome reveals extensive duplication of immune-related genes in bivalves. Mol. Ecol. Resour. 2020, 20, 980–994. [Google Scholar] [CrossRef]
- Song, H.; Xie, C.; Dong, M.; Zhang, Y.; Huang, H.; Han, Y.; Liu, Y.; Wei, L.; Wang, X. Effects of ambient UVB light on Pacific oyster Crassostrea gigas mantle tissue based on multivariate data. Ecotoxicol. Environ. Saf. 2024, 274, 116236. [Google Scholar] [CrossRef] [PubMed]
- Valavanidis, A.; Vlahogianni, T.; Dassenakis, M.; Scoullos, M. Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf. 2006, 64, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Björklund, Å.K.; Ekman, D.; Elofsson, A. Expansion of protein domain repeats. PLoS Comput. Biol. 2006, 2, e114. [Google Scholar] [CrossRef] [PubMed]






| Sample | Raw_Reads | Raw_Bases | Clean_Reads | Clean_Bases | Error_Rate | Q20 | Q30 | GC_pct |
|---|---|---|---|---|---|---|---|---|
| Hin1-S | 22,938,901 | 6.88 | 22,567,525 | 6.77 | 0.01 | 98.10 | 94.44 | 39.17 |
| Hin1-W | 22,733,450 | 6.82 | 22,161,418 | 6.65 | 0.01 | 98.38 | 95.17 | 39.40 |
| Hin1-B | 22,769,642 | 6.83 | 22,389,164 | 6.72 | 0.01 | 98.37 | 95.06 | 40.64 |
| Hin1-C | 22,880,343 | 6.86 | 22,430,060 | 6.73 | 0.01 | 98.07 | 94.18 | 40.94 |
| Hsp1-S | 23,648,413 | 7.09 | 23,224,331 | 6.97 | 0.01 | 98.26 | 94.84 | 38.52 |
| Hsp1-W | 23,424,910 | 7.03 | 23,131,650 | 6.94 | 0.01 | 98.59 | 95.78 | 38.87 |
| Hsp1-B | 21,968,086 | 6.59 | 21,616,279 | 6.48 | 0.01 | 98.09 | 94.23 | 40.26 |
| Hsp1-C | 21,172,853 | 6.35 | 20,776,969 | 6.23 | 0.01 | 98.50 | 95.76 | 41.88 |
| Hin2-S | 23,079,592 | 6.92 | 22,625,841 | 6.79 | 0.01 | 98.17 | 94.59 | 38.83 |
| Hin2-W | 23,123,036 | 6.94 | 22,787,435 | 6.84 | 0.01 | 98.12 | 94.45 | 39.62 |
| Hin2-B | 23,654,582 | 7.10 | 23,320,078 | 7.00 | 0.01 | 98.28 | 94.74 | 40.92 |
| Hin2-C | 23,907,955 | 7.17 | 23,509,157 | 7.05 | 0.01 | 98.56 | 95.91 | 42.32 |
| Hsi1-S | 24,393,919 | 7.32 | 23,774,686 | 7.13 | 0.01 | 98.08 | 94.35 | 39.35 |
| Hsi1-W | 22,652,736 | 6.80 | 22,170,153 | 6.65 | 0.01 | 98.17 | 94.62 | 40.53 |
| Hsi1-B | 25,280,014 | 7.58 | 25,021,203 | 7.51 | 0.01 | 98.25 | 94.62 | 41.03 |
| Hsi1-C | 23,009,172 | 6.90 | 22,629,224 | 6.79 | 0.01 | 98.42 | 95.05 | 42.55 |
| Hsi2-S | 23,669,501 | 7.10 | 23,156,523 | 6.95 | 0.01 | 98.48 | 95.88 | 38.43 |
| Hsi2-W | 23,362,518 | 7.01 | 22,844,009 | 6.85 | 0.01 | 98.13 | 94.50 | 39.11 |
| Hsi2-B | 23,528,698 | 7.06 | 22,924,594 | 6.88 | 0.01 | 98.41 | 95.06 | 42.39 |
| Hsi2-C | 23,625,965 | 7.09 | 23,049,423 | 6.91 | 0.01 | 98.13 | 94.23 | 44.36 |
| Sequence ID | Amino Acid | MW (kDa) | pI | Instability Index | Aliphatic Index | GRAVY | Cellular Location | Signal Peptide | Phosphorylation Sites |
