Genome-Wide Genetic Diversity and Population Structure of Sillago sinica (Perciformes, Sillaginidae) from the Coastal Waters of China: Implications for Phylogeographic Pattern and Fishery Management
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
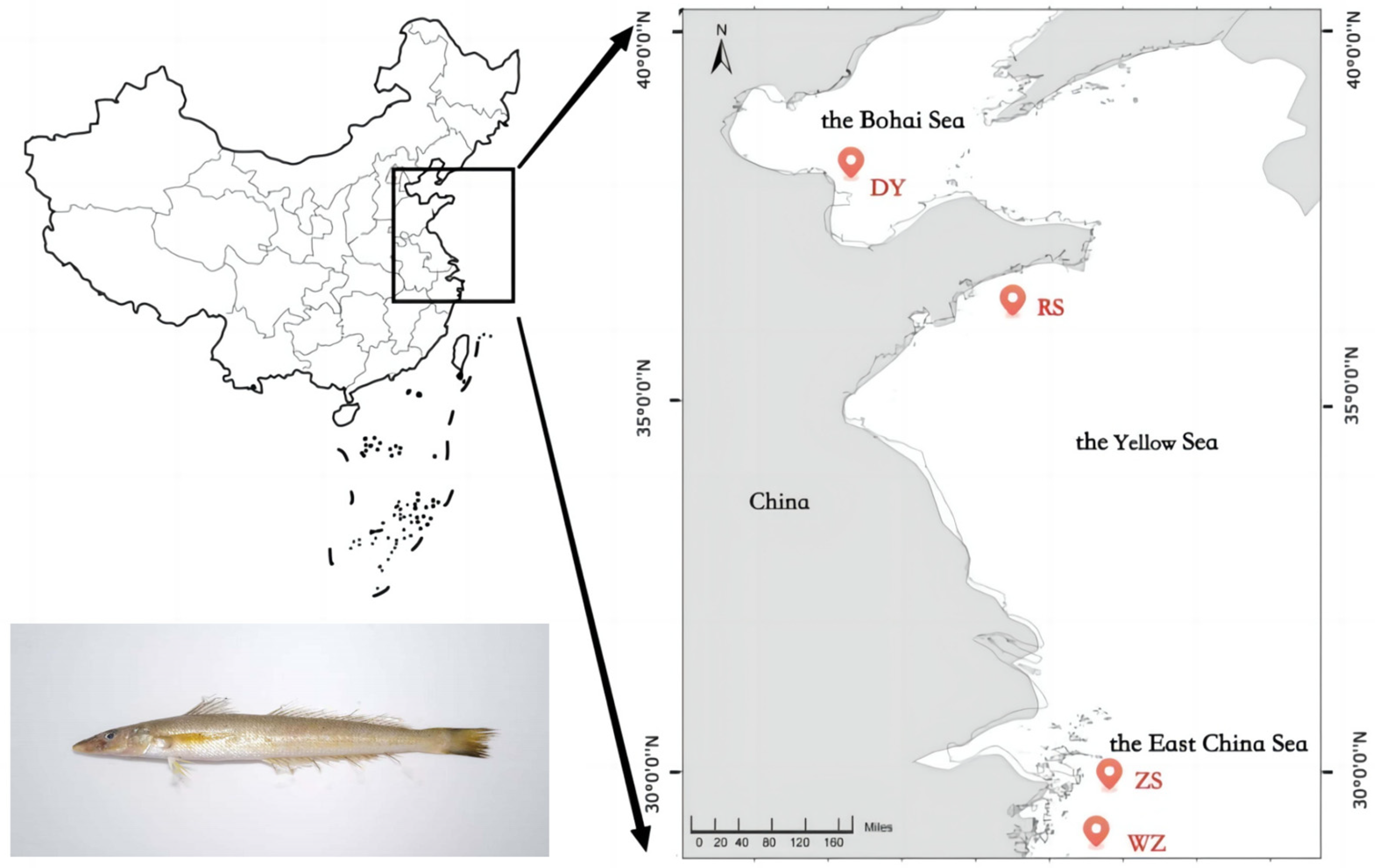
2.1. Sample Collection and Genomic DNA Extraction
2.2. Library Construction and High-Throughput Sequencing
2.3. Sequencing Data Filtering and Reads Mapping
2.4. SNP Calling and Annotation
2.5. Genetic Diversity, Population Structure, and Genetic Differentiation Analysis
2.6. Linkage Disequilibrium and Effective Population Size
3. Results
3.1. Sequencing Data Statistics and Alignment
3.2. Screening and Annotation of SNPs
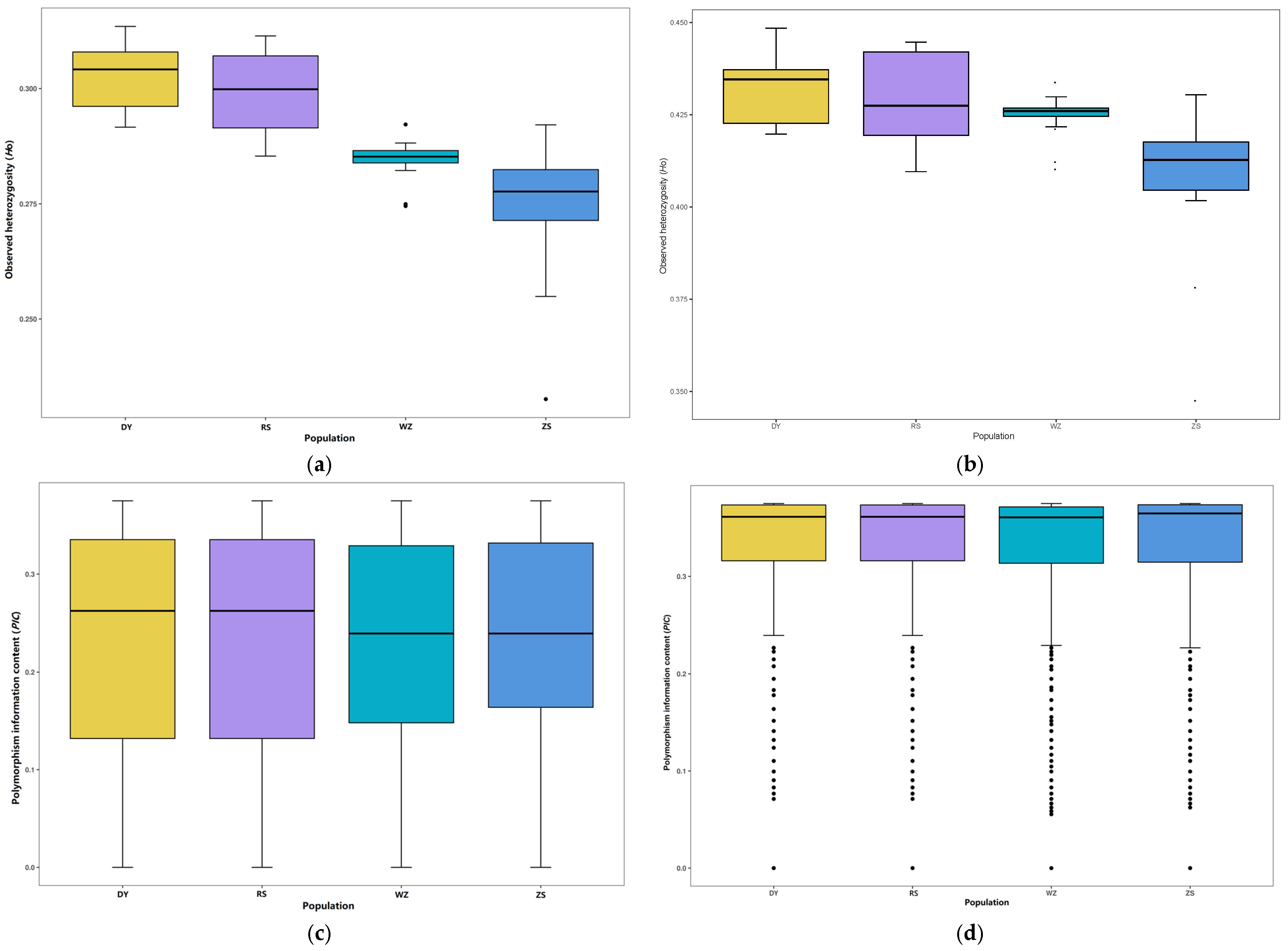
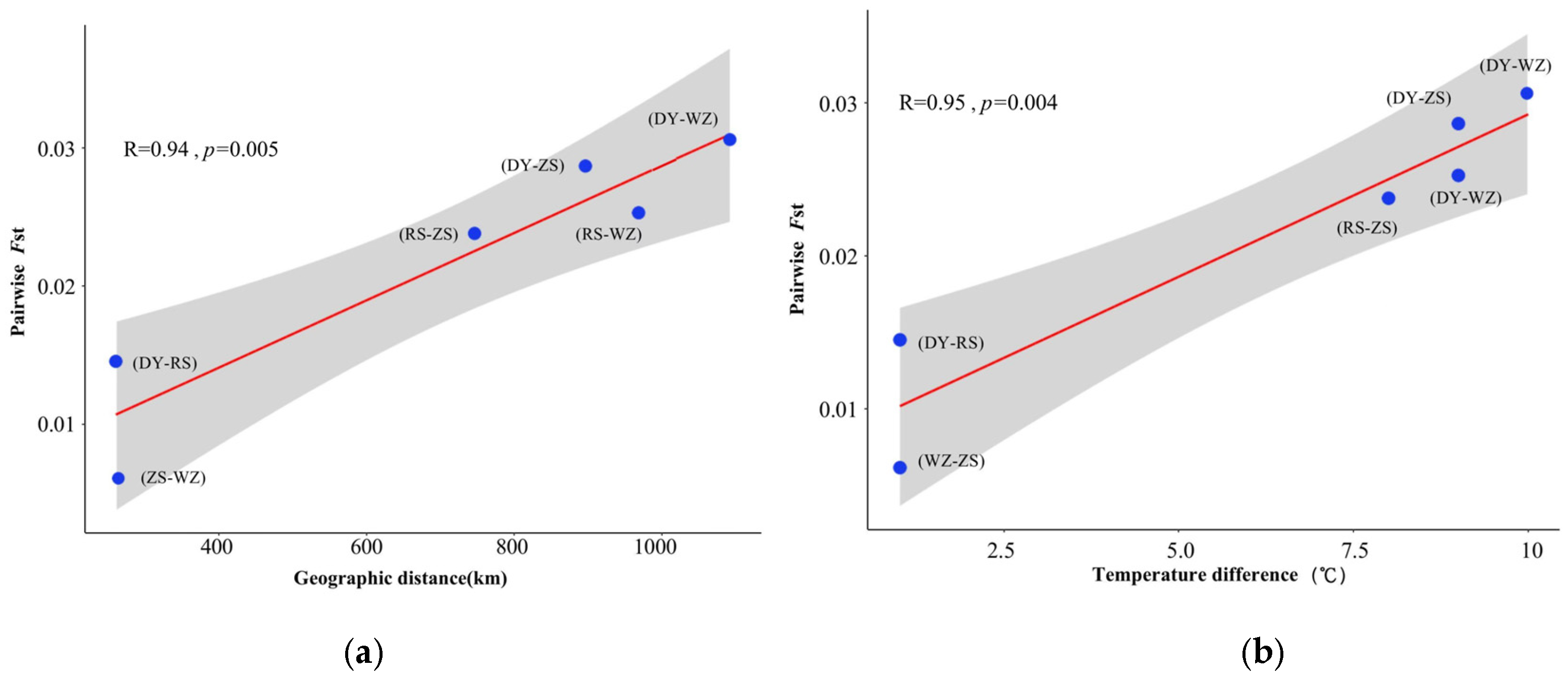
3.3. Genetic Diversity and Differentiation
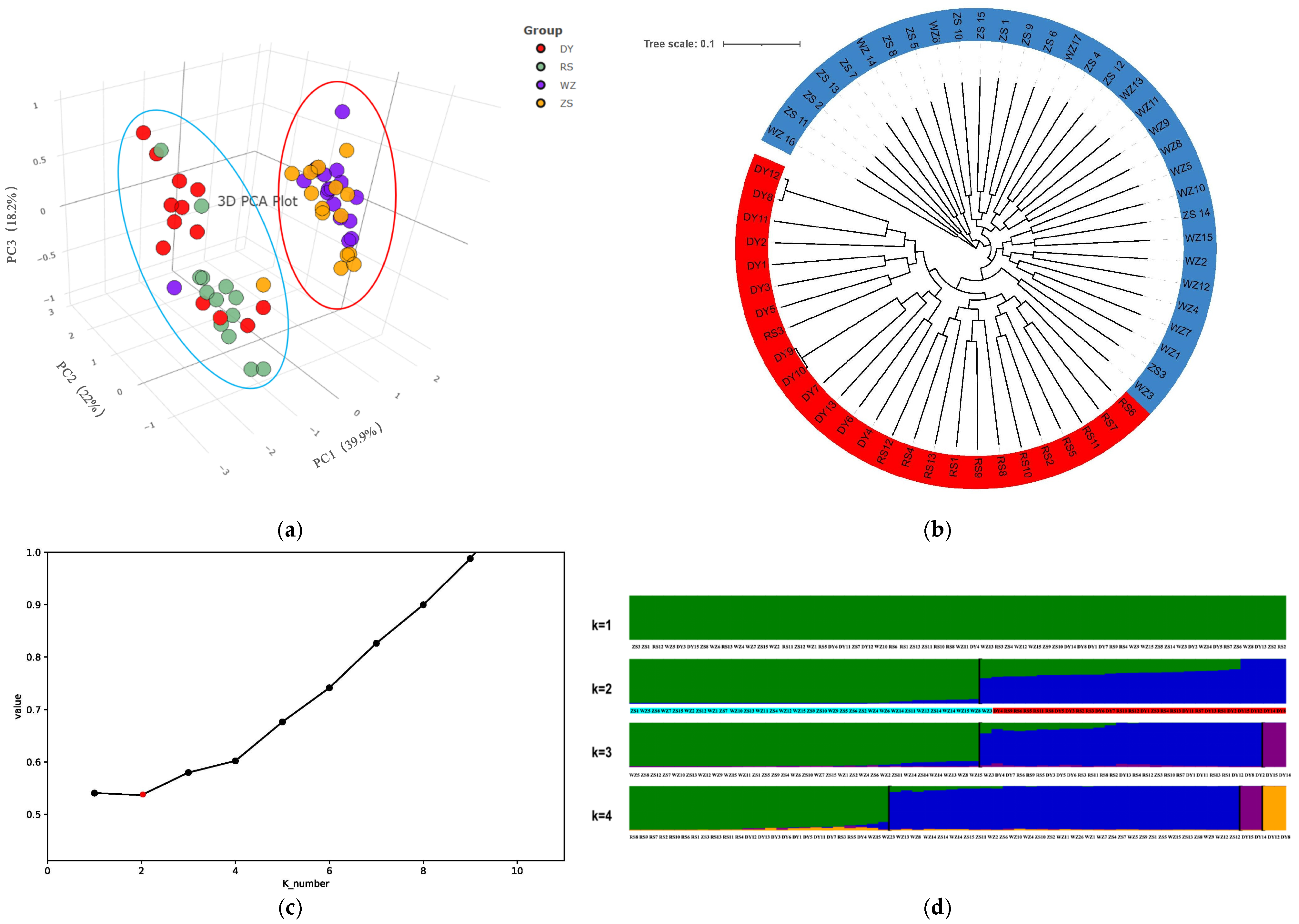
3.4. The Hierarchical Genetic Structure and Phylogenetic Relationships
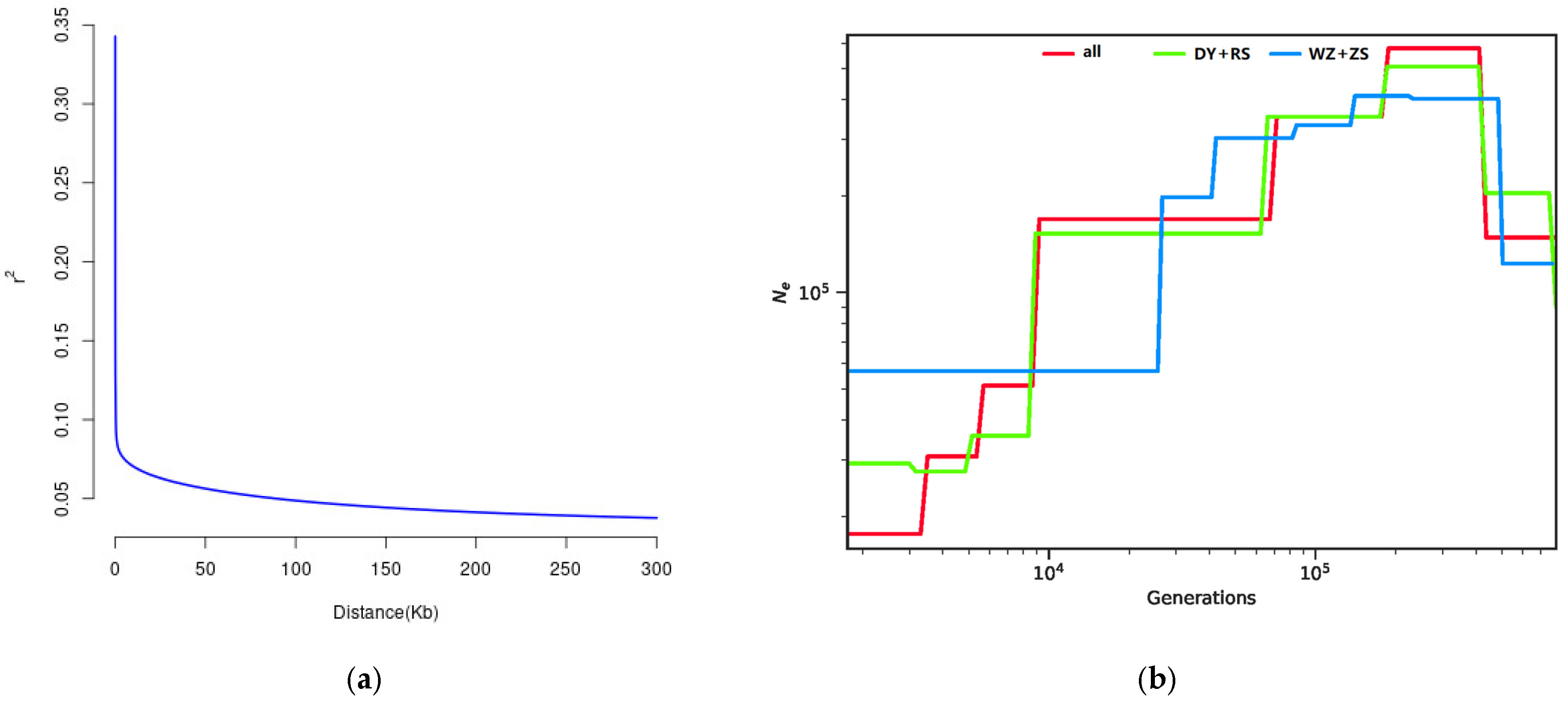
3.5. Demographic History Inference
4. Discussion
4.1. Genetic Diversity and Variation
4.2. Genetic Differentiation Caused by Historical and Contemporary Population Dynamics
4.3. Fishery Resource Protection and Utilization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mckay, R.J. Sillaginid fishes of the world (family Sillaginidae): An annotated and illustrated catalogue of the Sillago, smelt or Indo-Pacific whiting species known to date. In FAO Fisheries Synopsis; Food and Agriculture Organization of the United Nations: Rome, Italy, 1992. [Google Scholar]
- Konishi, H.; Nakabo, T. Color Guide to the Japanese Fishes for Sportfishermen; Enterbrain Inc.: Tokyo, Japan, 2007; pp. 400–402. [Google Scholar]
- Gao, T.X.; Ji, D.P.; Xiao, Y.S.; Xue, T.Q.; Yanagimoto, T.; Setoguma, T. Description and DNA barcoding of a new Sillago species, Sillago sinica (Perciformes: Sillaginidae), from coastal waters of China. Zool. Stud. 2011, 50, 254–263. [Google Scholar]
- Bae, S.E.; Kwun, H.J.; Kim, J.K.; Kweon, S.M.; Kang, C.B. New record of Sillago sinica (Pisces: Sillaginidae) in Korean waters, and re-identification of Sillago parvisquamis previously reported from Korea as S. sinica. Anim. Syst. Evol. Divers. 2013, 29, 288–293. [Google Scholar] [CrossRef]
