Evaluation of Bacterial Population Changes and Ecological Dynamics in Oil-Impacted Soils Using 16S rRNA Amplicon Sequencing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Experimental Design
2.2. Soil Sample Collection, DNA Extraction, and 16S rRNA Amplicon Sequencing
2.3. Bioinformatics and Statistical Analyses
3. Results
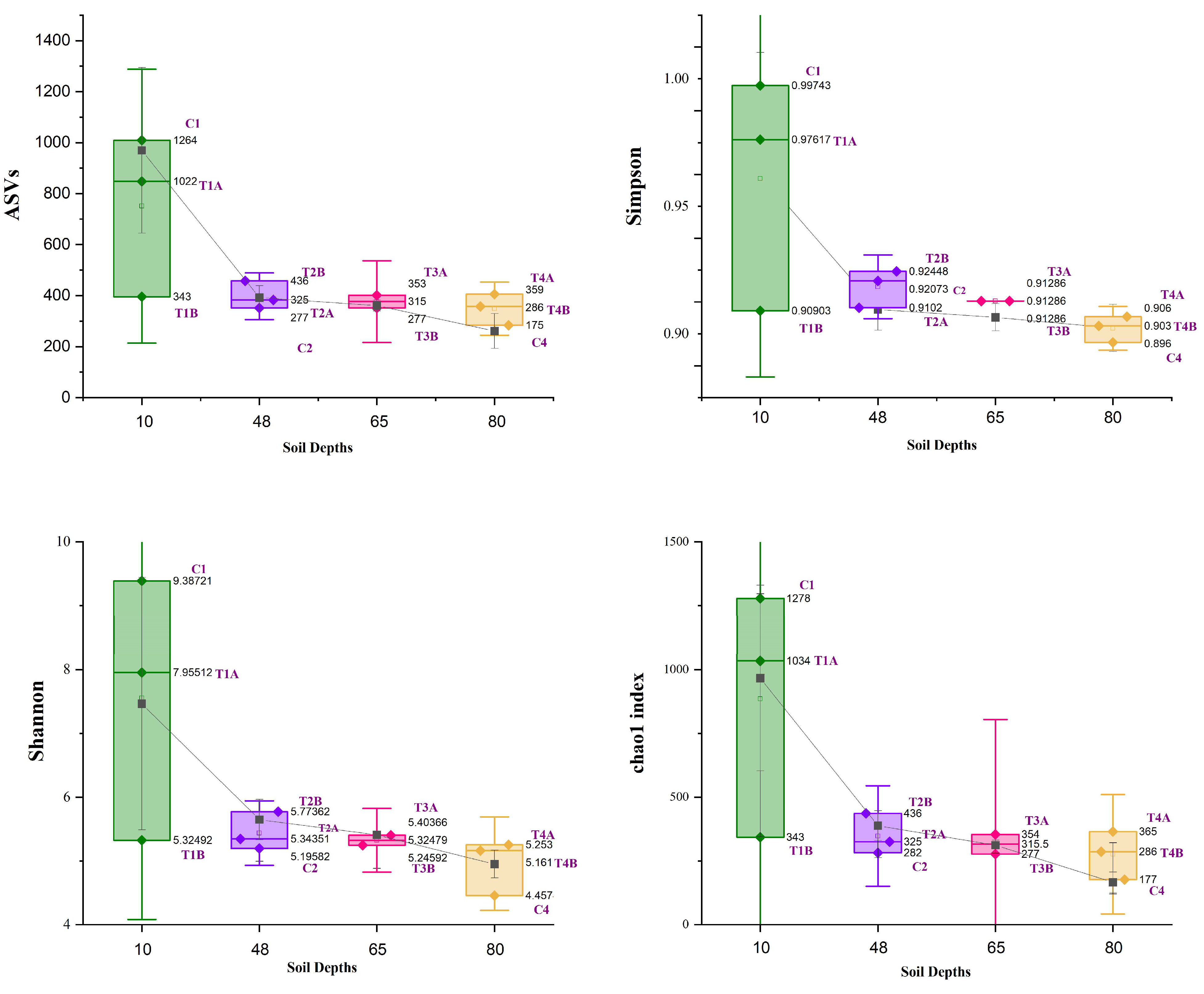
3.1. Bacterial Diversity Analysis
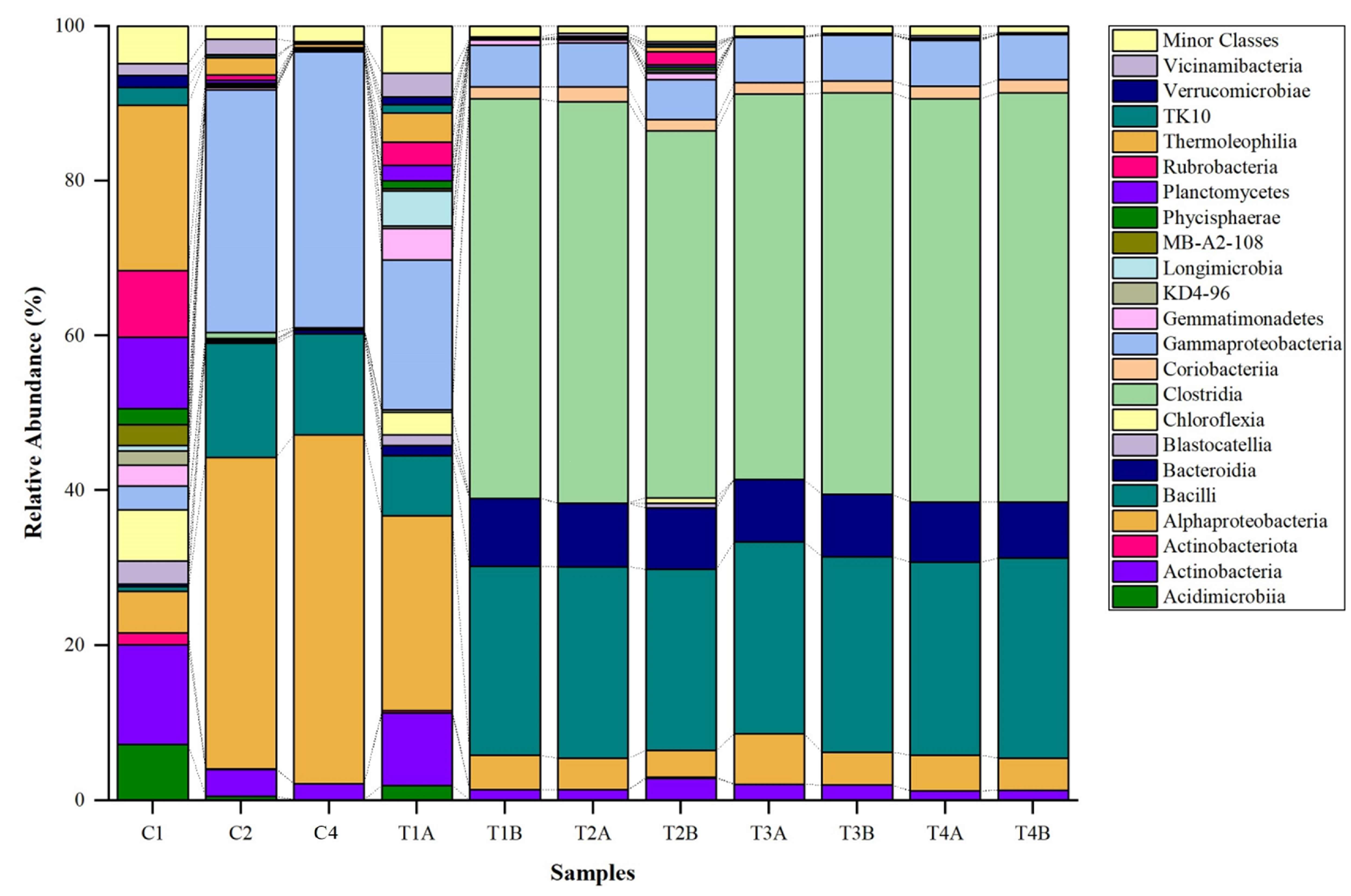
3.2. Bacterial Composition at Phylum Level
3.3. Bacterial Composition at Class Level
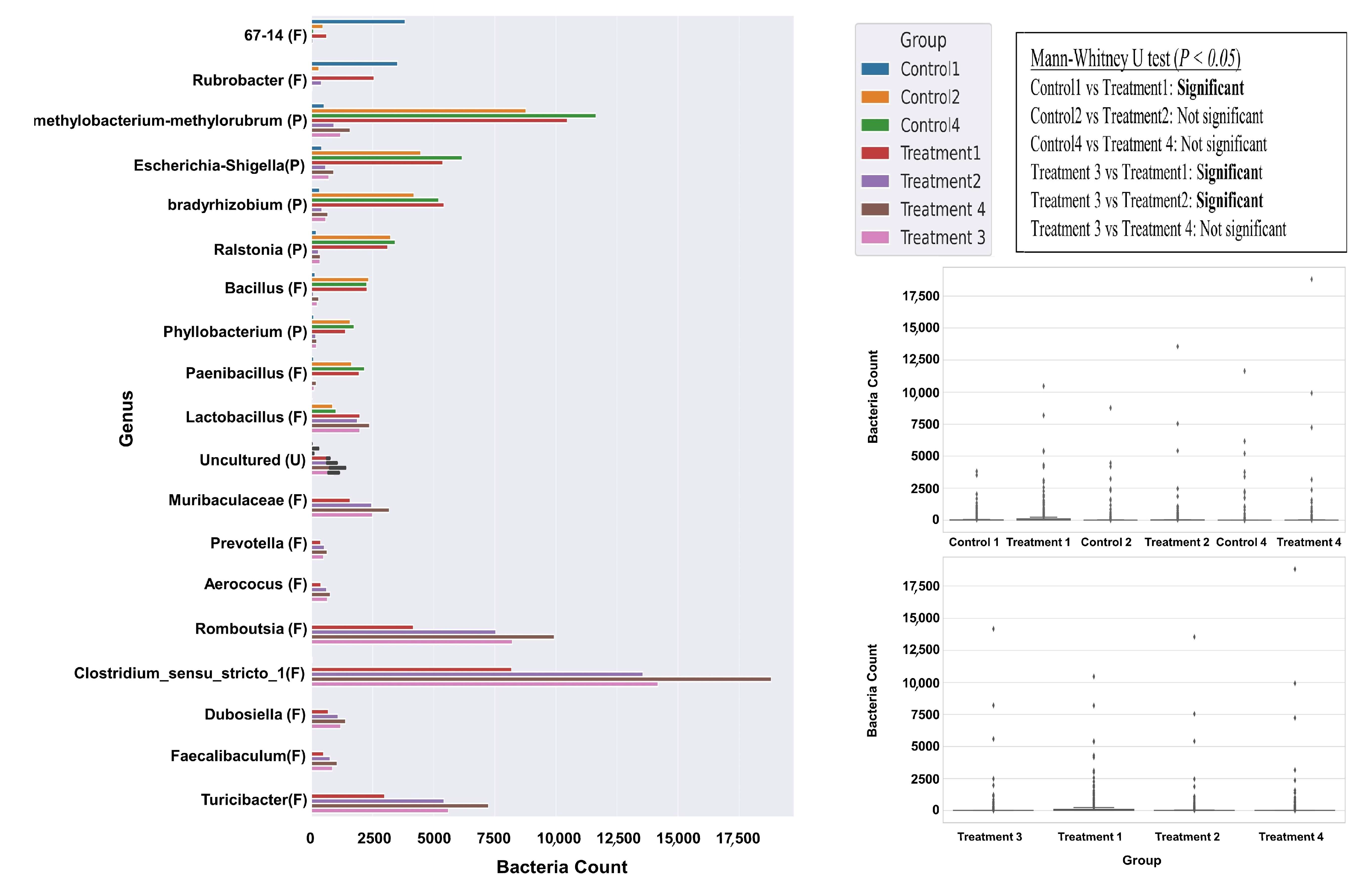
3.4. Bacterial Abundance at Genus Level
3.5. Bacterial Variation Between Control and Treatment
3.6. Co-Occurrence Network Analysis at the Bacterial Genus Level
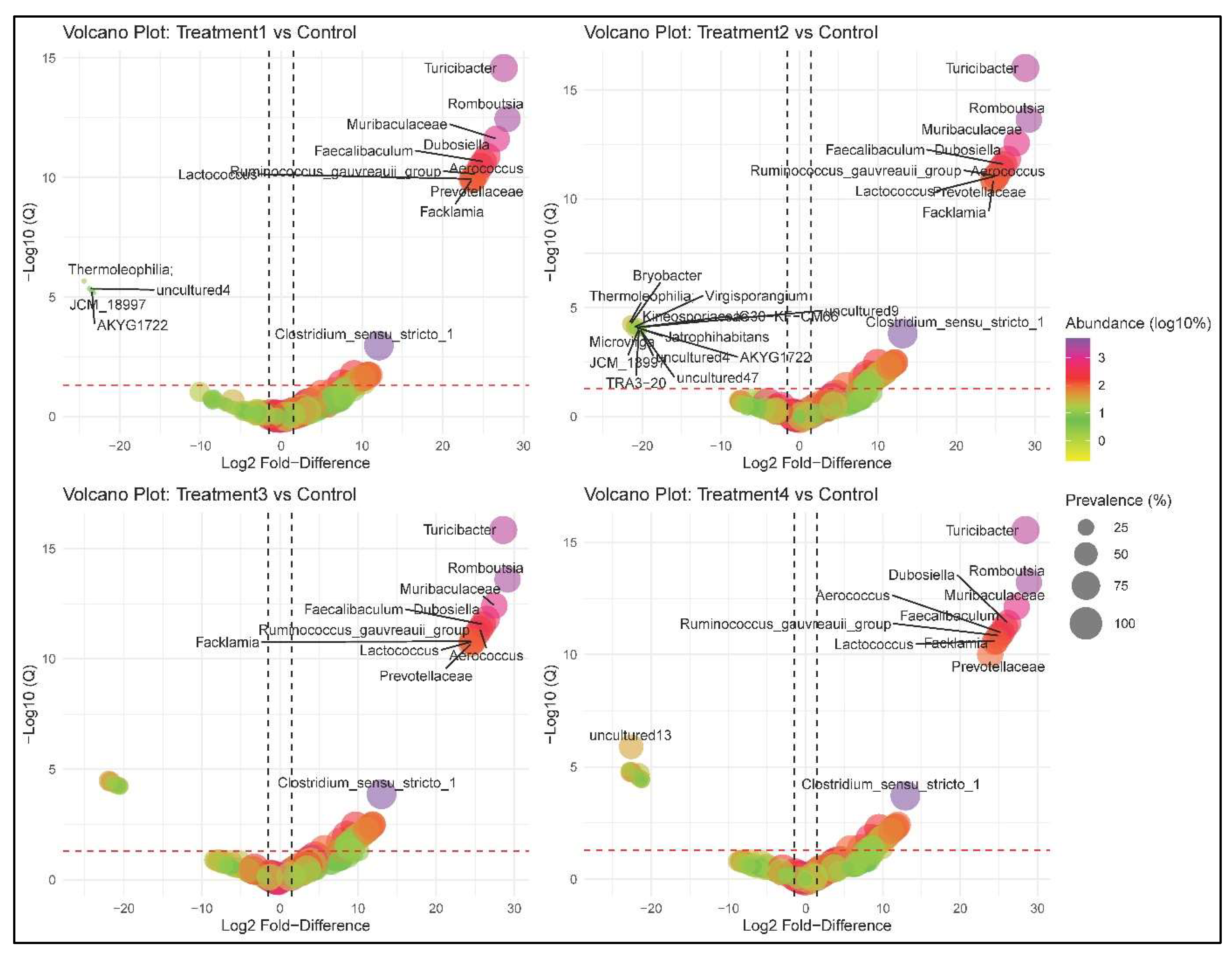
3.7. Differential Abundance Analysis
4. Discussion
4.1. Enrichment of Bacterial Genera: Implications for Soil Ecosystem Function and Health
4.2. The Loss of Bacterial Genera and Its Impact on Soil Ecological Functions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Needelman, B.A. What are soils? Nat. Educ. Knowl. 2013, 4, 2. [Google Scholar]
- De Deyn, G.B.; Kooistra, L. The role of soils in habitat creation, maintenance and restoration. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200170. [Google Scholar] [CrossRef]
- FAO; UNEP. Global Assessment of Soil Pollution: Report; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2021. [Google Scholar] [CrossRef]
- Akhtar, N.; Ishak, M.I.S.; Bhawani, S.A.; Umar, K. Various natural and anthropogenic factors responsible for water quality degradation: A review. Water 2021, 13, 2660. [Google Scholar] [CrossRef]
- Garbuio, F.J.; Howard, J.L.; dos Santos, L.M. Impact of human activities on soil contamination. Appl. Environ. Soil Sci. 2012, 2012, 619548. [Google Scholar] [CrossRef]
