HemN2 Regulates the Virulence of Pseudomonas donghuensis HYS through 7-Hydroxytropolone Synthesis and Oxidative Stress
Abstract
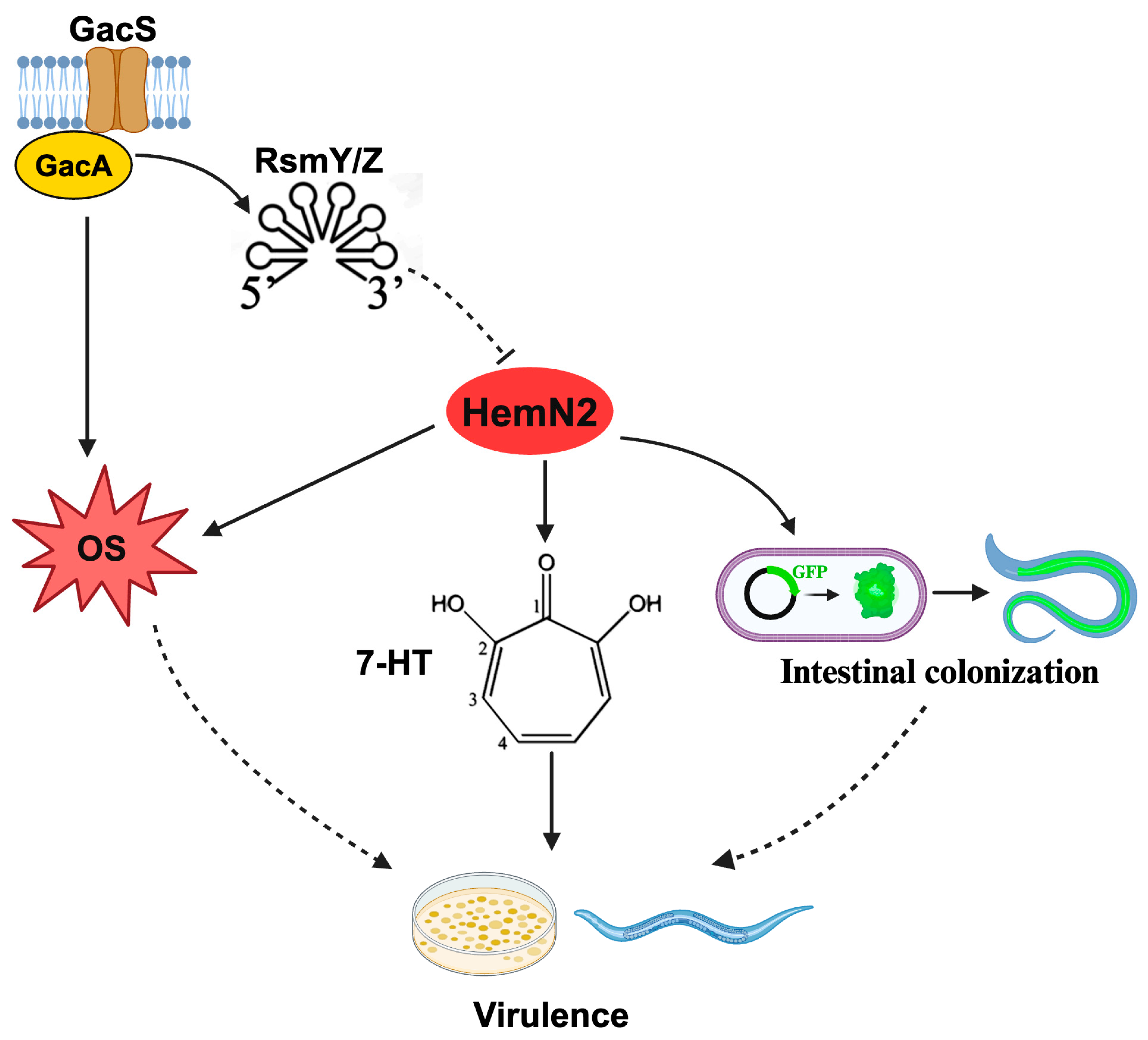
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, C. elegans, Zebrafish, and Growth Conditions
2.2. DNA Manipulation and Plasmid Construction
2.3. Slow-Killing Assay
2.4. Growth Curve Analysis
2.5. Statistics of Pharyngeal Pump Rate for C. elegans
2.6. Analysis of Bacterial Colonization of the C. elegans Gut
2.7. RNA Extraction and RT-qPCR
2.8. Quantification of 7-HT Siderophore Production
2.9. Quantification of Total Antioxidant Capacity
2.10. Quantification of Nitrite and Nitrate
2.11. P. donghuensis HYS Immersion Infection of Zebrafish Larvae
2.12. Statistical Analysis
3. Results
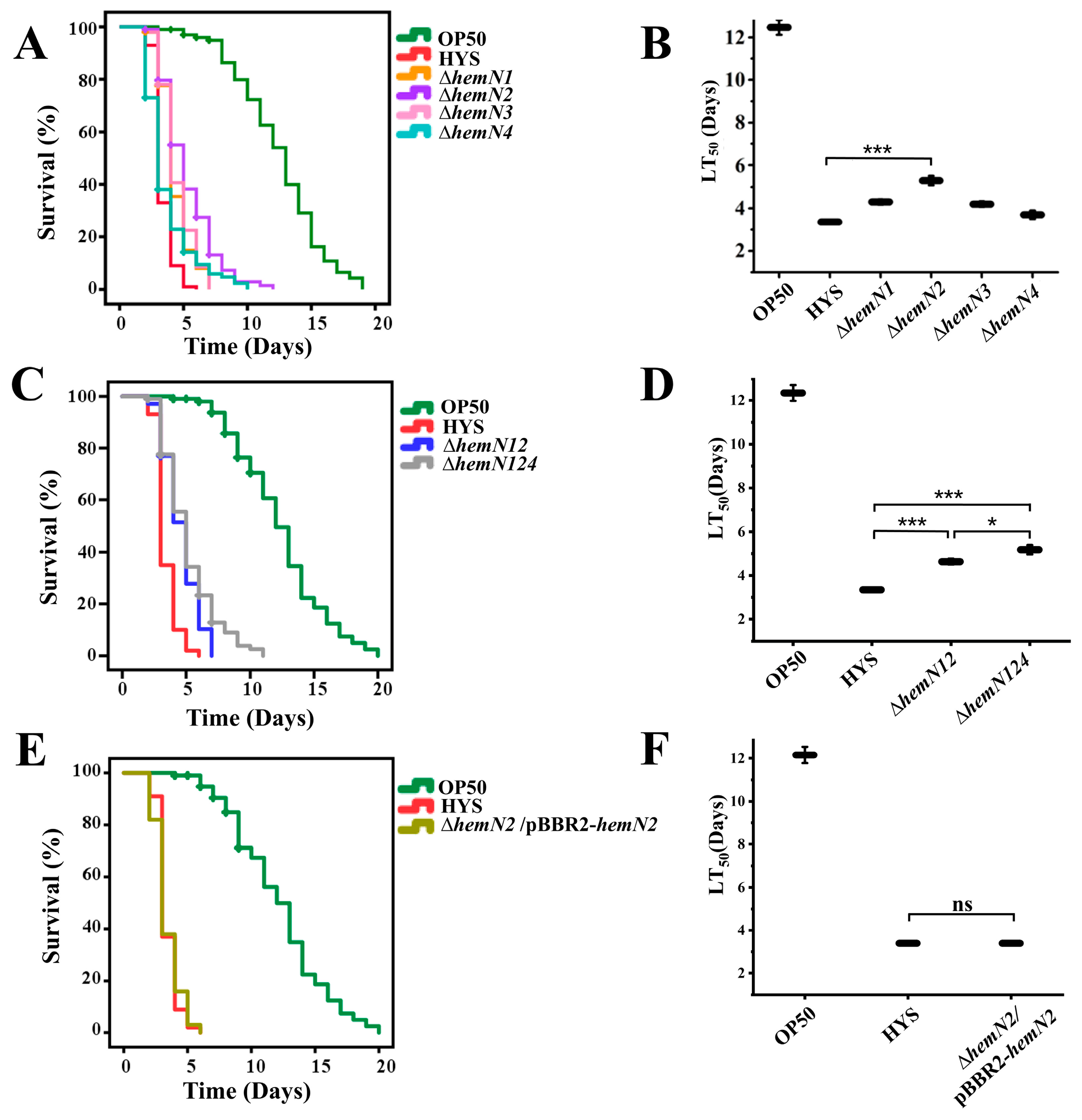
3.1. HemN Participates in the Virulence of P. donghuensis HYS to C. elegans
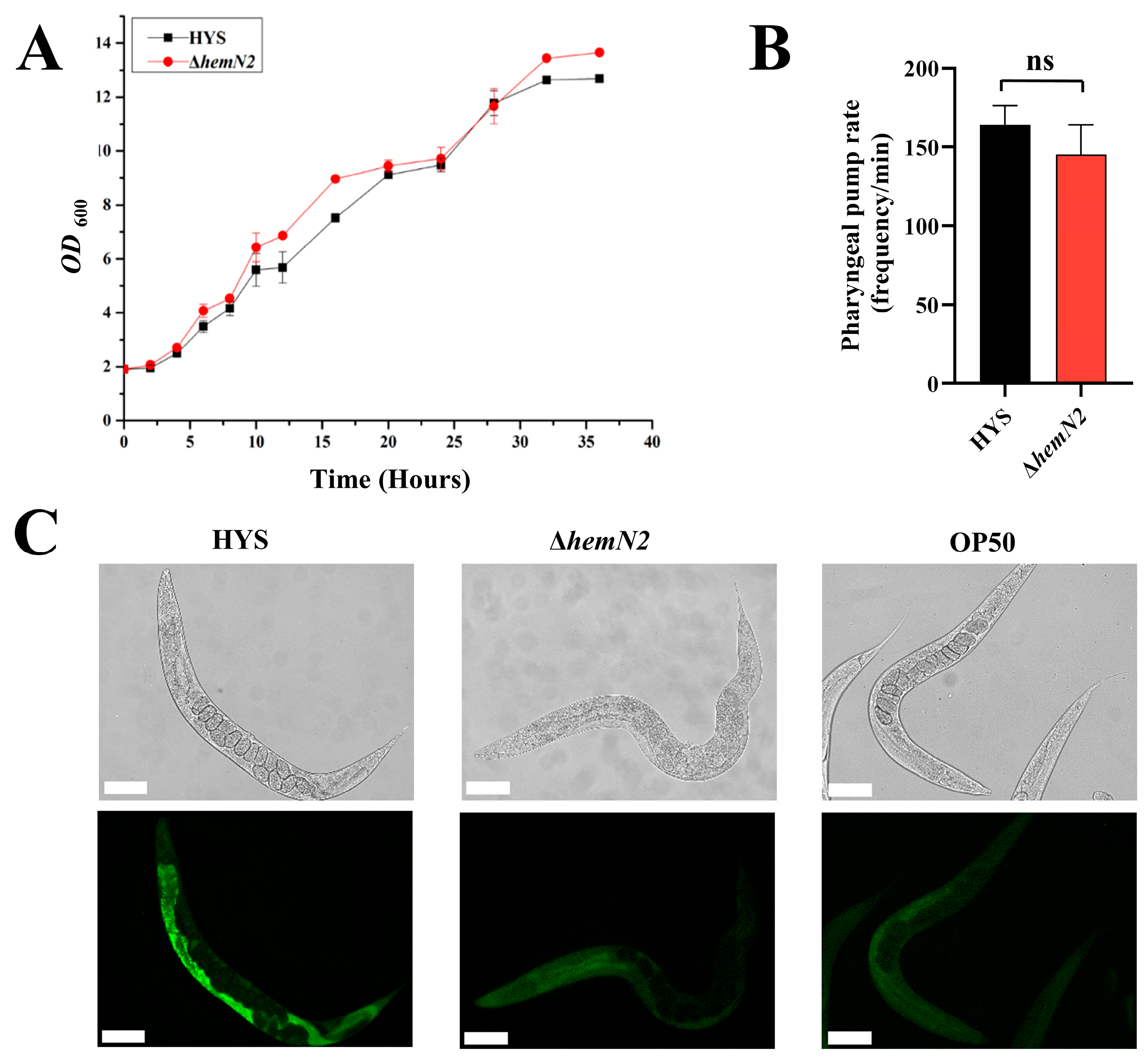
3.2. HemN2 Reduced the Colonization of P. donghuensis HYS in the C. elegans Gut
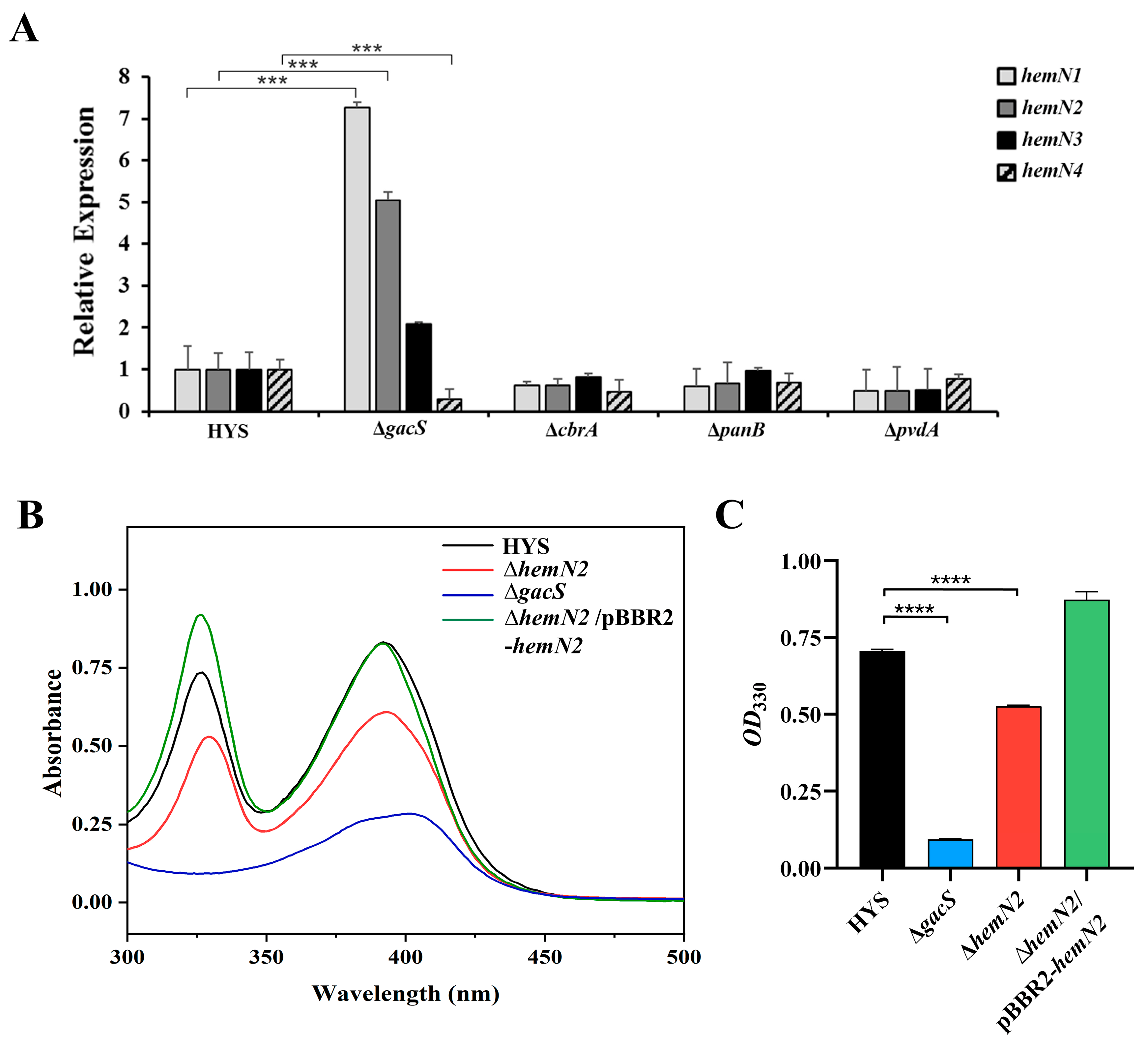
3.3. HemN2 Is Regulated Transcriptionally by GacS and Affects 7-HT Production
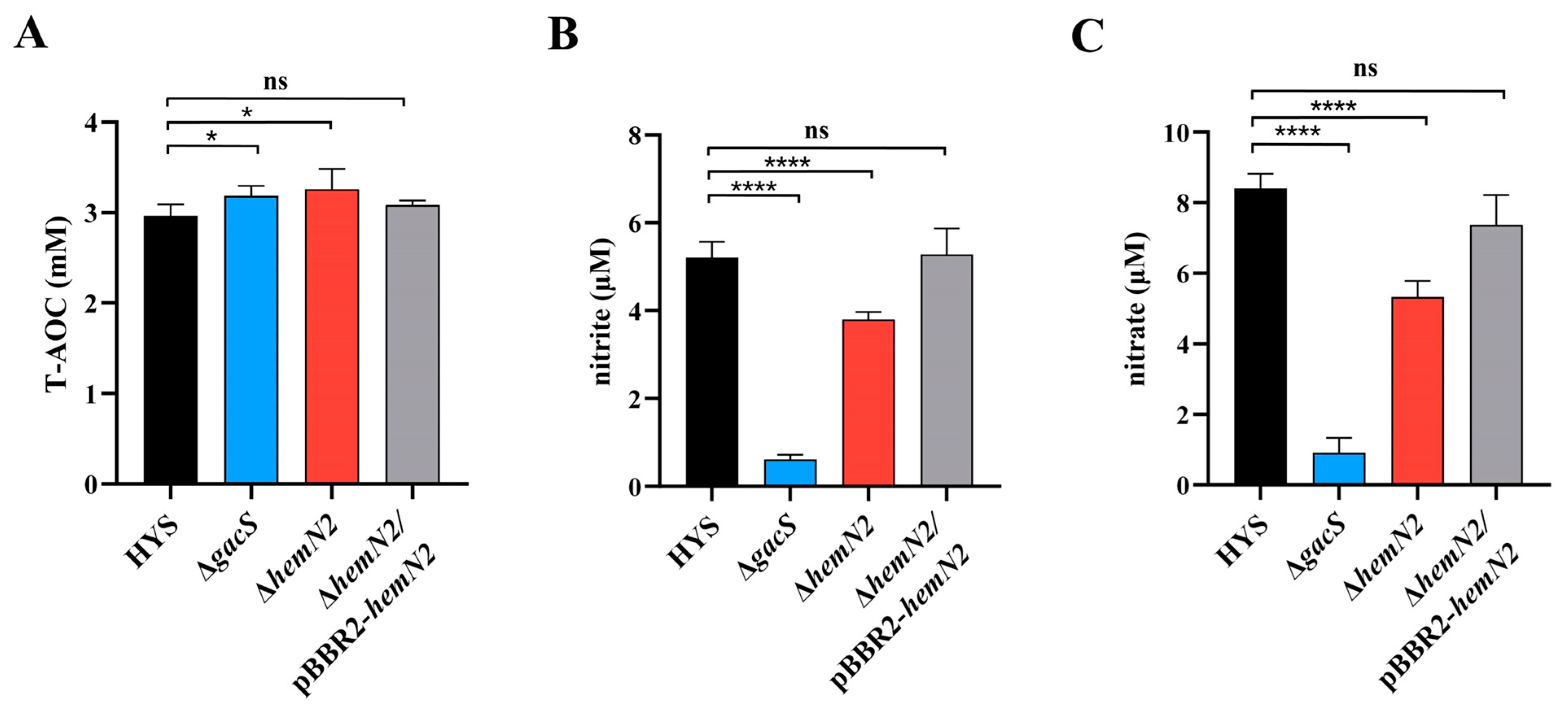
3.4. HemN2 Is Involved in Bacterial Virulence through Redox
3.5. HemN2 Is Involved in the Pathogenicity of P. donghuensis HYS to Zebrafish
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paliwal, D.; Rabiey, M.; Mauchline, T.H.; Hassani-Pak, K.; Nauen, R.; Wagstaff, C.; Andrews, S.; Bass, C.; Jackson, R.W. Multiple toxins and a protease contribute to the aphid-killing ability of Pseudomonas fluorescens PpR24. Environ. Microbiol. 2024, 26, e16604. [Google Scholar] [CrossRef]
- Corona, F.; Martínez, J.L.; Nikel, P.I. The global regulator Crc orchestrates the metabolic robustness underlying oxidative stress resistance in Pseudomonas aeruginosa. Environ. Microbiol. 2019, 21, 898–912. [Google Scholar] [CrossRef]
- Hernandez-Morales, A.; Alexis Rojas-Morales, J.; Reynoso-Lopez, M.; Bernardette Martinez-Rizo, A.; Bernardino Velazquez-Fernandez, J.; Lizzeta Arvizu-Gomez, J. Oxidative stress regulates the expression of the Pht cluster genes involved in phaseolotoxin synthesis in Pseudomonas syringae pv. phaseolicola NPS3121. J. Gen. Plant Pathol. 2018, 84, 137–141. [Google Scholar] [CrossRef]