- Chen, D.G.; Zhang, M.Z. Marine Fishes of China; China Ocean University Press: Qingdao, China, 2015; pp. 1060–1064. (In Chinese) [Google Scholar]
- Zhang, H.Y. Study on Morphology and Genetics of Sillago sinica. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2013. (In Chinese). [Google Scholar]
- Wang, L.Y. Population Genetics for Chinese Sillago and Japanese Sillago Based on Microsatellite Markers. Master’s Thesis, Ocean University of China, Qingdao, China, 2014. (In Chinese). [Google Scholar]
- Zhang, S.Q.; Xiao, J.G.; Deng, Z.C.; Han, Z.Q.; Gao, T.X. Preliminary study on the reproductive biology of Sillago sinica from the Wenzhou coastal waters. J. Zhejiang Ocean Univ. 2016, 35, 493–497. (In Chinese) [Google Scholar]
- Zhao, X.; Zheng, T.L.; Gao, T.X.; Song, N. Whole-genome resequencing reveals genetic diversity and selection signals in warm temperate and subtropical Sillago sinica populations. BMC Genom. 2023, 24, 547. [Google Scholar] [CrossRef]
- Yang, T.Y.; Xiao, P.Y.; Jiang, X.Z.; Zhang, Q.Y.; Zhao, Y. Otolith morphometrics and variations between two populations of Sillago sinica (Perciformes, Sillaginidae) in the East China Sea and the Yellow Sea. Thalassas 2024, 40, 1007–1017. [Google Scholar] [CrossRef]
- Tamaki, K.; Honza, E. Global tectonics and formation of marginal basins: Role of the western Pacific. Episodes 1991, 14, 224–230. [Google Scholar] [CrossRef]
- Wang, P.X. Response of western Pacific marginal seas to glacial cycles: Paleoceanographic and sedimentological features. Mar. Geol. 1999, 156, 5–39. [Google Scholar] [CrossRef]
- Ni, G.; Li, Q.; Kong, L.F.; Yu, H. Comparative phylogeography in marginal seas of the northwestern Pacific. Mol. Ecol. 2014, 23, 534–548. [Google Scholar] [CrossRef]
- Xue, T.Q. Study on Morphology and Genetics of Some Sillaginidae Species. Master’s Thesis, Ocean University of China, Qingdao, China, 2010. (In Chinese). [Google Scholar]
- Zhang, H.Y.; Gao, T.X.; Li, J.; Pan, X.Z.; Zhang, H.; Zhang, Y. Preliminary biological study on morphology of Sillago sinica populations. J. Shanghai Ocean Univ. 2013, 22, 17–22. (In Chinese) [Google Scholar]
