Mutational Signatures in Radiation-Induced Cancer: A Review of Experimental Animal and Human Studies
Simple Summary
Abstract
1. Introduction
2. Radiation Signatures in Animal Models of Cancer
2.1. Interstitial Chromosomal Deletions
2.1.1. Interstitial Chromosomal Deletions in Tumors of Wild-Type Animals
2.1.2. Interstitial Chromosomal Deletions in Tumors from Heterozygous Mutant Animals
2.1.3. Cancer-Risk Assessment Using Interstitial Deletion
2.2. Genome-Wide Mutational Signatures
3. Radiation Signatures in Human Cancers
3.1. Genomic Alterations
3.1.1. Genomic Alterations in Hematopoietic Neoplasms
3.1.2. Genomic Alterations in Thyroid Cancer
3.1.3. Genomic Alterations in Breast Cancer
3.2. Genome-Wide Mutational Signatures
3.3. Clinical Implications of Mutational Signatures
4. Discussion and Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Preston, D.L.; Ron, E.; Tokuoka, S.; Funamoto, S.; Nishi, N.; Soda, M.; Mabuchi, K.; Kodama, K. Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat. Res. 2007, 168, 1–64. [Google Scholar] [CrossRef] [PubMed]
- Ruhm, W.; Laurier, D.; Wakeford, R. Cancer risk following low doses of ionising radiation—Current epidemiological evidence and implications for radiological protection. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2022, 873, 503436. [Google Scholar] [CrossRef]
- McLean, A.R.; Adlen, E.K.; Cardis, E.; Elliott, A.; Goodhead, D.T.; Harms-Ringdahl, M.; Hendry, J.H.; Hoskin, P.; Jeggo, P.A.; Mackay, D.J.C.; et al. A restatement of the natural science evidence base concerning the health effects of low-level ionizing radiation. Proc. Biol. Sci. 2017, 284, 20171070. [Google Scholar] [CrossRef]
- UNSCEAR. Sources, Effects and Risk of Ionizing Radiation, UNSCEAR 2020/2021 Report to the General Assembly, with Scientific Annexes, VOLUME III, Scientific Annex C; United Nations: New York, NY, USA, 2022; Available online: https://www.unscear.org/unscear/uploads/documents/unscear-reports/UNSCEAR_2020_21_Report_Vol.III-CORR.pdf (accessed on 29 June 2025).
- Bouffler, S.; Auvinen, A.; Baiocco, G.; Candéias, S.; Cardis, E.; Galderisi, U.; Lundholm, L.; Madas, B.; Moertl, S.; Pazzaglia, S.; et al. Strategic Research Agenda of the Multidisciplinary European Low Dose Initiative (MELODI)-2022; Multidisciplinary European Low Dose Initiative; MELODI: Paris, France, 2022; Available online: https://melodi-online.eu/wp-content/uploads/2023/03/MELODI-SRA-2022-FINAL-post-consultation1.pdf (accessed on 29 June 2025).
- Yamada, Y.; Imaoka, T.; Iwasaki, T.; Kobayashi, J.; Misumi, M.; Sakai, K.; Sugihara, T.; Suzuki, K.; Tauchi, H.; Yasuda, H.; et al. Establishment and activity of the planning and acting network for low dose radiation research in Japan (PLANET): 2016–2023. J. Radiat. Res. 2024, 65, 561–574. [Google Scholar] [CrossRef]