- Agaras, B.C.; Iriarte, A.; Valverde, C.F. Genomic insights into the broad antifungal activity, plant-probiotic properties, and their regulation, in Pseudomonas donghuensis strain SVBP6. PLoS ONE 2018, 13, e0194088. [Google Scholar] [CrossRef] [PubMed]
- Zdorovenko, E.L.; Dmitrenok, A.S.; Masi, M.; Castaldi, S.; Muzio, F.M.; Isticato, R.; Valverde, C.; Knirel, Y.A.; Evidente, A. Structural studies on the O-specific polysaccharide of the lipopolysaccharide from Pseudomonas donghuensis strain SVBP6, with antifungal activity against the phytopathogenic fungus Macrophomina phaseolina. Int. J. Biol. Macromol. 2021, 182, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, P.; Zhu, X.; Xie, Z. Pseudomonas donghuensis HYS gtrA/B/II gene cluster contributes to its pathogenicity toward Caenorhabditis elegans. Int. J. Mol. Sci. 2021, 22, 10741. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zeng, M.; You, J.; Xie, Z. Pseudomonas donghuensis HYS virulence towards Caenorhabditis elegans is regulated by the Cbr/Crc system. Sci. Rep. 2019, 9, 8772. [Google Scholar] [CrossRef]
- Tan, M.W.; Rahme, L.G.; Sternberg, J.A.; Tompkins, R.G.; Ausubel, F.M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 1999, 96, 2408–2413. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Shao, Z.; Cai, M.; Zheng, L.; Li, G.; Huang, D.; Cheng, W.; Thomashow, L.S.; Weller, D.M.; Yu, Z.; et al. Multiple modes of nematode control by volatiles of Pseudomonas putida 1A00316 from antarctic soil against Meloidogyne incognita. Front. Microbiol. 2018, 9, 253. [Google Scholar] [CrossRef]
- Gui, Z.; You, J.; Xie, G.; Qin, Y.; Wu, T.; Xie, Z. Pseudomonas donghuensis HYS 7-hydroxytropolone contributes to pathogenicity toward Caenorhabditis elegans and is influenced by pantothenic acid. Biochem. Biophys. Res. Commun. 2020, 533, 50–56. [Google Scholar] [CrossRef]