- Gao, T.X.; Yang, T.Y.; Yanagimoto, T.; Xiao, Y.S. Levels and patterns of genetic variation in Japanese whiting (Sillago japonica) based on mitochondrial DNA control region. Mitochondrial DNA Part A 2019, 30, 172–183. [Google Scholar] [CrossRef]
- Yang, T.Y.; Gao, T.X.; Meng, W.; Jiang, Y.L. Genome-wide population structure and genetic diversity of Japanese whiting (Sillago japonica) inferred from genotyping-by-sequencing (GBS): Implications for fisheries management. Fish. Res. 2020, 225, 105501. [Google Scholar] [CrossRef]
- Han, Z.Q.; Guo, X.Y.; Liu, Q.; Liu, S.S.; Zhang, Z.X.; Xiao, S.J.; Gao, T.X. Whole-genome resequencing of Japanese whiting (Sillago japonica) provide insights into local adaptations. Zool. Res. 2021, 42, 548–561. [Google Scholar] [CrossRef]
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; Neigel, J.E.; Reeb, A.C.; Saunders, N.C. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Ann. Rev. Ecol. Syst. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Avise, J.C. Phylogeography: Retrospect and prospect. J. Biogeogr. 2009, 3, 3–15. [Google Scholar] [CrossRef]
- Nikula, R.; Spencer, H.G.; Waters, J.M. Passive rafting is a powerful driver of transoceanic gene flow. Biol. Lett. 2013, 9, 20120821. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.M.; Han, Z.Q.; Gao, T.X. AFLP markers suggest low population genetic differentiation of the black rockfish Sebastes schlegelii. Biochem. Syst. Ecol. 2015, 59, 325–330. [Google Scholar] [CrossRef]
- Liu, B.J.; Zhang, B.D.; Xue, D.X.; Gao, T.X.; Liu, J.X. Population structure and adaptive divergence in a high gene flow marine fish: The small yellow croaker (Larimichthys polyactis). PLoS ONE 2016, 11, e0154020. [Google Scholar] [CrossRef]
- Yi, M.R.; Hsu, K.C.; Wang, J.X.; Feng, B.; Lin, H.D.; Yan, Y.R. Genetic structure and diversity of the yellowbelly threadfin bream Nemipterus bathybius in the northern South China Sea. Diversity 2021, 13, 324. [Google Scholar] [CrossRef]
- Song, C.Y.; Tu, Z.; Song, N. Discordant patterns of genetic variation between mitochondrial and microsatellite markers in Acanthogobius ommaturus across the coastal areas of China. Acta Oceanol. Sin. 2023, 42, 72–80. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, S.Y.; Li, D.L. Recent advances in population genomics of marine fishes. Agri. Biotech. 2017, 6, 36–40. (In Chinese) [Google Scholar]