- Frankenberg-Schwager, M. Induction, repair and biological relevance of radiation-induced DNA lesions in eukaryotic cells. Radiat. Environ. Biophys. 1990, 29, 273–292. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef]
- Silver, A.; Moody, J.; Dunford, R.; Clark, D.; Ganz, S.; Bulman, R.; Bouffler, S.; Finnon, P.; Meijne, E.; Huiskamp, R.; et al. Molecular mapping of chromosome 2 deletions in murine radiation-induced AML localizes a putative tumor suppressor gene to a 1.0 cM region homologous to human chromosome segment 11p11-12. Genes Chromosomes Cancer 1999, 24, 95–104. [Google Scholar] [CrossRef]
- Shimada, Y.; Nishimura, M.; Kakinuma, S.; Okumoto, M.; Shiroishi, T.; Clifton, K.H.; Wakana, S. Radiation-associated loss of heterozygosity at the Znfn1a1 (Ikaros) locus on chromosome 11 in murine thymic lymphomas. Radiat. Res. 2000, 154, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Takabatake, T.; Kakinuma, S.; Amasaki, Y.; Nishimura, M.; Imaoka, T.; Yamauchi, K.; Shang, Y.; Miyoshi-Imamura, T.; Nogawa, H.; et al. Complicated biallelic inactivation of Pten in radiation-induced mouse thymic lymphomas. Mutat. Res. 2010, 686, 30–38. [Google Scholar] [CrossRef]
- Sunaoshi, M.; Amasaki, Y.; Hirano-Sakairi, S.; Blyth, B.J.; Morioka, T.; Kaminishi, M.; Shang, Y.; Nishimura, M.; Shimada, Y.; Tachibana, A.; et al. The effect of age at exposure on the inactivating mechanisms and relative contributions of key tumor suppressor genes in radiation-induced mouse T-cell lymphomas. Mutat. Res. 2015, 779, 58–67. [Google Scholar] [CrossRef]
- Tachibana, H.; Daino, K.; Ishikawa, A.; Morioka, T.; Shang, Y.; Ogawa, M.; Matsuura, A.; Shimada, Y.; Kakinuma, S. Genomic profile of radiation-induced early-onset mouse B-cell lymphoma recapitulates features of Philadelphia chromosome-like acute lymphoblastic leukemia in humans. Carcinogenesis 2022, 43, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, D.; Imaoka, T.; Takabatake, T.; Nishimura, M.; Kakinuma, S.; Nishimura, Y.; Shimada, Y. DNA copy number aberrations and disruption of the p16Ink4a/Rb pathway in radiation-induced and spontaneous rat mammary carcinomas. Radiat. Res. 2010, 174, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, H.; Daino, K.; Ishikawa, A.; Imaoka, T.; Nishimura, M.; Nishimura, Y.; Takabatake, M.; Morioka, T.; Inoue, K.; Fukushi, M.; et al. Exome of Radiation-induced Rat Mammary Carcinoma Shows Copy-number Losses and Mutations in Human-relevant Cancer Genes. Anticancer. Res. 2021, 41, 55–70. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Tanori, M.; Mancuso, M.; Gessi, M.; Pasquali, E.; Leonardi, S.; Oliva, M.A.; Rebessi, S.; Di Majo, V.; Covelli, V.; et al. Two-hit model for progression of medulloblastoma preneoplasia in Patched heterozygous mice. Oncogene 2006, 25, 5575–5580. [Google Scholar] [CrossRef]