- Gao, J.; Xie, G.; Peng, F.; Xie, Z. Pseudomonas donghuensis sp. nov., exhibiting high-yields of siderophore. Antonie Van Leeuwenhoek 2015, 107, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, M.; Jiang, Z.; Hu, Y.; Xie, Z. The two-component regulators GacS and GacA positively regulate a nonfluorescent siderophore through the Gac/Rsm signaling cascade in high-siderophore-yielding Pseudomonas sp. strain HYS. J. Bacteriol. 2014, 196, 3259–3270. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Mo, T.; Liu, W.Q.; Ding, W.; Deng, Z.; Zhang, Q. Revisiting the mechanism of the anaerobic coproporphyrinogen III oxidase HemN. Angew. Chem. Int. Ed. Engl. 2019, 58, 6235–6238. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.B.; Wu, S.; Xu, Y.F.; Yuan, H.; Tang, G.L. Recent advances in HemN-like radical S-adenosyl-l-methionine enzyme-catalyzed reactions. Nat. Prod. Rep. 2020, 37, 17–28. [Google Scholar] [CrossRef]
- Fischer, H.M.; Velasco, L.; Delgado, M.J.; Bedmar, E.J.; Schären, S.; Zingg, D.; Göttfert, M.; Hennecke, H. One of two hemN genes in Bradyrhizobium japonicum is functional during anaerobic growth and in symbiosis. J. Bacteriol. 2001, 183, 1300–1311. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.P. Metal uptake in host-pathogen interactions: Role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontol. 2000 2010, 52, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Jondle, C.N.; Gupta, K.; Mishra, B.B.; Sharma, J. Klebsiella pneumoniae infection of murine neutrophils impairs their efferocytic clearance by modulating cell death machinery. PLoS Pathog. 2018, 14, e1007338. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.; Youdim, M.B.; Riederer, P. Redox imbalance. Cell Tissue Res. 2004, 318, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Kajla, M.; Gupta, K.; Gupta, L.; Kumar, S. A fine-tuned management between physiology and immunity maintains the gut microbiota in insects. Biochem. Physiol. 2015, 4, 182. [Google Scholar]
- Domej, W.; Oettl, K.; Renner, W. Oxidative stress and free radicals in COPD–implications and relevance for treatment. Int. J. Chron. Obstruct. Pulmon. Dis. 2014, 9, 1207–1224. [Google Scholar] [CrossRef]
