Simple Summary
Milk production and its composition are important for the economy, and they depend on complex biological processes. By finding the key genes and causal mutations linked to milk yield, we can improve breeding strategies for dairy animals. Thanks to advanced bioinformatics tools, it is now easier to find the genetic factors that affect milk traits. In our study, we used these methods to explore the genetics of milk traits in Murciano-Granadina goats. Although we found distinct genes associated with each trait, the regulatory proteins showed shared-yet-dynamic roles in controlling gene activity across different traits. This helped us understand how genes work together in the mammary gland, which affects milk production and udder health.
Abstract
The Murciano-Granadina goat (MUG) is a renowned dairy breed, known for its adaptability and resilience, as well as for its exceptional milk traits characterized by high protein and fat content, along with low somatic cell counts. These traits are governed by complex biological processes, crucial in shaping phenotypic diversity. Thus, it is imperative to explore the factors regulating milk production and lactation for this breed. In this study, we investigated the genetic architecture of seven milk traits in MUGs, employing a two-step computational analysis to examine genotype–phenotype associations. Initially, a random forest algorithm identified the relative importance of each single-nucleotide polymorphism (SNP) in determining the traits of interest. The second step applied an information theory-based approach to exploring the complex genetic architecture of quantitative milk traits, focusing on epistatic interactions that may have been overlooked in the first step. These approaches allowed us to identify an almost distinct set of candidate genes for each trait. In contrast, by analyzing the promoter regions of these genes, we revealed common regulatory networks among the milk traits under study. These findings are crucial for understanding the molecular mechanisms underlying gene regulation, and they highlight the pivotal role of transcription factors (TFs) and their preferential interactions in the development of these traits. Notably, TFs such as DBP, HAND1E47, HOXA4, PPARA, and THAP1 were consistently identified for all traits, highlighting their important roles in immunity within the mammary gland and milk production during lactation.
1. Introduction
The Murciano-Granadina goat (MUG), a prominent dairy breed from Spain, is closely associated with the regions of Murcia and Granada, reflecting its historical origins. This breed has been introduced to various farming systems, potentially even beyond its native region, due to its resilience in semi-arid conditions and its ability to cope with fluctuating food availability [1]. Morphologically, MUGs typically have a medium build, weighing around 50 kg, and brown or black coats. They may either be horned or hornless. Furthermore, this breed consistently exhibits a polyestrous nature, with notable prolificacy, often giving birth to two or more kids and demonstrating longevity with up to six births [2,3]. To improve the morphology, milk production, and milk composition of MUGs, the National Association of Murciano-Granadina Goat Breeders (CAPRIGRAN) started a breeding program in 2012. As a result, the MUG can now produce an average of 584 kg of milk during the 287 days of lactation cycle, with average fat, protein, and lactose contents of 5.3, 3.6, and 4.7 percent, respectively [1]. The main use of goat milk in Spain is to manufacture cheese, and the MUG breed is the best choice for this purpose because of its high protein and fat content and its low somatic cell counts [4,5,6].
Several association studies have pinpointed genomic regions and the distribution of single-nucleotide polymorphisms (SNPs) affecting caprine milk production and composition [7,8,9,10,11,12,13]. Similarly, for the MUG breed, identifying the genetic variants impacting milk traits has been the subject of numerous studies [6,14,15,16,17,18,19,20,21,22,23,24,25], as presented in Table 1.

Table 1.
Representative studies on association analysis of milk traits in MUG goats.
In particular, these studies explored various genetic and molecular factors in MUGs, including the effects on milk quality, the impact of casein complex haplotype variants on production and composition, the relationship between genetic variations and dairy traits, and the use of genetic markers from artificial selection in milk trait associations. Predominantly, these studies focused on the additive effects of genotypes, analyzing associations between single SNPs and traits of interest. However, it is well known that epistasis among genes plays a critical role in shaping the complex inheritance mechanisms of quantitative traits, an aspect that single-SNP-based association studies may miss [26]. Consequently, a range of computational tools has been developed to detect and incorporate epistasis into analytical frameworks, thereby enhancing the efficacy of single-SNP models [27,28,29,30]. These tools improve predictive capabilities in association analysis and help decipher the complex genetic mechanisms underlying quantitative traits [27,31]. In this context, Liang et al. [32] demonstrated that including both additive and epistatic SNP effects significantly improved the accuracy of predicting residual feed intake in US Holstein cows. Furthermore, Prakapenka et al. [33] extended their initial single-SNP model [34] by emphasizing the role of epistasis in dairy production and fertility traits among first-lactation US Holstein cattle. Similarly, Pizarro et al. [18] showed in their study on MUGs that incorporating epistasis effects, particularly of SNPs related to the casein complex, resulted in better predictive accuracy for breeding-value estimations of dairy traits compared to models that only considered additive effects.