- Ishida, Y.; Takabatake, T.; Kakinuma, S.; Doi, K.; Yamauchi, K.; Kaminishi, M.; Kito, S.; Ohta, Y.; Amasaki, Y.; Moritake, H.; et al. Genomic and gene expression signatures of radiation in medulloblastomas after low-dose irradiation in Ptch1 heterozygous mice. Carcinogenesis 2010, 31, 1694–1701. [Google Scholar] [CrossRef][Green Version]
- Kokubo, T.; Kakinuma, S.; Kobayashi, T.; Watanabe, F.; Iritani, R.; Tateno, K.; Nishimura, M.; Nishikawa, T.; Hino, O.; Shimada, Y. Age dependence of radiation-induced renal cell carcinomas in an Eker rat model. Cancer Sci. 2010, 101, 616–623. [Google Scholar] [CrossRef]
- Inoue, T.; Kokubo, T.; Daino, K.; Yanagihara, H.; Watanabe, F.; Tsuruoka, C.; Amasaki, Y.; Morioka, T.; Homma-Takeda, S.; Kobayashi, T.; et al. Interstitial chromosomal deletion of the tuberous sclerosis complex 2 locus is a signature for radiation-associated renal tumors in Eker rats. Cancer Sci. 2020, 111, 840–848. [Google Scholar] [CrossRef]
- Yanagihara, H.; Morioka, T.; Yamazaki, S.; Yamada, Y.; Tachibana, H.; Daino, K.; Tsuruoka, C.; Amasaki, Y.; Kaminishi, M.; Imaoka, T.; et al. Interstitial deletion of the Apc locus in beta-catenin-overexpressing cells is a signature of radiation-induced intestinal tumors in C3B6F1 ApcMin/+ micedagger. J. Radiat. Res. 2023, 64, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Bouffler, S.D.; Meijne, E.I.; Morris, D.J.; Papworth, D. Chromosome 2 hypersensitivity and clonal development in murine radiation acute myeloid leukaemia. Int. J. Radiat. Biol. 1997, 72, 181–189. [Google Scholar] [CrossRef]
- Daino, K.; Ishikawa, A.; Suga, T.; Amasaki, Y.; Kodama, Y.; Shang, Y.; Hirano-Sakairi, S.; Nishimura, M.; Nakata, A.; Yoshida, M.; et al. Mutational landscape of T-cell lymphoma in mice lacking the DNA mismatch repair gene Mlh1: No synergism with ionizing radiation. Carcinogenesis 2019, 40, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Hahn, H.; Wojnowski, L.; Zimmer, A.M.; Hall, J.; Miller, G.; Zimmer, A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med. 1998, 4, 619–622. [Google Scholar] [CrossRef]
- Moser, A.R.; Luongo, C.; Gould, K.A.; McNeley, M.K.; Shoemaker, A.R.; Dove, W.F. ApcMin: A mouse model for intestinal and mammary tumorigenesis. Eur. J. Cancer 1995, 31A, 1061–1064. [Google Scholar] [CrossRef]
- Okamoto, M.; Yonekawa, H. Intestinal tumorigenesis in Min mice is enhanced by X-irradiation in an age-dependent manner. J. Radiat. Res. 2005, 46, 83–91. [Google Scholar] [CrossRef]
- Tsuruoka, C.; Blyth, B.J.; Morioka, T.; Kaminishi, M.; Shinagawa, M.; Shimada, Y.; Kakinuma, S. Sensitive Detection of Radiation-Induced Medulloblastomas after Acute or Protracted Gamma-Ray Exposures in Ptch1 Heterozygous Mice Using a Radiation-Specific Molecular Signature. Radiat. Res. 2016, 186, 407–414. [Google Scholar] [CrossRef]