|---|---|---|---|---|---|---|---|---|---|
| Hin1-DN145157c0g1i1.p1 | 161 | 16.57 | 5.94 | 25.20 | 77.45 | −0.278 | cyto | Other | 11 |
| Hin1-DN1483c1g1i7.p1 | 1035 | 111.91 | 6.41 | 35.72 | 80.09 | −0.174 | plas | Other | 130 |
| Hin1-DN12320c0g1i1.p1 | 351 | 37.04 | 6.70 | 32.90 | 63.56 | −0.452 | E.R. | SP (Sec/SPI) | 21 |
| Hin1-DN2804c1g1i4.p2 | 254 | 27.54 | 5.50 | 54.06 | 83.15 | −0.393 | extr | Other | 21 |
| Hin1-DN21488c0g1i1.p1 | 174 | 17.51 | 7.78 | 40.19 | 50.00 | −0.563 | cytonucl | Other | 17 |
| Hin1-DN636c0g1i2.p1 | 225 | 25.31 | 6.44 | 33.25 | 88.00 | −0.288 | mito | Other | 21 |
| Hsp1-DN3819c0g1i1.p1 | 161 | 16.62 | 5.94 | 25.13 | 75.03 | −0.342 | cyto | Other | 11 |
| Hsp1-DN109130c0g1i1.p1 | 154 | 15.94 | 5.70 | 21.62 | 78.44 | −0.344 | cyto | Other | 7 |
| Hsp1-DN9279c0g1i1.p1 | 1035 | 112.32 | 6.26 | 36.00 | 79.81 | −0.204 | plas | Other | 129 |
| Hsp1-DN4908c2g1i6.p1 | 247 | 26.84 | 5.66 | 50.45 | 82.75 | −0.432 | cyto | Other | 21 |
| Hsp1-DN4908c2g1i3.p1 | 254 | 27.56 | 5.36 | 53.34 | 82.01 | −0.423 | extr | Other | 21 |
| Hsp1-DN3853c0g1i1.p1 | 225 | 25.31 | 6.44 | 33.25 | 88.00 | −0.288 | mito | Other | 21 |
| Hin2-DN8896c0g1i4.p1 | 161 | 16.55 | 5.94 | 24.01 | 79.88 | −0.232 | cyto | Other | 11 |
| Hin2-DN27385c0g2i1.p1 | 154 | 15.94 | 5.70 | 21.62 | 78.44 | −0.344 | cyto | Other | 7 |
| Hin2-DN27385c0g1i1.p1 | 154 | 15.88 | 5.88 | 24.94 | 72.73 | −0.421 | cyto | Other | 7 |
| Hin2-DN252c1g4i1.p1 | 1035 | 111.97 | 6.28 | 37.21 | 80.10 | −0.190 | plas | SP (Sec/SPI) | 129 |
| Hin2-DN8896c0g1i3.p1 | 139 | 14.60 | 5.76 | 30.00 | 79.14 | −0.156 | cyto | Other | 11 |
| Hin2-DN20518c0g1i1.p1 | 351 | 36.96 | 6.70 | 34.66 | 62.19 | −0.459 | plas | SP (Sec/SPI) | 21 |
| Hin2-DN5802c0g1i2.p2 | 254 | 27.54 | 5.50 | 54.06 | 83.15 | −0.393 | extr | Other | 21 |
| Hin2-DN30196c0g1i3.p1 | 236 | 25.90 | 6.29 | 34.63 | 76.78 | −0.371 | cyto | Other | 22 |
| Hin2-DN19943c0g1i4.p1 | 174 | 17.51 | 7.78 | 40.19 | 50.00 | −0.563 | cyto | Other | 17 |
| Hin2-DN64c0g1i15.p1 | 225 | 25.31 | 6.44 | 33.25 | 88.00 | −0.288 | mito | Other | 21 |
| Hsi1-DN3539c0g2i1.p1 | 160 | 16.51 | 6.10 | 14.29 | 77.94 | −0.293 | cyto | Other | 10 |
| Hsi1-DN28797c0g1i1.p1 | 944 | 104.43 | 6.29 | 40.35 | 81.42 | −0.293 | plas | SP (Sec/SPI) | 88 |
| Hsi1-DN2474c0g1i7.p1 | 985 | 106.72 | 6.30 | 38.64 | 79.11 | −0.234 | plas | Other | 124 |
| Hsi1-DN2474c0g1i1.p1 | 1005 | 109.03 | 6.36 | 38.44 | 81.21 | −0.195 | plas | Other | 126 |
| Hsi1-DN2474c0g1i8.p1 | 1014 | 110.15 | 6.36 | 37.53 | 79.64 | −0.209 | plas | Other | 128 |
| Hsi1-DN28797c0g1i3.p1 | 907 | 100.56 | 6.05 | 40.16 | 78.83 | −0.348 | plas | SP (Sec/SPI) | 86 |
| Hsi1-DN2474c0g1i4.p1 | 682 | 73.65 | 6.09 | 37.49 | 84.68 | −0.210 | plas | Other | 89 |