Complementing these genetic studies, another crucial aspect in unraveling the complex genetic architecture of milk-related traits involves investigating their transcriptional gene regulatory mechanisms. Although these mechanisms are often understudied, they are essential for controlling the biological processes underlying quantitative traits in several organisms [35,36]. To address the limited knowledge available about the biological functions of milk traits, it is essential to consider TFs and their cooperation. To date, only a few studies have focused on the specific role of individual TFs within the MUG genome. In pursuit of this purpose, Yahyaoui et al. [37] conducted base excision sequence scanning and pinpointed an SNP in the promoter region of the beta-lactoglobulin gene linked to milk protein that did not affect the binding site of any TF in their study. Additionally, Zidi et al. [25] focused on scrutinizing the coding region of the sterol regulatory element-binding transcription factor 1, uncovering its connection with milk fat content and composition traits. More recently, Luigi-Sierra et al. [19] explored the genetic architecture of the udder in MUGs, detecting the presence of the ATF3 gene in proximity to the SNP rs268273468, which was found to be correlated with the development of mammary gland tissue.
Previous studies on milk traits have primarily identified variants using conventional SNP-based analyses, focusing on additive effects and often overlooking the role of SNP–SNP interactions, or epistasis, in shaping these complex traits [26]. Furthermore, while transcriptional regulation plays a crucial role in quantitative traits, the specific functions of TFs within the MUG goat genome remain largely unexamined. To address these gaps, our study investigated the genetic architecture governing seven key milk traits in MUGs, using a genotype–phenotype dataset of 822 animals. Our approach integrated a two-step analysis pipeline: the first step used the random forest (RF) algorithm to evaluate the relative importance of each SNP in determining the traits of interest [38,39,40,41]; in the second step, we applied an information theory-based epistasis method, mutual information-based detection of epistatic SNP pairs (MIDESP) [27], to uncover complex genetic interactions that single-SNP analyses might overlook. Our analyses revealed distinct candidate genes associated with each trait, despite high phenotypic correlations among some traits. To further understand the regulatory mechanisms driving these gene–trait relationships, we employed the potentially collaborating transcription factor finder (PC-TraFF) algorithm [42,43], focusing on promoter regions to identify cooperative TFs specific to each trait. Although only a few genes are commonly associated with all traits, the PC-TraFF results suggest that a large number of TFs and their cooperative partners consistently participate in the regulatory networks underlying these milk traits in MUGs.
2. Materials and Methods
This section describes the genotype–phenotype dataset that was analyzed, as well as the research methodology.
2.1. Murciano-Granadina Goat Dataset
To elucidate the role of regulatory elements, such as TFs and their interactions, that could be essential in orchestrating the development of the seven milk traits, we analyzed a genotype–phenotype dataset previously set up by Guan et al. [24] to study the genetic basis of milk yield and composition in MUGs. The dataset contains the phenotypic data for 1023 MUGs, which includes dry matter percentage (DM), length of lactation/milk production days (DPM), fat percentage (FP), protein percentage (PP), lactose percentage (LP), milk yield at 210 days (MY), and somatic cell count (SCC). These traits were recorded across 15 dairy farms affiliated with CAPRIGRAN during the goats’ first lactation, where artificial insemination was the breeding method. While all the traits were standardized for a first lactation length of 210 days, DPM was an exception. A total of 822 goats were included in our analysis, after excluding those with insufficient or missing phenotypic data. Genotypic data for the animals were generated using the Goat SNP50 BeadChip, which contains 53,347 SNP markers, following the instructions provided by Illumina [44]. Following the criteria set out in [24], our analysis only included SNPs that (i) mapped to the autosomes; (ii) displayed a minor allele frequency of >0.01; (iii) did not deviate significantly from the Hardy–Weinberg expectation (p-value ); (iv) had genotype call rates percent; and (v) had SNP call rates percent. After filtering, we used 822 animals and 49,272 SNPs for our analyses.