- Wang, J.; Mei, F.; Bai, L.; Zhou, S.; Liu, D.; Yao, L.; Ahluwalia, A.; Ghiladi, R.A.; Su, L.; Shu, T.; et al. Serum nitrite and nitrate: A potential biomarker for post-COVID-19 complications? Free. Radic. Biol. Med. 2021, 175, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.Y.; Yang, X.Z.; Teng, X.; Gao, S.Q.; Wen, G.B.; Lin, Y.W. Myoglobin mutant with enhanced nitrite reductase activity regulates intracellular oxidative stress in human breast cancer cells. Arch. Biochem. Biophys. 2022, 730, 109399. [Google Scholar] [CrossRef] [PubMed]
- Vatassery, G.T.; SantaCruz, K.S.; DeMaster, E.G.; Quach, H.T.; Smith, W.E. Oxidative stress and inhibition of oxidative phosphorylation induced by peroxynitrite and nitrite in rat brain subcellular fractions. Neurochem. Int. 2004, 45, 963–970. [Google Scholar] [CrossRef]
- Ilyasov, I.R.; Beloborodov, V.L.; Selivanova, I.A.; Terekhov, R.P. ABTS/PP decolorization assay of antioxidant capacity reaction pathways. Int. J. Mol. Sci. 2020, 21, 1131. [Google Scholar] [CrossRef] [PubMed]
- Muller, M. Premature cellular senescence induced by pyocyanin, a redox-active Pseudomonas aeruginosa toxin. Free. Radic. Biol. Med. 2006, 41, 1670–1677. [Google Scholar] [CrossRef] [PubMed]
- Mahajan-Miklos, S.; Tan, M.W.; Rahme, L.G.; Ausubel, F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 1999, 96, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Hassett, D.J.; Ma, J.F.; Elkins, J.G.; McDermott, T.R.; Ochsner, U.A.; West, S.E.; Huang, C.T.; Fredericks, J.; Burnett, S.; Stewart, P.S.; et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 1999, 34, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.W.; Schurr, M.J.; Yu, H.; Deretic, V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: Relationship to sigma E and stress response. J. Bacteriol. 1994, 176, 6688–6696. [Google Scholar] [CrossRef] [PubMed]
- Shatalin, K.; Gusarov, I.; Avetissova, E.; Shatalina, Y.; McQuade, L.E.; Lippard, S.J.; Nudler, E. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. USA 2008, 105, 1009–1013. [Google Scholar] [CrossRef]
- van Sorge, N.M.; Beasley, F.C.; Gusarov, I.; Gonzalez, D.J.; von Köckritz-Blickwede, M.; Anik, S.; Borkowski, A.W.; Dorrestein, P.C.; Nudler, E.; Nizet, V. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J. Biol. Chem. 2013, 288, 6417–6426. [Google Scholar] [CrossRef]