- Lee, C.L.; Brock, K.D.; Hasapis, S.; Zhang, D.; Sibley, A.B.; Qin, X.; Gresham, J.S.; Caraballo, I.; Luo, L.; Daniel, A.R.; et al. Whole-Exome Sequencing of Radiation-Induced Thymic Lymphoma in Mouse Models Identifies Notch1 Activation as a Driver of p53 Wild-Type Lymphoma. Cancer Res. 2021, 81, 3777–3790. [Google Scholar] [CrossRef]
- Sherborne, A.L.; Davidson, P.R.; Yu, K.; Nakamura, A.O.; Rashid, M.; Nakamura, J.L. Mutational Analysis of Ionizing Radiation Induced Neoplasms. Cell Rep. 2015, 12, 1915–1926. [Google Scholar] [CrossRef]
- Davidson, P.R.; Sherborne, A.L.; Taylor, B.; Nakamura, A.O.; Nakamura, J.L. A pooled mutational analysis identifies ionizing radiation-associated mutational signatures conserved between mouse and human malignancies. Sci. Rep. 2017, 7, 7645. [Google Scholar] [CrossRef]
- Rose Li, Y.; Halliwill, K.D.; Adams, C.J.; Iyer, V.; Riva, L.; Mamunur, R.; Jen, K.Y.; Del Rosario, R.; Fredlund, E.; Hirst, G.; et al. Mutational signatures in tumours induced by high and low energy radiation in Trp53 deficient mice. Nat. Commun. 2020, 11, 394. [Google Scholar] [CrossRef]
- Nakanishi, M.; Tanaka, K.; Shintani, T.; Takahashi, T.; Kamada, N. Chromosomal instability in acute myelocytic leukemia and myelodysplastic syndrome patients among atomic bomb survivors. J. Radiat. Res. 1999, 40, 159–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zharlyganova, D.; Harada, H.; Harada, Y.; Shinkarev, S.; Zhumadilov, Z.; Zhunusova, A.; Tchaizhunusova, N.J.; Apsalikov, K.N.; Kemaikin, V.; Zhumadilov, K.; et al. High frequency of AML1/RUNX1 point mutations in radiation-associated myelodysplastic syndrome around Semipalatinsk nuclear test site. J. Radiat. Res. 2008, 49, 549–555. [Google Scholar] [CrossRef]
- Ojha, J.; Dyagil, I.; Finch, S.C.; Reiss, R.F.; de Smith, A.J.; Gonseth, S.; Zhou, M.; Hansen, H.M.; Sherborne, A.L.; Nakamura, J.; et al. Genomic characterization of chronic lymphocytic leukemia (CLL) in radiation-exposed Chornobyl cleanup workers. Environ. Health 2018, 17, 43. [Google Scholar] [CrossRef]
- Poluben, L.; Puligandla, M.; Neuberg, D.; Bryke, C.R.; Hsu, Y.; Shumeiko, O.; Yuan, X.; Voznesensky, O.; Pihan, G.; Adam, M.; et al. Characteristics of myeloproliferative neoplasms in patients exposed to ionizing radiation following the Chernobyl nuclear accident. Am. J. Hematol. 2019, 94, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.; Thomas, G.; Braselmann, H.; Bauer, V.; Bogdanova, T.; Wienberg, J.; Zitzelsberger, H.; Unger, K. Gain of chromosome band 7q11 in papillary thyroid carcinomas of young patients is associated with exposure to low-dose irradiation. Proc. Natl. Acad. Sci. USA 2011, 108, 9595–9600. [Google Scholar] [CrossRef] [PubMed]
- Selmansberger, M.; Feuchtinger, A.; Zurnadzhy, L.; Michna, A.; Kaiser, J.C.; Abend, M.; Brenner, A.; Bogdanova, T.; Walch, A.; Unger, K.; et al. CLIP2 as radiation biomarker in papillary thyroid carcinoma. Oncogene 2015, 34, 3917–3925. [Google Scholar] [CrossRef] [PubMed]