2.2. Association Analysis
To identify SNPs potentially associated with the traits of interest in this study, we applied two distinct approaches: (i) a machine learning algorithm—specifically, RF—to assess the relative importance of each SNP (attribute) in predicting response variables (phenotype values); and (ii) an information theory-based methodology, the MIDESP algorithm [27], to detect epistatic interactions between pairs of SNPs and their corresponding phenotype values. In line with previous studies [45,46], we considered genes located within a 25 kb range around the identified SNPs as potential candidates for further analysis.
2.2.1. Association Analysis Using Random Forest
In assessing the importance of SNPs for the prediction of the phenotype of interest, we applied an association analysis method anchored in RF-based feature selection, as described in previous studies [38,39,40,41]. Specifically, we used the Boruta algorithm [47], a tool tailored for RF-based feature selection, to calculate the importance scores of each SNP. These scores were instrumental in determining the relevance of the SNPs, to subsequently facilitate their ranking in phenotype prediction.
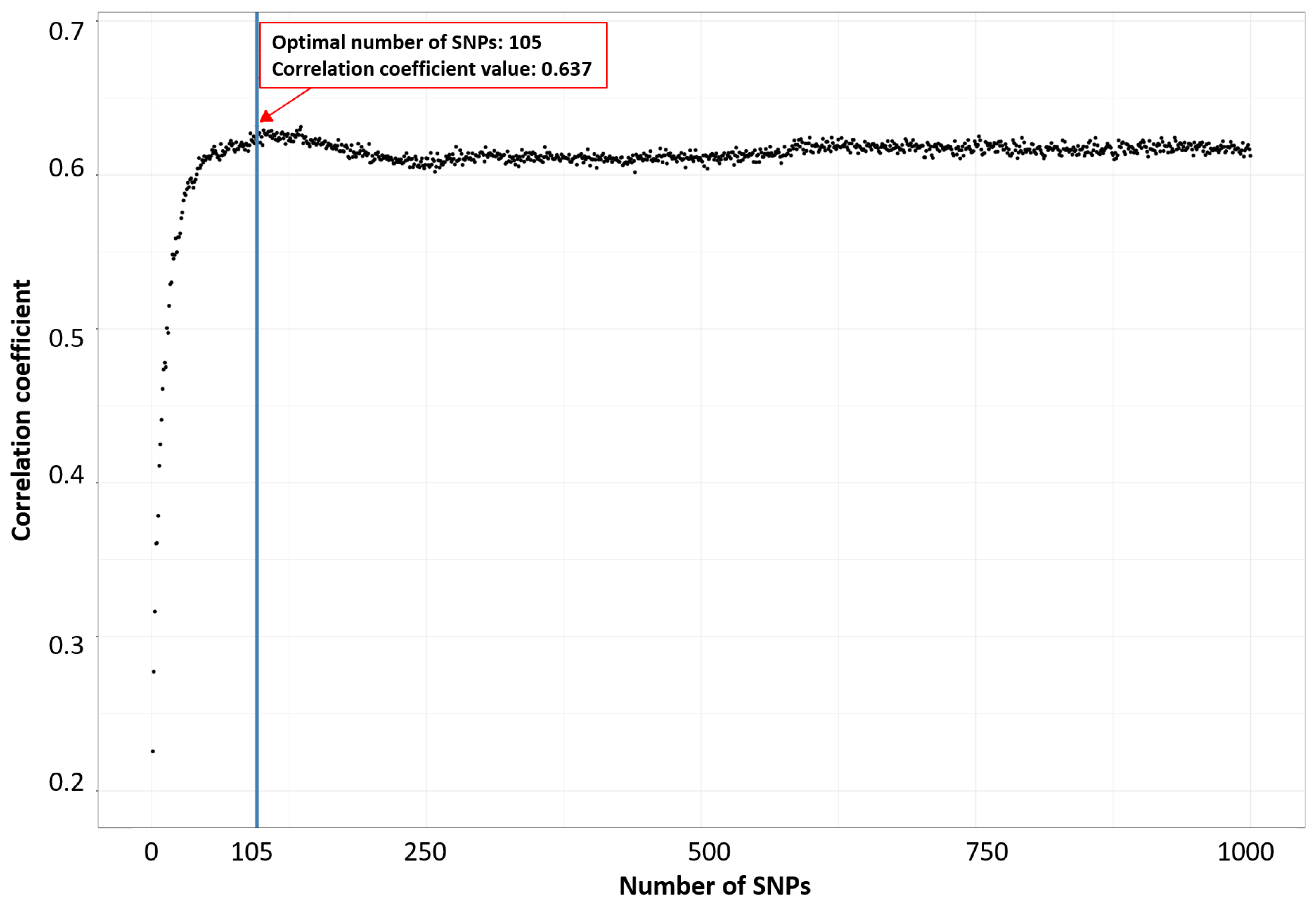
Secondly, we utilized the incremental feature selection (IFS) technique, which prioritized SNPs based on their rankings in an RF model. This approach was used to identify the optimal number of features (SNPs) associated with a particular phenotype, as shown in Figure 1 for milk yield at 210 days, following the methodology suggested in [41,48,49,50]. During the application of IFS, SNPs were progressively added in descending order of their ranks within the feature set. Subsequently, an RF classifier was built, and its predictive performance was assessed based on the correlation coefficient. Supplementary Figures S1 and S2 provide an extended view of Figure 1 with 3000 and 5000 SNPs, enabling a comparative analysis of the impact of different SNP numbers.
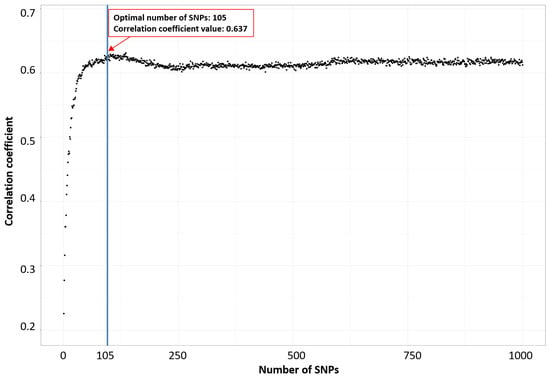
Figure 1.
Identification of the optimal number of SNPs associated with milk yield at 210 days through using incremental feature selection.
2.2.2. Epistasis Analysis Using MIDESP Algorithm