- Fuentes-Pardo, A.P.; Ruzzante, D.E. Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol. 2017, 26, 5369–5406. [Google Scholar] [CrossRef]
- Huang, X.H.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.P.; Fan, D.L.; Weng, Q.J.; Huang, T.; et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Kon, T.; Pei, L.Y.; Ichikawa, R.; Chen, C.Y.; Wang, P.; Takemura, I.; Ye, Y.Y.; Yan, X.J.; Guo, B.Y.; Li, W.Y.; et al. Whole-genome resequencing of large yellow croaker (Larimichthys crocea) reveals the population structure and signatures of environmental adaptation. Sci. Rep. 2021, 11, 11235. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Liu, H.Y.; Zhai, D.D.; Duan, X.B.; Tian, H.W.; Chen, D.Q. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing. Biodivers. Sci. 2023, 31, 22391. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Liu, X.M.; Huang, W.J.; Wang, Y.; Anwarullah, K.; Luo, L.F.; Gao, Z.X. Whole-genome resequencing reveals genetic diversity and signatures of selection in mono-female grass carp (Ctenopharyngodon idella). Aquaculture 2023, 575, 739816. [Google Scholar] [CrossRef]
- Xu, S.Y.; Xiao, S.J.; Zhu, S.L.; Zeng, X.F.; Luo, J.; Liu, J.Q.; Gao, T.X.; Chen, N.S. A draft genome assembly of the Chinese sillago (Sillago sinica), the first reference genome for Sillaginidae fishes. GigaScience 2018, 7, giy108. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning—A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Cuddapah, S.; Barski, A.; Cui, K.R.; Schones, D.E.; Wang, Z.B.; Wei, G.; Zhao, K.J. Native Chromatin Preparation and Illumina/Solexa Library Construction. Cold Spring Harb. Protoc. 2009, 6, pdb.prot5237. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2001, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.Y.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; Depristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Xia, X.H. Maximum parsimony method in phylogenetics. In A Mathematical Primer of Molecular Phylogenetics, 1st ed.; Xia, X.H., Ed.; Apple Academic Press: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Jensen, J.L.; Bohonak, A.J.; Kelley, S.T. Isolation by distance, web service. BMC Genet. 2005, 6, 13. [Google Scholar] [CrossRef]