- Holden, J.K.; Kang, S.; Beasley, F.C.; Cinelli, M.A.; Li, H.; Roy, S.G.; Dejam, D.; Edinger, A.L.; Nizet, V.; Silverman, R.B.; et al. Nitric oxide synthase as a target for methicillin-resistant Staphylococcus aureus. Chem. Biol. 2015, 22, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ji, P.C.; Qi, Y.H.; Chen, S.J.; Wang, C.Y.; Yang, Y.J.; Zhao, X.Y.; Zhou, J.W. Inactivation of Pseudomonas aeruginosa biofilms by thymoquinone in combination with nisin. Front. Microbiol. 2022, 13, 1029412. [Google Scholar] [CrossRef] [PubMed]
- Stiernagle, T. Maintenance of C. elegans. WormBook 2006, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Ausubel, F.M.; Perfect, J.R.; Heitman, J.; Calderwood, S.B. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 15675–15680. [Google Scholar] [CrossRef] [PubMed]
- van Tilburg Bernardes, E.; Charron-Mazenod, L.; Reading, D.J.; Reckseidler-Zenteno, S.L.; Lewenza, S. Exopolysaccharide-repressing small molecules with antibiofilm and antivirulence activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61, e01997-16. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., 2nd; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Ali, M.; Sun, Y.; Xie, L.; Yu, H.; Bashir, A.; Li, L. The Pathogenicity of Pseudomonas syringae MB03 against Caenorhabditis elegans and the transcriptional response of nematicidal genes upon different nutritional conditions. Front. Microbiol. 2016, 7, 805. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nogaret, P.; El Garah, F.; Blanc-Potard, A.B. A novel infection protocol in zebrafish embryo to assess Pseudomonas aeruginosa virulence and validate efficacy of a quorum sensing inhibitor in vivo. Pathogens 2021, 10, 401. [Google Scholar] [CrossRef] [PubMed]
- Troup, B.; Hungerer, C.; Jahn, D. Cloning and characterization of the Escherichia coli hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase. J. Bacteriol. 1995, 177, 3326–3331. [Google Scholar] [CrossRef]
- Xu, K.; Elliott, T. Cloning, DNA sequence, and complementation analysis of the Salmonella typhimurium hemN gene encoding a putative oxygen-independent coproporphyrinogen III oxidase. J. Bacteriol. 1994, 176, 3196–3203. [Google Scholar] [CrossRef] [PubMed]