While single-SNP-based association analysis offers valuable insights into genotype–phenotype associations, it is now widely recognized that many complex traits arise from the interactions of multiple factors [27,51,52,53]. Therefore, the incorporation of epistasis could yield a more comprehensive understanding of these interactions by providing novel insights into the genetic architecture of complex traits that might be overlooked in a standard single-SNP analysis. To elucidate further the biological processes underlying milk traits in MUGs, we incorporated the MIDESP algorithm [27] in this study. Central to the MIDESP algorithm is the use of the mutual information (MI) metric, a powerful measure for assessing the strength of association between two or more variables. By considering both the marginal and joint probability distribution of the variables, the MI metric can account for linear as well as non-linear dependencies. Leveraging the MI metric, we applied the MIDESP algorithm to identify pairwise epistatic interactions between SNPs and the milk traits being analyzed. Consequently, the IFS technique was applied to identify the optimal number of SNP pairs associated with the phenotype of interest.
2.3. Identification of Transcription Factor Cooperation
In this study, we utilized the PC-TraFF algorithm [42] to identify cooperation between TF pairs by focusing mainly on the co-occurrence of their TF binding sites (TFBSs) in the promoter sequences of the candidate genes found for each trait. We let TFBSA and TFBSB, etc., be the binding sites of TFs TFA and TFB, respectively. The PC-TraFF algorithm computed the pointwise mutual information (PMI) score in six steps, using the joint probability and the marginal probabilities and , to yield a PMI(TFA; TFB) value for each TF pair TFA and TFB. In the final phase, the algorithm transformed the PMI(TFA; TFB) values into z-scores. The cooperation between TFs TFA and TFB was considered to be significant if the z-score was ≥3. The application of the PC-TraFF algorithm required pre-defined distance preferences of the TFBSs, position weight matrices (PWMs), and promoter sequences as input parameters. We implemented the algorithm with the recommended distance values, setting the minimum distance to ≥5 and the maximum distance to ≤20. For identifying putative TFBSs, we first extracted the promoter sequences (ranging from −500 bp to +100 bp relative to the transcription start site) of the candidate genes, using gene annotation and the ARS1 genome assembly of goats from the Ensembl database [54]. We then employed a non-redundant vertebrate PWM library from the TRANSFAC database [55], along with the Match [56] program, to scan these sequences for predicting the TFBSs.