- Weir, B.S. Inferences about linkage disequilibrium. Biometrics 1979, 35, 235–254. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Charlesworth, B. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205. [Google Scholar] [CrossRef]
- Terhorst, J.; Kamm, J.A.; Song, Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2016, 49, 303–309. [Google Scholar] [CrossRef]
- Wilcox, R. Some multivariate methods. In Introduction to Robust Estimation and Hypothesis Testing, 3rd ed.; Wilcox, R., Ed.; Academic Press: New York, NY, USA, 2012; pp. 215–289. [Google Scholar] [CrossRef]
- Ardlie, K.G.; Kruglyak, L.; Seielstad, M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002, 3, 299. [Google Scholar] [CrossRef]
- McKay, S.D.; Schnabel, R.D.; Murdoch, B.M.; Matukumalli, L.K.; Aerts, J.; Coppieters, W.; Crews, D.; Neto, E.D.; Gill, C.A.; Gao, C.; et al. Whole genome linkage disequilibrium maps in cattle. BMC Genet. 2007, 8, 74. [Google Scholar] [CrossRef]
- Hoban, S.; Bruford, M.; Jackson, J.D.; Lopes-Fernandes, M.; Heuertz, M.; Hohenlohe, P.A.; Paz-Vinas, I.; Sjögren-Gulve, P.; Segelbacher, G.; Vernesi, C.; et al. Genetic diversity targets and indicators in the CBD post-2020 global biodiversity framework must be improved. Biol. Conserv. 2020, 248, 108654. [Google Scholar] [CrossRef]
- Li, Y.; Lou, F.R.; Song, P.Q.; Liu, S.G.; Siyal, F.K.; Lin, L.S. New perspective on the genetic structure and habitat adaptation of Pampus minor off the coast of China based on RAD-seq. Comp. Biochem. Physiol. D 2021, 39, 100865. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Huang, X.X.; Jiang, Y.L. Reveal the population genetic characteristics of Bombay duck (Harpadon nehereus) in coastal waters of China with genotyping-by-sequencing technique. J. Ocean Univ. China 2022, 21, 1373–1380. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, Y.F.; Li, J.S.; Zhang, K.; Jin, X.; Zheng, S.X.; Wang, Y.P.; Lü, Z.M.; Liu, L.Q.; Gong, L.; et al. New perspectives on the genetic structure of dotted gizzard shad (Konosirus punctatus) based on RAD-seq. Mar. Life Sci. Technol. 2024, 6, 50–67. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.X.; Yang, Q.L.; Zong, S.B.; Gao, T.X.; Liu, J.X. Genetic variation within and among range-wide populations of three ecotypes of the Japanese grenadier anchovy Coilia nasus with implications to its conservation and management. J. Ocean Limnol. 2020, 38, 851–861. [Google Scholar] [CrossRef]