- Demie, G. Analyzing soil contamination status in garage and auto mechanical workshops of Shashemane City: Implication for hazardous waste management. Environ. Syst. Res. 2015, 4, 15. [Google Scholar] [CrossRef]
- Abdel-Shafy, H.I.; Mansour, M.S.M. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egypt. J. Pet. 2016, 25, 107–123. [Google Scholar] [CrossRef]
- Akintunde, W.O.; Olugbenga, O.A.; Olufemi, O.O. Some adverse effects of used engine oil (common waste pollutant) on reproduction of male sprague dawley rats. Open Access Maced. J. Med. Sci. 2015, 3, 46–51. (In English) [Google Scholar] [CrossRef]
- Shahid, M.J.; Al-Surhanee, A.A.; Kouadri, F.; Ali, S.; Nawaz, N.; Afzal, M.; Rizwan, M.; Ali, B.; Soliman, M.H. Role of microorganisms in the remediation of wastewater in floating treatment wetlands: A review. Sustainability 2020, 12, 5559. [Google Scholar] [CrossRef]
- Raynaud, X.; Nunan, N. Spatial ecology of bacteria at the microscale in soil. PLoS ONE 2014, 9, e87217. [Google Scholar] [CrossRef] [PubMed]
- Wade, W. Unculturable bacteria-the uncharacterized organisms that cause oral infections. J. R. Soc. Med. 2002, 95, 81–83. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gomez-Valero, L.; Buchrieser, C. Metagenomic approaches in microbial ecology: An update on whole-genome and marker gene sequencing analyses. Microb. Genom. 2020, 6, e000409. [Google Scholar] [CrossRef]
- Sonthiphand, P.; Ruangroengkulrith, S.; Mhuantong, W.; Charoensawan, V.; Chotpantarat, S.; Boonkaewwan, S. Metagenomic insights into microbial diversity in a groundwater basin impacted by a variety of anthropogenic activities. Environ. Sci. Pollut. Res. 2019, 26, 26765–26781. [Google Scholar] [CrossRef] [PubMed]