- Selmansberger, M.; Kaiser, J.C.; Hess, J.; Guthlin, D.; Likhtarev, I.; Shpak, V.; Tronko, M.; Brenner, A.; Abend, M.; Blettner, M.; et al. Dose-dependent expression of CLIP2 in post-Chernobyl papillary thyroid carcinomas. Carcinogenesis 2015, 36, 748–756. [Google Scholar] [CrossRef]
- Miura, S.; Nakashima, M.; Ito, M.; Kondo, H.; Meirmanov, S.; Hayashi, T.; Soda, M.; Matsuo, T.; Sekine, I. Significance of HER2 and C-MYC oncogene amplifications in breast cancer in atomic bomb survivors: Associations with radiation exposure and histologic grade. Cancer 2008, 112, 2143–2151. [Google Scholar] [CrossRef]
- Oikawa, M.; Yoshiura, K.; Kondo, H.; Miura, S.; Nagayasu, T.; Nakashima, M. Significance of genomic instability in breast cancer in atomic bomb survivors: Analysis of microarray-comparative genomic hybridization. Radiat. Oncol. 2011, 6, 168. [Google Scholar] [CrossRef]
- Wilke, C.M.; Braselmann, H.; Hess, J.; Klymenko, S.V.; Chumak, V.V.; Zakhartseva, L.M.; Bakhanova, E.V.; Walch, A.K.; Selmansberger, M.; Samaga, D.; et al. A genomic copy number signature predicts radiation exposure in post-Chernobyl breast cancer. Int. J. Cancer 2018, 143, 1505–1515. [Google Scholar] [CrossRef]
- Lesluyes, T.; Baud, J.; Perot, G.; Charon-Barra, C.; You, A.; Valo, I.; Bazille, C.; Mishellany, F.; Leroux, A.; Renard-Oldrini, S.; et al. Genomic and transcriptomic comparison of post-radiation versus sporadic sarcomas. Mod. Pathol. 2019, 32, 1786–1794. [Google Scholar] [CrossRef]
- Behjati, S.; Gundem, G.; Wedge, D.C.; Roberts, N.D.; Tarpey, P.S.; Cooke, S.L.; Van Loo, P.; Alexandrov, L.B.; Ramakrishna, M.; Davies, H.; et al. Mutational signatures of ionizing radiation in second malignancies. Nat. Commun. 2016, 7, 12605. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Han, D.J.; Kim, B.H.; Yoo, J.; Kim, H.J.; Wu, H.G.; Kim, K.S.; Kim, H.S.; Han, I.; Moon, K.C.; et al. Whole-Genome Sequencing Reveals Mutational Signatures Related to Radiation-Induced Sarcomas and DNA-Damage-Repair Pathways. Mod. Pathol. 2023, 36, 100004. [Google Scholar] [CrossRef] [PubMed]
- Goerlitz, D.S.; Blancato, J.; Ramesh, A.; Islam, M.; Graham, G.T.; Revina, V.; Kallakury, B.; Zeck, J.; Kirillova, E.; Loffredo, C.A. Somatic mutation signatures in primary liver tumors of workers exposed to ionizing radiation. Sci. Rep. 2019, 9, 18199. [Google Scholar] [CrossRef] [PubMed]
- Morton, L.M.; Karyadi, D.M.; Stewart, C.; Bogdanova, T.I.; Dawson, E.T.; Steinberg, M.K.; Dai, J.; Hartley, S.W.; Schonfeld, S.J.; Sampson, J.N.; et al. Radiation-related genomic profile of papillary thyroid carcinoma after the Chernobyl accident. Science 2021, 372, eabg2538. [Google Scholar] [CrossRef]
- Deng, M.Y.; Sturm, D.; Pfaff, E.; Sill, M.; Stichel, D.; Balasubramanian, G.P.; Tippelt, S.; Kramm, C.; Donson, A.M.; Green, A.L.; et al. Radiation-induced gliomas represent H3-/IDH-wild type pediatric gliomas with recurrent PDGFRA amplification and loss of CDKN2A/B. Nat. Commun. 2021, 12, 5530. [Google Scholar] [CrossRef]