| Hsi1-DN22897c0g1i1.p1 | 351 | 36.93 | 6.70 | 33.95 | 61.08 | −0.465 | plas | SP (Sec/SPI) | 20 |
| Hsi1-DN90c2g2i1.p1 | 292 | 32.67 | 6.79 | 44.89 | 65.75 | −0.822 | extr | SP (Sec/SPI) | 27 |
| Hsi1-DN28603c0g3i2.p1 | 296 | 31.16 | 8.75 | 29.60 | 52.40 | −0.551 | nucl | Other | 30 |
| Hsi1-DN28603c0g3i1.p1 | 525 | 54.59 | 9.75 | 41.41 | 47.39 | −0.673 | mito | SP (Sec/SPI) | 24 |
| Hsi1-DN15006c0g1i2.p1 | 197 | 21.26 | 5.15 | 46.29 | 91.32 | −0.268 | cyto | Other | 13 |
| Hsi1-DN129556c0g1i2.p1 | 192 | 21.14 | 5.10 | 42.66 | 72.71 | −0.453 | plas | SP (Sec/SPI) | 12 |
| Hsi1-DN2474c0g1i4.p2 | 298 | 32.57 | 6.74 | 41.25 | 66.38 | −0.276 | cyto | Other | 34 |
| Hsi1-DN3599c0g1i4.p1 | 225 | 25.29 | 6.63 | 36.78 | 88.44 | −0.287 | mito | Other | 21 |
| Hsi1-DN51085c0g1i2.p1 | 95 | 11.06 | 6.26 | 50.81 | 78.00 | −0.612 | cyto | Other | 5 |
| Hsi2-DN6176c0g1i1.p1 | 160 | 16.51 | 6.10 | 14.29 | 77.94 | −0.293 | cyto | Other | 10 |
| Hsi2-DN7547c0g1i12.p1 | 1032 | 112.09 | 6.42 | 35.77 | 82.68 | −0.157 | plas | Other | 128 |
| Hsi2-DN7547c0g1i8.p1 | 1012 | 109.79 | 6.36 | 35.91 | 80.66 | −0.194 | plas | Other | 126 |
| Hsi2-DN4644c0g2i4.p1 | 292 | 32.62 | 6.96 | 45.10 | 65.41 | −0.809 | extr | SP (Sec/SPI) | 27 |
| Hsi2-DN7796c0g1i1.p1 | 525 | 54.52 | 9.75 | 42.28 | 46.84 | −0.677 | mito | SP (Sec/SPI) | 42 |
| Hsi2-DN4644c0g2i2.p1 | 292 | 32.71 | 6.33 | 47.72 | 61.44 | −0.847 | extr | SP (Sec/SPI) | 27 |
| Hsi2-DN39370c0g1i1.p1 | 254 | 27.55 | 5.50 | 46.37 | 84.69 | −0.404 | extr | Other | 20 |
| Hsi2-DN2531c0g1i1.p1 | 225 | 25.29 | 6.63 | 36.78 | 88.44 | −0.287 | mito | Other | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Kong, X.; Liu, S.; Zhang, S.; Liu, Y.; Lin, Z.; Xue, Q. Transcriptome Assembly and Comparative Analysis of the Superoxide Dismutase (SOD) Gene Family in Three Hyotissa Species. Biology 2026, 15, 4. https://doi.org/10.3390/biology15010004
Kong X, Liu S, Zhang S, Liu Y, Lin Z, Xue Q. Transcriptome Assembly and Comparative Analysis of the Superoxide Dismutase (SOD) Gene Family in Three Hyotissa Species. Biology. 2026; 15(1):4. https://doi.org/10.3390/biology15010004
Chicago/Turabian StyleKong, Xiangjie, Sheng Liu, Shan Zhang, Youli Liu, Zhihua Lin, and Qinggang Xue. 2026. "Transcriptome Assembly and Comparative Analysis of the Superoxide Dismutase (SOD) Gene Family in Three Hyotissa Species" Biology 15, no. 1: 4. https://doi.org/10.3390/biology15010004
APA StyleKong, X., Liu, S., Zhang, S., Liu, Y., Lin, Z., & Xue, Q. (2026). Transcriptome Assembly and Comparative Analysis of the Superoxide Dismutase (SOD) Gene Family in Three Hyotissa Species. Biology, 15(1), 4. https://doi.org/10.3390/biology15010004