- James, G.L.; Latif, M.T.; Isa, M.N.M.; Abu Bakar, M.F.; Yusuf, N.Y.M.; Broughton, W.; Murad, A.M.; Abu Bakar, F.D. Metagenomic datasets of air samples collected during episodes of severe smoke-haze in Malaysia. Data Brief 2021, 36, 107124. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Rakan, M.A.; Ibrahim, A.A. The role of metagenomic approaches in the analysis of microbial community in extreme environment. In Life in Extreme Environments; Afef, N., Ed.; IntechOpen: London, UK, 2023. [Google Scholar]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Nthaba, B.; Shemang, E.; Gareutlwane, O.; Molwalefhe, L. Application of electrical resistivity imaging (ERI) to investigate an oil contaminated experimental site. Environ. Eng. Manag. J. 2019, 18, 2623–2634. [Google Scholar] [CrossRef]
- Li, P.; Karunanidhi, D.; Subramani, T.; Srinivasamoorthy, K. Sources and consequences of groundwater contamination. Arch. Environ. Contam. Toxicol. 2021, 80, 1–10. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Aguado-Norese, C.; Cárdenas, V.; Gaete, A.; Mandakovic, D.; Vasquez-Dean, J.; Hodar, C.; Pfeiffer, M.; Gonzalez, M. Topsoil and subsoil bacterial community assemblies across different drainage conditions in a mountain environment. Biol. Res. 2023, 56, 35. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Shahi, A.; Aydin, S.; Ince, B.; Ince, O. Reconstruction of bacterial community structure and variation for enhanced petroleum hydrocarbons degradation through biostimulation of oil contaminated soil. Chem. Eng. J. 2016, 306, 60–66. [Google Scholar] [CrossRef]