- Kocakavuk, E.; Anderson, K.J.; Varn, F.S.; Johnson, K.C.; Amin, S.B.; Sulman, E.P.; Lolkema, M.P.; Barthel, F.P.; Verhaak, R.G.W. Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer. Nat. Genet. 2021, 53, 1088–1096. [Google Scholar] [CrossRef]
- Ahire, V.; Bidakhvidi, N.A.; Boterberg, T.; Chaudhary, P.; Chevalier, F.; Daems, N.; Delbart, W.; Baatout, S.; Deroose, C.M.; Fernandez-Palomo, C.; et al. Radiobiology of Combining Radiotherapy with Other Cancer Treatment Modalities. In Radiobiology Textbook; Baatout, S., Ed.; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Nagashima, H.; Shiraishi, K.; Ohkawa, S.; Sakamoto, Y.; Komatsu, K.; Matsuura, S.; Tachibana, A.; Tauchi, H. Induction of somatic mutations by low-dose X-rays: The challenge in recognizing radiation-induced events. J. Radiat. Res. 2018, 59, ii11–ii17. [Google Scholar] [CrossRef]
- Tsuruoka, C.; Kaminishi, M.; Shinagawa, M.; Shang, Y.; Amasaki, Y.; Shimada, Y.; Kakinuma, S. High Relative Biological Effectiveness of 2 MeV Fast Neutrons for Induction of Medulloblastoma in Ptch1+/– Mice with Radiation-specific Deletion on Chromosome 13. Radiat. Res. 2021, 196, 225–234. [Google Scholar] [CrossRef]
- Tsuruoka, C.; Shinagawa, M.; Shang, Y.; Amasaki, Y.; Sunaoshi, M.; Imaoka, T.; Morioka, T.; Shimada, Y.; Kakinuma, S. Relative Biological Effectiveness of Carbon Ion Beams for Induction of Medulloblastoma with Radiation-specific Chromosome 13 Deletion in Ptch1+/– Mice. Radiat. Res. 2024, 202, 503–509. [Google Scholar] [CrossRef]
- De Saint-Hubert, M.; Saldarriaga Vargas, C.; Van Hoey, O.; Schoonjans, W.; De Smet, V.; Mathot, G.; Stichelbaut, F.; Manessi, G.; Dinar, N.; Aza, E.; et al. Secondary Neutron Doses in a Proton Therapy Centre. Radiat. Prot. Dosimetry 2016, 170, 336–341. [Google Scholar] [CrossRef]
- Yonai, S.; Furukawa, T.; Inaniwa, T. Measurement of neutron ambient dose equivalent in carbon-ion radiotherapy with an active scanned delivery system. Radiat. Prot. Dosim. 2014, 161, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Howell, R.M.; Ferenci, M.S.; Hertel, N.E.; Fullerton, G.D.; Fox, T.; Davis, L.W. Measurements of secondary neutron dose from 15 MV and 18 MV IMRT. Radiat. Prot. Dosim. 2005, 115, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, F.; Du, Y.; Shi, H.; Zhou, W. Hypoxic microenvironment in cancer: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Uchimura, A.; Satoh, Y.; Kato, N.; Toshishige, M.; Kajimura, J.; Hamasaki, K.; Yoshida, K.; Hayashi, T.; Noda, A.; et al. Spectra and characteristics of somatic mutations induced by ionizing radiation in hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2216550120. [Google Scholar] [CrossRef]