- Rompf, A.; Hungerer, C.; Hoffmann, T.; Lindenmeyer, M.; Römling, U.; Gross, U.; Doss, M.O.; Arai, H.; Igarashi, Y.; Jahn, D. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 1998, 29, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Lieb, C.; Siddiqui, R.A.; Hippler, B.; Jahn, D.; Friedrich, B. The Alcaligenes eutrophus hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase, is required for heme biosynthesis during anaerobic growth. Arch. Microbiol. 1998, 169, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, X.; Cai, M.; Lv, C.; Zhao, Y.; Wei, D.; Zhu, H. The heme transporter HtsABC of group A Streptococcus contributes to virulence and innate immune evasion in murine skin infections. Front. Microbiol. 2018, 9, 1105. [Google Scholar] [CrossRef] [PubMed]
- Śmiga, M.; Ślęzak, P.; Olczak, T. Comparative analysis of Porphyromonas gingivalis A7436 and ATCC 33277 strains reveals differences in the expression of heme acquisition systems. Microbiol. Spectr. 2024, 12, e0286523. [Google Scholar] [CrossRef] [PubMed]
- Olczak, T.; Sosicka, P.; Olczak, M. HmuY is an important virulence factor for Porphyromonas gingivalis growth in the heme-limited host environment and infection of macrophages. Biochem. Biophys. Res. Commun. 2015, 467, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Kusmierek, M.; Dersch, P. Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system. Curr. Opin. Microbiol. 2018, 41, 58–67. [Google Scholar] [CrossRef]
- O’Donnell, M.P.; Fox, B.W.; Chao, P.H.; Schroeder, F.C.; Sengupta, P. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature 2020, 583, 415–420. [Google Scholar] [CrossRef]
- Ferreiro, M.D.; Gallegos, M.T. Distinctive features of the Gac-Rsm pathway in plant-associated Pseudomonas. Environ. Microbiol. 2021, 23, 5670–5689. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Humair, B.; Dénervaud, V.; Riedel, K.; Spahr, S.; Eberl, L.; Valverde, C.; Haas, D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6026–6033. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Ceri, H.; Storey, D.G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 2001, 40, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Chen, G.; Xu, X.; Wang, T.; Liang, H. Iron facilitates the RetS-Gac-Rsm cascade to inversely regulate protease IV (piv) expression via the sigma factor PvdS in Pseudomonas aeruginosa. Environ. Microbiol. 2020, 22, 5402–5413. [Google Scholar] [CrossRef] [PubMed]