- Liu, H.; Gao, H.; Wu, M.; Ma, C.; Wu, J.; Ye, X. Distribution characteristics of bacterial communities and hydrocarbon degradation dynamics during the remediation of petroleum-contaminated soil by enhancing moisture content. Microb. Ecol. 2020, 80, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.P.; Jani, K.; Sharma, A.; Jha, D.K. Bacterial communities and their bioremediation capabilities in oil-contaminated agricultural soils. Environ. Monit. Assess. 2021, 194, 9. [Google Scholar] [CrossRef]
- Sharma, S.K.; Ramesh, A.; Sharma, M.P.; Joshi, O.P.; Govaerts, B.; Steenwerth, K.L.; Karlen, D.L. Microbial community structure and diversity as indicators for evaluating soil quality. In Biodiversity, Biofuels, Agroforestry and Conservation Agriculture; Lichtfouse, E., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 2011; pp. 317–358. [Google Scholar]
- Mukjang, N.; Chitov, T.; Mhuantong, W.; Champreda, V.; Pathom-Aree, W.; Sattayawat, P.; Bovonsombut, S. Bacterial communities associated with crude oil bioremediation through composting approaches with indigenous bacterial isolate. Life 2022, 12, 1712. [Google Scholar] [CrossRef]
- Biswas, T.; Banerjee, S.; Saha, A.; Bhattacharya, A.; Chanda, C.; Gantayet, L.M.; Bhadury, P.; Chaudhuri, S.R. Bacterial consortium based petrochemical wastewater treatment: From strain isolation to industrial effluent treatment. Environ. Adv. 2022, 7, 100132. [Google Scholar] [CrossRef]
- Dey, P.; Das, R.; Kazy, S.K. Benzene biodegradation during growth by aerococcus sp. isolated from oil sludge. In Urban Ecology, Water Quality and Climate Change; Sarma, A.K., Singh, V.P., Bhattacharjya, R.K., Kartha, S.A., Eds.; Springer International Publishing: Cham, Germany, 2018; pp. 305–319. [Google Scholar]