- Youk, J.; Kwon, H.W.; Lim, J.; Kim, E.; Kim, T.; Kim, R.; Park, S.; Yi, K.; Nam, C.H.; Jeon, S.; et al. Quantitative and qualitative mutational impact of ionizing radiation on normal cells. Cell Genom. 2024, 4, 100499. [Google Scholar] [CrossRef]
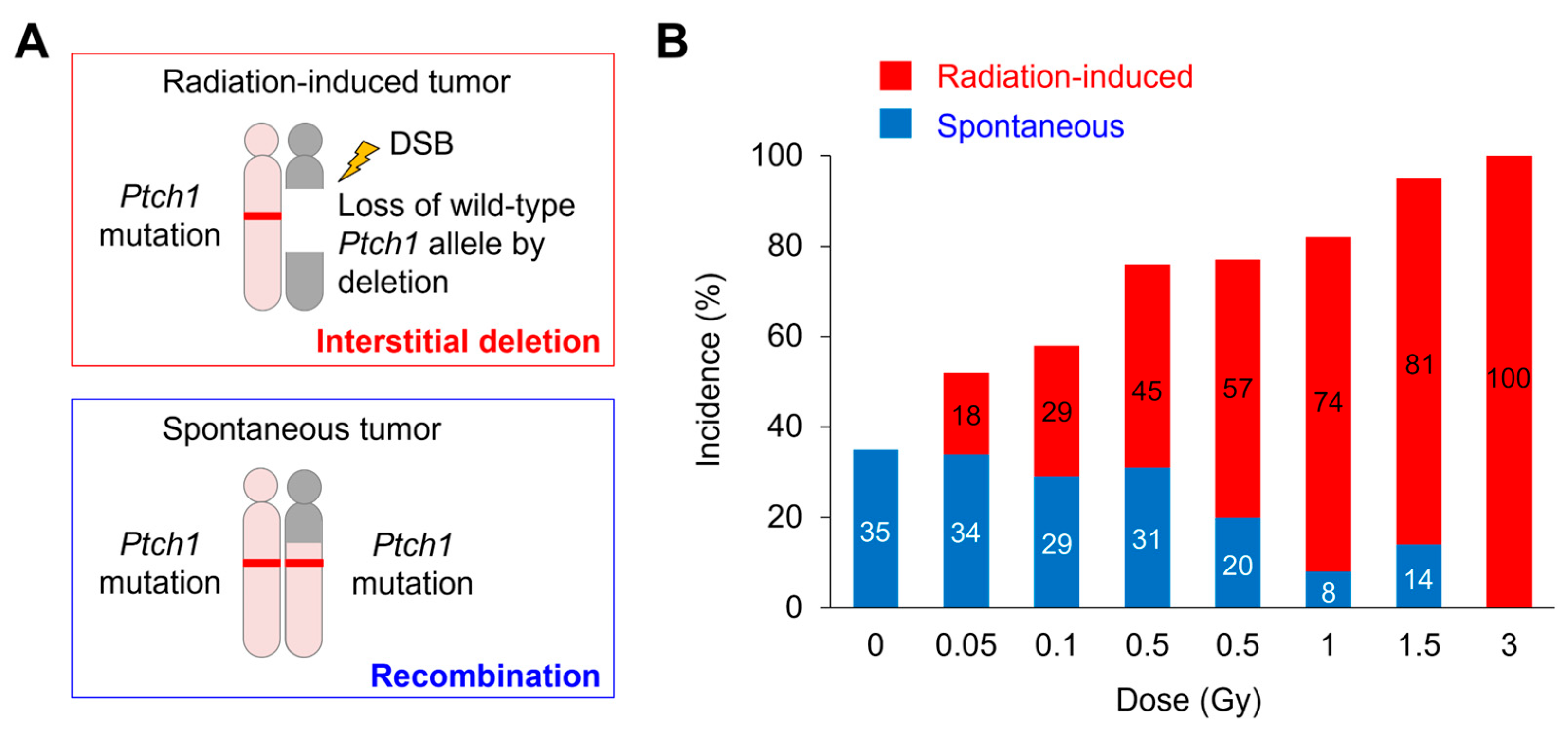
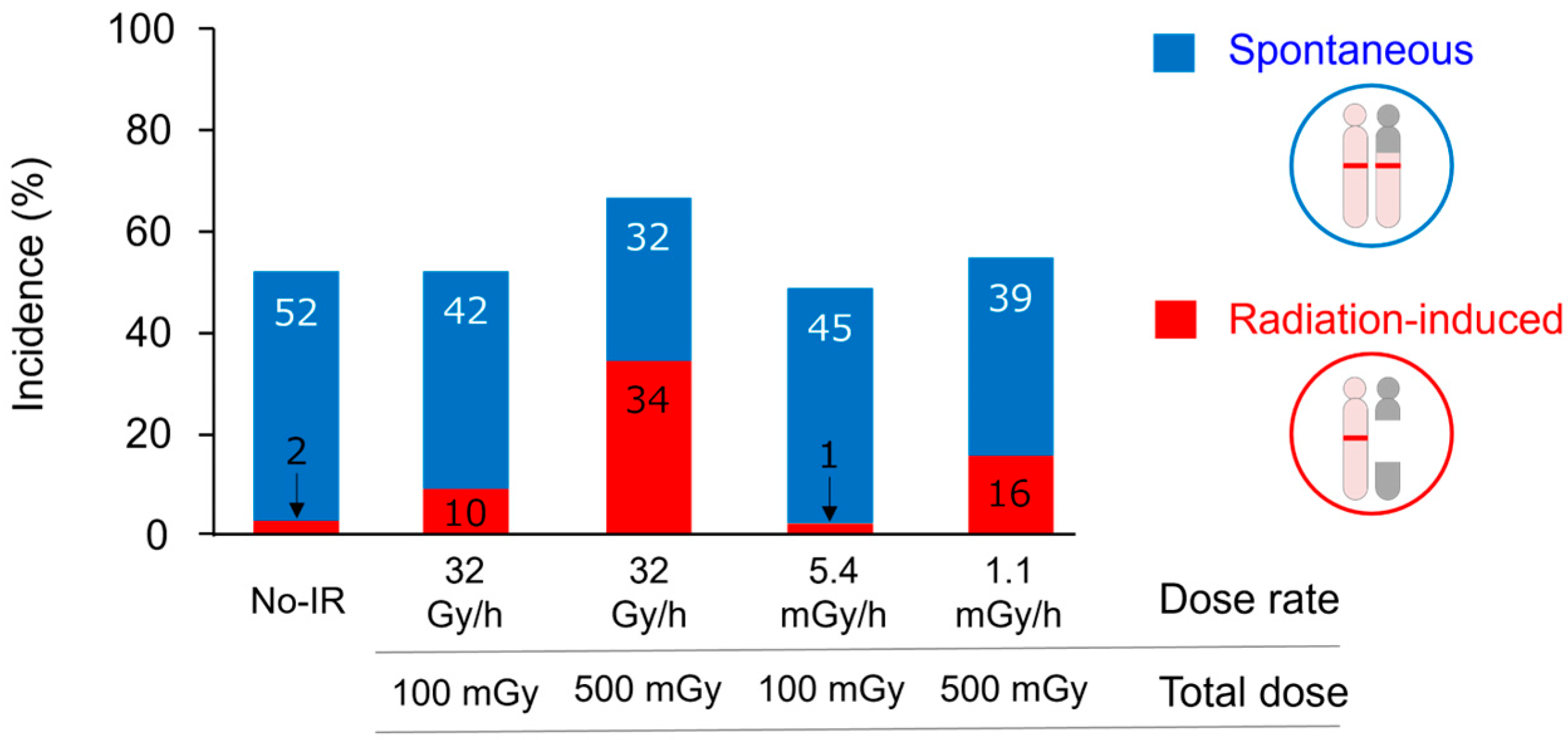
| Tumor Type | Animal Model | Genotype | Method | Dose | Deletion Target Gene (chr) | Reference(s) |
|---|---|---|---|---|---|---|
| AML | (CBA/H × C57BL/Lia) F1 mouse | Wild type | LOH | 3 Gy | PU.1/Spi1 (2) | [10] |
| Thymic lymphoma | (C57BL/6 × C3H/He) F1 mouse | Wild type | LOH and array-CGH | 6.4 Gy (1.6 Gy × 4 weekly fractionated X-ray) | Cdkn2a (4), Ikzf1 (11), Pten (19) | [11,12,13] |
| B-cell lymphoma | (C57BL/6 × C3H/He) F1 mouse | Wild type | LOH, array-CGH, and WES | 4 Gy | Pax5 (4) | [14] |
| Mammary carcinoma | Sprague-Dawley rat | Wild type | Array-CGH and WES | 2 or 4 Gy, 1 Gy (fast neutron) | Cdkn2a (5) | [15,16] |
| Medulloblastoma | Ptch1+/− mouse | Heterozygous loss-of-function mutation | LOH and array-CGH | 0.05 Gy to 3 Gy | Ptch1 (13) | [17,18] |
| Renal carcinoma | Tsc2Eker/+ rat | Heterozygous loss-of-function mutation | LOH and array-CGH | 2 Gy | Tsc2 (10) | [19,20] |
| Intestinal carcinoma | ApcMin/+ mouse | Heterozygous loss-of-function mutation | LOH and array-CGH | 2 Gy | Apc (18) | [21] |
| Tumor Type(s) | Animal Model | Method | Dose | Characteristic Alterations | Reference(s) |
|---|---|---|---|---|---|
| Sarcoma, squamous cell carcinoma, mammary carcinoma, and hematopoietic neoplasm | Nf1+/− and wild-type mice | WES | 30 Gy (3 Gy × 5, daily fractionated X-ray) | DNA copy-number loss (Nf1+/− mice) and mutational signatures enriched for C-to-T or T-to-G substitutions | [29,30] |
| Mammary carcinoma | Trp53+/− and Trp53ΔP mice | WGS | 0.5 Gy (Fe-ion or gamma-ray) | Focal SVs and Met (chr 6) amplification (Fe-ion), SVs and COSMIC signature SBS18 associated with ROS (gamma-ray) | [31] |
| Tumor Type | Cohort | Method | Dose | Characteristic Alterations | Reference(s) |
|---|---|---|---|---|---|
| AML | Atomic-bomb survivors | FISH | N.A. | Monosomy 7 and chromosomal deletion at 20q13.2 | [32] |
| MDS/AML | Residents near Semipalatinsk Nuclear Test Site | PCR-SSCP 7 | N.A. | AML1/RUNX1 (chr 21) point mutations | [33] |
| CLL | Chornobyl clean-up workers | Targeted deep sequencing of 538 cancer-relevant genes and off-target reads mapping | Median bone marrow dose, 40.5 mGy; range, 0.4 to 1536.2 mGy | POT1 (chr 7) and ATM (chr 11) mutations and longer telomere length | [34] |
| MPN | Chornobyl clean-up workers and residents | WES | 20 to 500 mSv (cleanup workers) and 5.9 to 31 mSv (residents) | JAK2 (chr 9), CALR (chr 19), ATM (chr 11), EZH2 (chr 7), and SUZ12 (chr 17) mutations | [35] |
| Papillary thyroid cancer | Residents after Chornobyl accident | Array-CGH, real-time qPCR, and IHC | Mean dose, 0.15 to 1.2 Gy | DNA copy-number gain of 7q11.22-11.23 and CLIP2 (chr 7) overexpression | [36,37,38] |