- Naito, T.; Nakayama, K.; Takeshima, H.; Hashiguchi, Y.; Akita, T.; Yamasaki, Y.Y.; Mishina, T.; Takeshita, N.; Nagano, A.J.; Takahashi, H. The detailed population genetic structure of the rare endangered latid fish akame Lates japonicus with extremely low genetic diversity revealed from single-nucleotide polymorphisms. Conserv. Genet. 2023, 24, 523–535. [Google Scholar] [CrossRef]
- Kimura, M. The neutral theory of molecular evolution. Sci. Am. 1979, 241, 98–126. [Google Scholar] [CrossRef]
- Hague, M.T.J.; Routman, E.J. Does population size affect genetic diversity? A test with sympatric lizard species. Heredity 2016, 116, 92–98. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Berry, O.; Ryman, N. So long to genetic diversity, and thanks for all the fish. Mol. Ecol. 2014, 23, 23–25. [Google Scholar] [CrossRef]
- Ni, G.; Kern, E.; Dong, Y.W.; Li, Q.; Park, J.K. More than meets the eye: The barrier effect of the Yangtze River outflow. Mol. Ecol. 2017, 26, 4591–4602. [Google Scholar] [CrossRef] [PubMed]
- Mitsuzawa, K.; Holloway, G. Characteristics of deep currents along trenches in the northwest Pacific. J. Geophys. Res. Ocean. 1998, 103, 13085–13092. [Google Scholar] [CrossRef]
- Sandel, B.; Arge, L.; Dalsgaard, B.; Davies, R.G.; Gaston, K.J.; Sutherland, W.J.; Svenning, J.C. The influence of late Quaternary climate-change velocity on species endemism. Science 2011, 334, 660–664. [Google Scholar] [CrossRef]
- He, L.J.; Zhang, A.B.; Zhu, C.D.; Weese, D.; Qiao, Z.G. Phylogeography of the mud crab (Scylla serrata) in the Indo-West Pacific reappraised from mitochondrial molecular and oceanographic clues: Transoceanic dispersal and coastal sequential colonization. Mar. Ecol. 2011, 32, 52–64. [Google Scholar] [CrossRef]
- Wang, J.; Tsang, L.M.; Dong, Y.W. Causations of phylogeographic barrier of some rocky shore species along the Chinese coastline. BMC Evol. Biol. 2015, 15, 114. [Google Scholar] [CrossRef]