3. Results
In this study, we analyzed a genotype–phenotype dataset of 822 MUGs to identify candidate genes and their regulatory mechanisms associated with seven milk-related traits. To achieve this, an RF-based feature selection algorithm in conjunction with the MIDESP approach was used to identify candidate SNPs, SNP pairs, and their corresponding genes for each trait (Table 2). The complete list of identified SNPs, SNP pairs, their corresponding genes, and gene pairs are listed in Supplementary Tables S1 and S2.

Table 2.
Results of single-SNP-based GWAS and SNP–SNP interaction epistasis-based GWAS.
Although both methods aim to reveal important associations and their significance within the genotype–phenotype dataset, Table 2 shows that each method identified a considerably different number of associated SNPs and genes. The disparity can be attributed to the difference between their algorithms. Specifically, the RF-based feature selection method focuses primarily on assessing the importance of individual SNPs, whereas the MIDESP algorithm performs a stringent association analysis between SNP pairs and the traits of interest. The MIDESP algorithm also includes an additional filtering step that eliminates expected background associations, resulting in a dramatic reduction in the number of significant pairs identified. A closer examination of the identified candidate gene lists highlights the complementary nature of both association analysis methods, as they provide different types of information, resulting in a small overlap between the gene lists. Moreover, given the quantitative nature of the seven milk-related traits under study, it is likely that multiple genes with both additive and epistatic effects influenced them. Therefore, an integrated consideration of the candidate genes obtained from both analyses for each trait could be quintessential for understanding the genetic mechanisms controlling milk-related traits in MUGs.
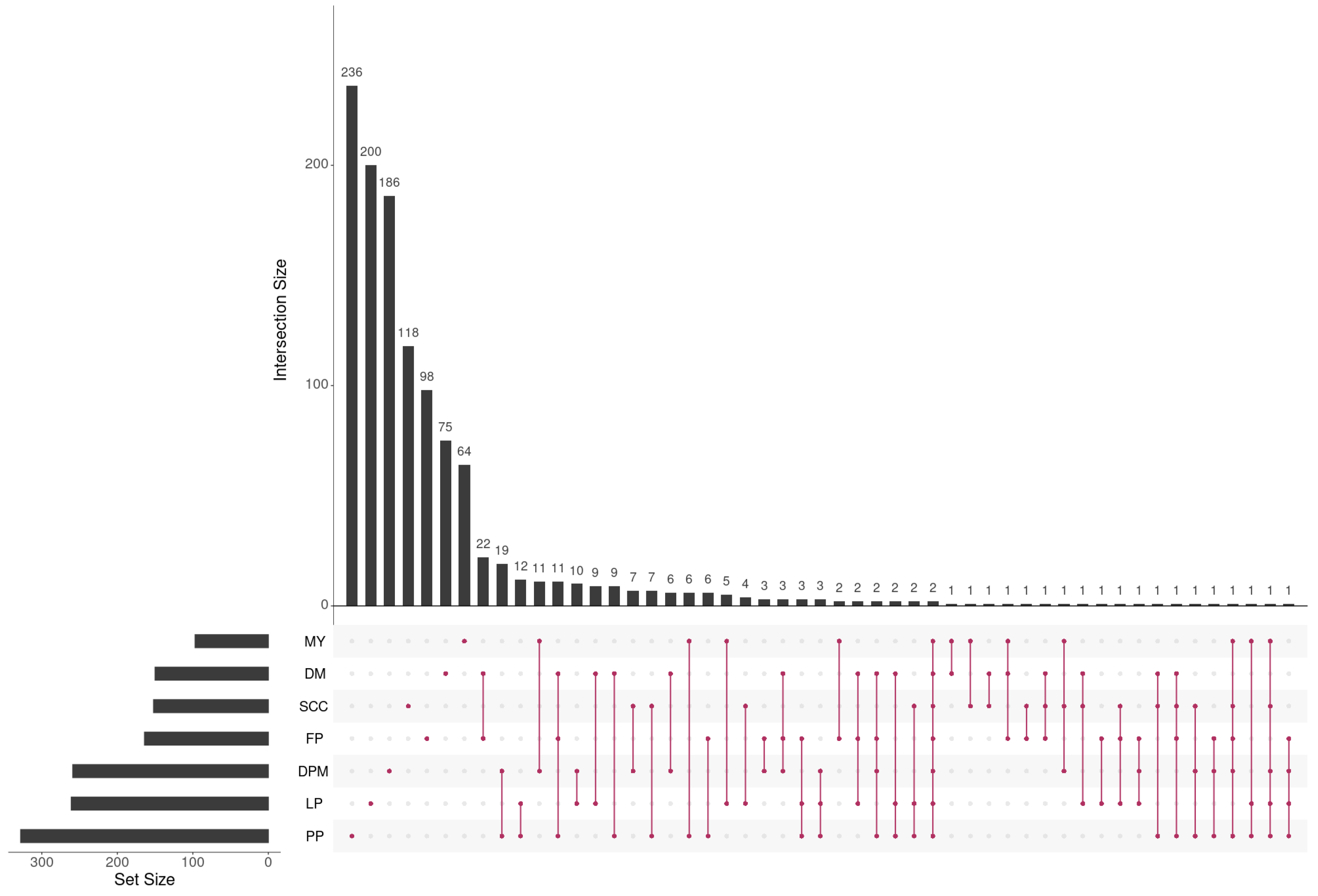
Figure 2 shows that the number of candidate genes, as well as the overlap between them, varied considerably across different traits. Interestingly, only two genes linked to U6 and 5S-rRNA were identified as common across all dairy traits, as depicted in Figure 2. It is well known that U6 and 5S-rRNA play crucial roles in regulating gene expression and protein synthesis as part of the ribosomal RNA family. Their presence suggests potential involvement in the regulation of milk production and composition, which is consistent with prior research highlighting their impact on milk traits [57].
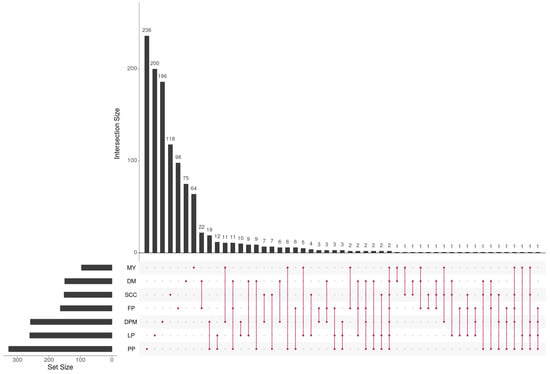
Figure 2.
The number of genes and their overlap across the seven dairy traits represented in matrix layouts using the UpSet technique [58]. The maroon circles in the matrix layout are related to the traits that are part of the intersection. For the sake of clarity, not all intersections are displayed.
Following the association analysis, we compiled a set of candidate genes for each trait and, subsequently, we extracted their promoter sequences. This enabled us to identify cooperative TFs regulating the genes that are associated with a particular trait.