- Huang, J.; Weng, L.; Zhang, X.; Long, K.; An, X.; Bao, J.; Wu, H.; Zhou, X.; Zhang, S.; Claesen, J. Trypoxylus dichotomus gut bacteria provides an effective system for bamboo lignocellulose degradation. Microbiol. Spectr. 2022, 10, e0214722. [Google Scholar] [CrossRef]
- Jacoline, G.; Hornung, B.; Ritari, J.; Paulin, L.; Rijkers, G.T.; Schaap, P.J.; de Vos, W.M.; Smidt, H. A comparative and functional genomics analysis of the genus Romboutsia provides insight into adaptation to an intestinal lifestyle. BioRxiv 2019, 845511. [Google Scholar] [CrossRef]
- Zuo, W.F.; Pang, Q.; Yao, L.-P.; Zhang, Y.; Peng, C.; Huang, W.; Han, B. Gut microbiota: A magical multifunctional target regulated by medicine food homology species. J. Adv. Res. 2023, 52, 151–170. [Google Scholar] [CrossRef]
- Li, L.L.; Wang, Y.T.; Zhu, L.M.; Liu, Z.Y.; Ye, C.Q.; Qin, S. Inulin with different degrees of polymerization protects against diet-induced endotoxemia and inflammation in association with gut microbiota regulation in mice. Sci. Rep. 2020, 10, 978. [Google Scholar] [CrossRef]
- Li, C.J.; Zhang, Z.; Zhan, P.-C.; Lv, A.-P.; Li, P.-P.; Liu, L.; Li, W.-J.; Yang, L.-L.; Zhi, X.-Y. Comparative genomic analysis and proposal of Clostridium yunnanense sp. nov., Clostridium rhizosphaerae sp. nov., and Clostridium paridis sp. nov., three novel Clostridium sensu stricto endophytes with diverse capabilities of acetic acid and ethanol production. Anaerobe 2023, 79, 102686. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Cao, X.; Huang, R.; Zeng, J.; Wu, Q.L. Variation of bacterial communities in water and sediments during the decomposition of microcystis biomass. PLoS ONE 2017, 12, e0176397. [Google Scholar] [CrossRef]