| Breast cancer | Atomic-bomb survivors | FISH and array-CGH | N.A. | C-MYC (chr 8) and HER2 (chr 17) amplification | [39,40] |
| Breast cancer | Chornobyl clean-up workers and evacuees | Array-CGH | Median dose, 13.0 mGy; range, 0.06 to 582.9 mGy (clean-up workers) and median dose, 18.4 mGy; range, 5.72 to 36.6 mGy (evacuees) | CNA signature consists of chromosome regions at 7q11.22–11.23, 7q21.3, 16q24.3, 17q21.31, 20p11.23–11.21, 1p21.1, 2q35, and 6p22.2 | [41] |
| Sarcoma | Patients who received radiotherapy | Array-CGH | N.A. | C-MYC (chr 8) amplification and CDKN2A (chr 9) and CDKN2B (chr 9) losses | [42] |
| Tumor Type | Cohort | Method | Dose | Characteristic Alterations | Reference |
|---|---|---|---|---|---|
| Sarcoma and breast cancer | Patients who received radiotherapy | WGS | N.A. | Small deletions and balanced chromosomal inversions | [43] |
| Sarcoma | Patients who received radiotherapy | WGS | 45 to 54 Gy (25 to 30 fractionated), including unknown cases | Small deletions, COSMIC signature ID8 and SVs | [45] |
| Liver tumor | Mayak employees | WES | N.A. | Small deletions and clustered mutations | [46] |
| Papillary thyroid cancer | Residents after Chornobyl accident | WGS, mRNA/microRNA-seq, DNA methylation profiling | Mean dose, 250 mGy; range, 11 to 8800 mGy | Small deletions and simple/balanced SVs, COSMIC signature ID8, and fusion driver mutations | [47] |
| Glioma | Patients who received radiotherapy | WES, mRNA-seq, DNA methylation profiling | N.A. | PDGFRA (chr 4) amplification and CDKN2A (chr 9) and CDKN2B (chr 9) losses | [48] |
| Glioma | Patients who received radiotherapy | WGS | N.A. | Small deletions, COSMIC signature ID8, and chromosomal deletions and inversions, and CDKN2A (chr 9) loss | [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daino, K.; Tsuruoka, C.; Ishikawa, A.; Kakinuma, S.; Imaoka, T. Mutational Signatures in Radiation-Induced Cancer: A Review of Experimental Animal and Human Studies. Biology 2025, 14, 1142. https://doi.org/10.3390/biology14091142
Daino K, Tsuruoka C, Ishikawa A, Kakinuma S, Imaoka T. Mutational Signatures in Radiation-Induced Cancer: A Review of Experimental Animal and Human Studies. Biology. 2025; 14(9):1142. https://doi.org/10.3390/biology14091142
Chicago/Turabian StyleDaino, Kazuhiro, Chizuru Tsuruoka, Atsuko Ishikawa, Shizuko Kakinuma, and Tatsuhiko Imaoka. 2025. "Mutational Signatures in Radiation-Induced Cancer: A Review of Experimental Animal and Human Studies" Biology 14, no. 9: 1142. https://doi.org/10.3390/biology14091142
APA StyleDaino, K., Tsuruoka, C., Ishikawa, A., Kakinuma, S., & Imaoka, T. (2025). Mutational Signatures in Radiation-Induced Cancer: A Review of Experimental Animal and Human Studies. Biology, 14(9), 1142. https://doi.org/10.3390/biology14091142