- Zha, D.; Xu, L.; Zhang, H.; Yan, Y. The two-component GacS-GacA system activates lipA translation by RsmE but not RsmA in Pseudomonas protegens Pf-5. Appl. Environ. Microbiol. 2014, 80, 6627–6637. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef]
- LeRoux, M.; Kirkpatrick, R.L.; Montauti, E.I.; Tran, B.Q.; Peterson, S.B.; Harding, B.N.; Whitney, J.C.; Russell, A.B.; Traxler, B.; Goo, Y.A.; et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 2015, 4, e05701. [Google Scholar] [CrossRef] [PubMed]
- Barahona, E.; Navazo, A.; Martínez-Granero, F.; Zea-Bonilla, T.; Pérez-Jiménez, R.M.; Martín, M.; Rivilla, R. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 2011, 77, 5412–5419. [Google Scholar] [CrossRef]
- Kay, E.; Dubuis, C.; Haas, D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. USA 2005, 102, 17136–17141. [Google Scholar] [CrossRef]
- Kong, H.S.; Roberts, D.P.; Patterson, C.D.; Kuehne, S.A.; Heeb, S.; Lakshman, D.K.; Lydon, J. Effect of overexpressing rsmA from Pseudomonas aeruginosa on virulence of select phytotoxin-producing strains of P. syringae. Phytopathology 2012, 102, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, P.; Xie, Z. A complex mechanism involving LysR and TetR/AcrR that regulates iron scavenger biosynthesis in Pseudomonas donghuensis HYS. J. Bacteriol. 2018, 200, e00087-18. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, J.A.; Mikkelsen, H.; Heeb, S.; Williams, P.; Filloux, A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011, 13, 3128–3138. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Choi, H.Y.; Kim, W.G. The Nitrite transporter facilitates biofilm formation via suppression of nitrite reductase and is a new antibiofilm target in Pseudomonas aeruginosa. mBio 2020, 11, e00878-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, H.; Yan, C.; Han, W.; Peng, L.; Xu, J.; Chen, X.; Langford, P.R.; Bei, W.; Huang, Q.; et al. The metabolic adaptation in response to nitrate is critical for Actinobacillus pleuropneumoniae growth and pathogenicity under the regulation of NarQ/P. Infect. Immun. 