- Dréan, P.; McAuley, C.M.; Moore, S.C.; Fegan, N.; Fox, E.M. Characterization of the spore-forming Bacillus cereus sensu lato group and Clostridium perfringens bacteria isolated from the Australian dairy farm environment. BMC Microbiol. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Bodor, A.; Bounedjoum, N.; Vincze, G.E.; Kis, Á.E.; Laczi, K.; Bende, G.; Szilágyi, Á.; Kovács, T.; Perei, K.; Rákhely, G. Challenges of unculturable bacteria: Environmental perspectives. Rev. Environ. Sci. Bio/Technol. 2020, 19, 1–22. [Google Scholar] [CrossRef]
- Huber, K.J.; Geppert, A.M.; Wanner, G.; Fösel, B.U.; Wüst, P.K.; Overmann, J. The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 2971–2979. [Google Scholar] [CrossRef]
- Chiba, A.; Uchida, Y.; Kublik, S.; Vestergaard, G.; Buegger, F.; Schloter, M.; Schulz, S. Soil bacterial diversity is positively correlated with decomposition rates during early phases of maize litter decomposition. Microorganisms 2021, 9, 357. [Google Scholar] [CrossRef]
- Truskewycz, A.; Gundry, T.D.; Khudur, L.S.; Kolobaric, A.; Taha, M.; Aburto-Medina, A.; Shahsavari, E. Petroleum hydrocarbon contamination in terrestrial ecosystems—Fate and microbial responses. Molecules 2019, 24, 3400. [Google Scholar] [CrossRef]
- Arora, J.; Kumari, A.; Ranjan, A.; Rajput, V.D.; Shende, S.; Prazdnova, E.V.E.; Jindal, T. Microbial adaptation in different extreme environmental conditions and its usefulness in differently polluted soil. In Extremophiles for Sustainable Agriculture and Soil Health Improvement; Springer Nature: Cham, Switzerland, 2024; pp. 47–62. [Google Scholar]
- Joye, S.B.; Kleindienst, S.; Gilbert, J.A.; Handley, K.M.; Weisenhorn, P.; Overholt, W.A.; Kostka, J.E. Responses of microbial communities to hydrocarbon exposures. Oceanography 2016, 29, 136–149. [Google Scholar] [CrossRef]