- Xiao, Y.S.; Ma, D.Y.; Dai, M.; Liu, Q.H.; Xiao, Z.Z.; Li, J.; Xu, S.H.; Cheng, Q.Q. The impact of Yangtze River discharge on the genetic structure of a population of the rock bream, Oplegnathus fasciatus. Mar. Biol. Res. 2016, 12, 426–434. [Google Scholar] [CrossRef]
- Muhammad, F.; Chen, W.; Liu, L.Q.; Gong, L.; Du, X.; Shafi, M.; Lü, Z.M. Genetic structure of Amphioctopus fangsiao (Mollusca, Cephalopoda) in Chinese waters inferred from variation in three mtDNA genes (ATPase 6, ND2, and ND5). Hydrobiologia 2019, 838, 111–119. [Google Scholar] [CrossRef]
- Aitken, S.N.; Whitlock, M.C. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367–388. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef]
- Grant, K.M.; Rohling, E.J.; Ramsey, C.B.; Cheng, H.; Edwards, R.L.; Florindo, F.; Heslop, D.; Marra, F.; Roberts, A.P.; Tamisiea, M.E.; et al. Sea-level variability over five glacial cycles. Nat. Commun. 2014, 5, 5076. [Google Scholar] [CrossRef] [PubMed]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Leber, K.M.; Kitada, S.; Blankenship, H.L.; Svasand, T. Stock Enhancement and Sea Ranching: Developments, Pitfalls and Opportunities, 2nd ed.; John Wiley & Sons: New York, NY, USA, 2008. [Google Scholar]
- Rojanapittayakul, S.; Vatanakul, V.; Yashiro, R. Breeding and Larval Rearing of Sand Whiting, Sillago sihama; AGRIS, FAO: Rome, Italy, 1999. [Google Scholar]
- Huang, Y.; Du, T.; Huang, H.L. A study on artificial breeding of Sillago sihama Forskál. J. Guangdong Ocean Univ. 2013, 33, 15–21. (In Chinese) [Google Scholar]

| Population | Sampling Date | Number | Body Length (cm) | Body Weight (g) |
|---|---|---|---|---|
| DY | 16 September 2022 | 13 | 13.40–18.20 | 17.88–39.59 |
| RS | 7 October 2022 | 13 | 12.25–16.61 | 14.67–47.59 |
| WZ | 5 September 2022 | 17 | 15.62–20.30 | 20.45–67.38 |
| ZS | 20 September 2022 | 15 | 16.40–20.70 | 24.95–73.05 |
| Population | Number | Average Raw Reads | Average Clean Reads | Average Mapping Rate (%) | Average Coverage (%) | Average Sequencing Depth (×) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|---|
| DY | 13 | 83,880,884 | 81,054,146 | 99.27 | 98.80 | 22.11 | 96.30 | 90.84 |
| RS | 13 | 88,775,733 | 85,728,715 | 99.34 | 98.84 | 23.37 | 96.18 | 90.59 |
| WZ | 17 | 92,952,993 | 90,221,569 | 98.74 | 98.81 | 24.25 | 95.90 | 89.82 |
| ZS | 15 | 124,514,599 | 124,459,799 | 96.16 | 98.51 | 25.69 | 93.29 | 82.83 |