2022, 90, e0023922. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Nicastro, L.K.; Bessho, S.; Grando, K.; White, A.P.; Zhang, Y.; Queisser, G.; Buttaro, B.A.; Tükel, Ç. Nitrate is an environmental cue in the gut for Salmonella enterica serovar Typhimurium biofilm dispersal through curli repression and flagellum activation via cyclic-di-GMP signaling. mBio 2021, 13, e0288621. [Google Scholar] [CrossRef] [PubMed]
- Ventre, I.; Goodman, A.L.; Vallet-Gely, I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar] [CrossRef]
- Lavado-Benito, C.; Murillo, J.; Martínez-Gil, M.; Ramos, C.; Rodríguez-Moreno, L. GacA reduces virulence and increases competitiveness in planta in the tumorigenic olive pathogen Pseudomonas savastanoi pv. savastanoi. Front. Plant Sci. 2024, 15, 1347982. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Xiang, W.; Ma, X.; Gao, D.; Bayram, H.; Lorimer, G.H.; Ghiladi, R.A.; Xie, Z.; Wang, J. HemN2 Regulates the Virulence of Pseudomonas donghuensis HYS through 7-Hydroxytropolone Synthesis and Oxidative Stress. Biology 2024, 13, 373. https://doi.org/10.3390/biology13060373
Xiao Y, Xiang W, Ma X, Gao D, Bayram H, Lorimer GH, Ghiladi RA, Xie Z, Wang J. HemN2 Regulates the Virulence of Pseudomonas donghuensis HYS through 7-Hydroxytropolone Synthesis and Oxidative Stress. Biology. 2024; 13(6):373. https://doi.org/10.3390/biology13060373
Chicago/Turabian StyleXiao, Yaqian, Wang Xiang, Xuerui Ma, Donghao Gao, Hasan Bayram, George H. Lorimer, Reza A. Ghiladi, Zhixiong Xie, and Jun Wang. 2024. "HemN2 Regulates the Virulence of Pseudomonas donghuensis HYS through 7-Hydroxytropolone Synthesis and Oxidative Stress" Biology 13, no. 6: 373. https://doi.org/10.3390/biology13060373
APA StyleXiao, Y., Xiang, W., Ma, X., Gao, D., Bayram, H., Lorimer, G. H., Ghiladi, R. A., Xie, Z., & Wang, J. (2024). HemN2 Regulates the Virulence of Pseudomonas donghuensis HYS through 7-Hydroxytropolone Synthesis and Oxidative Stress. Biology, 13(6), 373. https://doi.org/10.3390/biology13060373