- Dhar, K.; Panneerselvan, L.; Venkateswarlu, K.; Megharaj, M. Efficient bioremediation of PAHs-contaminated soils by a methylotrophic enrichment culture. Biodegradation 2022, 33, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Giri, D.D.; Singh, S.K.; Giri, A.; Dwivedi, H.; Kumar, A. Bioremediation potential of methylotrophic bacteria. In Microbe Mediated Remediation of Environmental Contaminants; Kumar, A., Singh, V.K., Singh, P., Mishra, V.K., Eds.; Woodhead Publishing: Cambridge, UK, 2021; pp. 199–207. [Google Scholar]
- Leducq, J.B.; Sneddon, D.; Santos, M.; Condrain-Morel, D.; Bourret, G.; Martinez-Gomez, N.C.; A Lee, J.; A Foster, J.; Stolyar, S.; Shapiro, B.J.; et al. Comprehensive phylogenomics of Methylobacterium reveals four evolutionary distinct groups and underappreciated phyllosphere diversity. Genome Biol. Evol. 2022, 14, evac123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, M.Y.; Khan, N.; Tan, L.L.; Yang, S. Potentials, utilization, and bioengineering of plant growth-promoting Methylobacterium for sustainable agriculture. Sustainability 2021, 13, 3941. [Google Scholar] [CrossRef]
- Rojas-Gätjens, D.; Fuentes-Schweizer, P.; Rojas-Jiménez, K.; Pérez-Pantoja, D.; Avendaño, R.; Alpízar, R.; Coronado-Ruíz, C.; Chavarría, M. Methylotrophs and hydrocarbon-degrading bacteria are key players in the microbial community of an abandoned century-old oil exploration well. Microb. Ecol. 2022, 83, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Le, H.T.Q.; Mai, D.H.A.; Na, J.G.; Lee, E.Y. Development of Methylorubrum extorquens AM1 as a promising platform strain for enhanced violacein production from co-utilization of methanol and acetate. Metab. Eng. 2022, 72, 150–160. [Google Scholar] [CrossRef]