Identification of Cooperative Transcription Factors Regulating Seven Milk-Related Traits
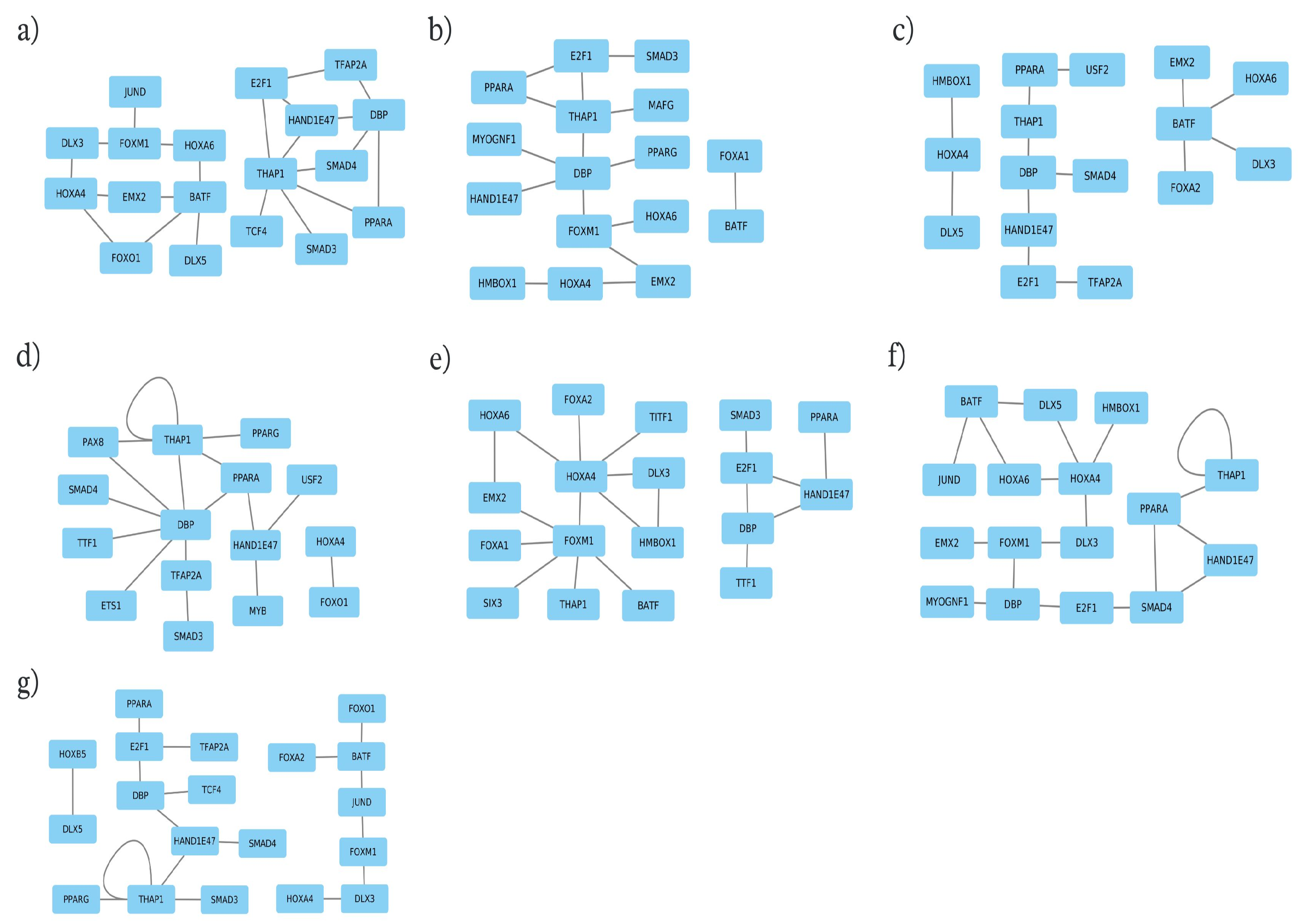
Applying the PC-TraFF algorithm [42,43] to promoter sequences of candidate genes revealed significant cooperative TF pairs for each trait. To further elucidate the gene regulatory mechanisms of the underlying biological processes for the seven milk-related traits in the MUGs, we extended our analysis by constructing trait-specific cooperation networks based on the identified TF pairs, as described in our previous studies [43,59,60,61,62]. As illustrated in Figure 3, the nodes and edges in these networks represented the TFs and their cooperative interactions, respectively. A closer look at these networks reveals that they comprised only 32 unique TFs participating in the formation of 118 TF pairs, which suggests that TFs switch partners according to their functional roles in different traits [60,61]. Additionally, to highlight the activity of these TFs during the lactation period, their gene expression levels, similar to those reported by Zeidler et al. [62], are provided in Supplementary File S1, Table S3 and Figure S3.

Figure 3.
Cooperation networks for the transcription factor pairs of: (a) dry matter percentage; (b) milk production days; (c) fat percentage; (d) lactose percentage; (e) milk yield at 210 days; (f) protein percentage, and (g) somatic cell count traits.
The number of individual TFs and TF pairs is given in Table 3 for each trait. Remarkably, the TFs DBP, HAND1E47, HOXA4, PPARA, and THAP1 were consistently identified across all milk traits. This overlap of TFs in cooperation networks for all milk traits suggests common regulatory processes in mammary gland tissue.