| Total | 58 | 98,145,791 | 96,014,497 | 98.33 | 98.74 | 23.96 | 95.38 | 88.41 |
| Category | Number | Percentage (%) | |
|---|---|---|---|
| Total | 7,409,691 | 100.00 | |
| Upstream (1 kp) | 285,912 | 3.86 | |
| Downstream (1 kb) | 246,947 | 3.33 | |
| Upstream/downstream (1 kb) | 47,169 | 0.64 | |
| 3′UTRs | 145,709 | 1.97 | |
| 5′UTRs | 46,762 | 0.63 | |
| Splicing | 1372 | 0.02 | |
| Intergenic | 2,849,683 | 38.46 | |
| Intronic | 3,461,507 | 46.71 | |
| Exonic | 324,630 | 4.38 | |
| Nonsynonymous SNV | 106,666 | 32.86 | |
| Synonymous SNV | 216,335 | 66.64 | |
| Stop gain | 1443 | 0.44 | |
| Stop loss | 186 | 0.06 | |
| DY | RS | WZ | ZS | |
|---|---|---|---|---|
| DY | ||||
| RS | 0.0145 ± 0.0215 (0.0145 ± 0.0228) | |||
| WZ | 0.0306 ± 0.0293 * (0.0291 ± 0.0307) * | 0.0253 ± 0.0263 (0.0256 ± 0.0302) | ||
| ZS | 0.0287 ± 0.0275 (0.0287 ± 0.0310) | 0.0238 ± 0.0256 (0.0246 ± 0.0295) | 0.0061 ± 0.0124 (0.0077 ± 0.0164) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, T.; Sun, Y.; Xiao, P. Genome-Wide Genetic Diversity and Population Structure of Sillago sinica (Perciformes, Sillaginidae) from the Coastal Waters of China: Implications for Phylogeographic Pattern and Fishery Management. Biology 2025, 14, 1329. https://doi.org/10.3390/biology14101329
Yang T, Sun Y, Xiao P. Genome-Wide Genetic Diversity and Population Structure of Sillago sinica (Perciformes, Sillaginidae) from the Coastal Waters of China: Implications for Phylogeographic Pattern and Fishery Management. Biology. 2025; 14(10):1329. https://doi.org/10.3390/biology14101329
Chicago/Turabian StyleYang, Tianyan, Yan Sun, and Peiyi Xiao. 2025. "Genome-Wide Genetic Diversity and Population Structure of Sillago sinica (Perciformes, Sillaginidae) from the Coastal Waters of China: Implications for Phylogeographic Pattern and Fishery Management" Biology 14, no. 10: 1329. https://doi.org/10.3390/biology14101329
APA StyleYang, T., Sun, Y., & Xiao, P. (2025). Genome-Wide Genetic Diversity and Population Structure of Sillago sinica (Perciformes, Sillaginidae) from the Coastal Waters of China: Implications for Phylogeographic Pattern and Fishery Management. Biology, 14(10), 1329. https://doi.org/10.3390/biology14101329