- Wongdee, J.; Piromyou, P.; Songwattana, P.; Greetatorn, T.; Teaumroong, N.; Boonkerd, N.; Giraud, E.; Nouwen, N.; Tittabutr, P. Role of two RpoN in Bradyrhizobium sp. strain DOA9 in symbiosis and free-living growth. Front. Microbiol. 2023, 14, 1131860. [Google Scholar] [CrossRef] [PubMed]
- Breitkreuz, C.; Buscot, F.; Tarkka, M.; Reitz, T. Shifts between and among populations of wheat rhizosphere pseudomonas, streptomyces and phyllobacterium suggest consistent phosphate mobilization at different wheat growth stages under abiotic stress. Front. Microbiol. 2020, 10, 3109. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahube, T.O.; Selvarajan, R.; Nduna, B.; Nthaba, B.; Molwalefhe, L.; Shemang, E. Evaluation of Bacterial Population Changes and Ecological Dynamics in Oil-Impacted Soils Using 16S rRNA Amplicon Sequencing. Biology 2025, 14, 1074. https://doi.org/10.3390/biology14081074
Rahube TO, Selvarajan R, Nduna B, Nthaba B, Molwalefhe L, Shemang E. Evaluation of Bacterial Population Changes and Ecological Dynamics in Oil-Impacted Soils Using 16S rRNA Amplicon Sequencing. Biology. 2025; 14(8):1074. https://doi.org/10.3390/biology14081074
Chicago/Turabian StyleRahube, Teddie O., Ramganesh Selvarajan, Batendi Nduna, Bokani Nthaba, Loago Molwalefhe, and Elisha Shemang. 2025. "Evaluation of Bacterial Population Changes and Ecological Dynamics in Oil-Impacted Soils Using 16S rRNA Amplicon Sequencing" Biology 14, no. 8: 1074. https://doi.org/10.3390/biology14081074
APA StyleRahube, T. O., Selvarajan, R., Nduna, B., Nthaba, B., Molwalefhe, L., & Shemang, E. (2025). Evaluation of Bacterial Population Changes and Ecological Dynamics in Oil-Impacted Soils Using 16S rRNA Amplicon Sequencing. Biology, 14(8), 1074. https://doi.org/10.3390/biology14081074