Table 3.
Significant TFs, cooperative TF pairs, and the top-three pairs for each trait, as identified by PC-TraFF. The pairs are sorted in ascending order, based on their z-scores provided by PC-TraFF.
The D-site binding protein (DBP) is a member of the proline and acidic amino acid-rich basic leucine zipper (PAR bZip) TF family, which includes other members, such as the hepatic leukemia factor and the thyrotroph embryonic factor [63,64]. DBP plays a pivotal role in regulating the expression of clock genes, which are essential for controlling circadian rhythms [65] and influence the activity of various enzymes [66]. Circadian rhythms, representing adaptations to daily environmental variations, are under the regulatory control of PAR bZip factors [63]. Consequently, this circadian system governs metabolic processes and physiological activities, which are crucial determinants of milk quality and quantity in mammals [67,68].
The basic helix–loop–helix TF family member, HAND1E47, plays crucial roles in the development of the mammary gland by regulating essential processes, such as cell proliferation, differentiation, and apoptosis [69,70]. Furthermore, Kumar et al. [71] revealed the participation of HAND1E47 in the promoter region of the S100A8 gene, elucidating its role in the immune function within mammary glands. This is further substantiated by the expression of S100A8 in milk somatic cells, particularly following infections in goat mammary glands [72]. Another interesting factor is HOXA4, a member of the homeobox (HOX) protein family characterized by its DNA-binding homeobox domain, which plays a crucial role in regulating early embryo development patterns and subsequent developmental processes [73,74]. HOXA4 plays a role in various physiological and pathological processes in organisms, mediated by well-established signaling pathways [73,75,76,77]. It is also involved in processes like cell proliferation, differentiation, apoptosis, signal transduction, and tumor suppression [78,79]. Furthermore, its regulatory role has been observed in controlling milk traits, aligning with the findings of Pellacani et al. [80], who identified this factor as an active enhancer for multiple genes in mammary gland epithelial cells. In our analysis, we also identified the PPARA factor, a member of the peroxisome proliferator-activated receptors (PPARs), which are well-recognized for their crucial role in governing lipid metabolism [81] and regulating cell proliferation and the cell cycle [82]. Moreover, PPARA has been identified as a key regulator of activities in both epithelial and stromal cells within mammary glands [83], influencing the synthesis of milk fat content.
Furthermore, we identified THAP1, a member of the Thanatos-associated proteins (THAP) family. THAP1 features a simplified proline-rich region, a coiled-coil domain, and a binary nuclear localization sequence [84]. It is involved in cell division, apoptosis, and transcription control [85,86], and it is broadly expressed in various tissues, including the brain, as well as extra-neural tissues such as the thyroid gland, prostate, liver, kidney, and blood [86,87]. Mutations in THAP1 can result in loss of function, leading to transcription dysregulation and dystonia-like conditions in humans [85]. Interestingly, THAP1 is also implicated in clock-controlled gene regulation [60,88], affecting physiological processes that influence mammary gland growth and lactation competence [89].
A closer look at the cooperation networks in Figure 3 further reveals that they frequently contain members of the DLX, FOX, HOX, PPAR, and SMAD families. In particular, the members of the distal-less homeobox (DLX) family play a vital role in embryonic ectoderm development and the formation of structures such as teeth, hair follicles, and mammary glands [90]. Exhibiting dual functionalities, they can act as either activators or repressors, depending on their partner choice [91]. Additionally, DLX members have been studied for their role in regulating mammary branching morphogenesis [92], as well as for their involvement in the immune response in the udder [93]. Interestingly, our analysis revealed cooperation between the DLX and HOX family members (e.g., the DLX3–HOXA4 pair for DM, MY, PP, and SCC, and DLX5–HOXA4 cooperation for FP and PP traits) that are essential for governing both body segmentation and developmental processes [94,95,96].
In this study, we identified several forkhead box (FOX) family members (FOXA1, FOXA2, FOXM1, and FOXO1), which are known to play pivotal roles in various aspects of mammalian development and disease, including immunological regulation, glucose and lipid metabolism, and the biological aging process [97,98]. Specifically, FOXA1, FOXM1, and FOXO1 have been recognized as key regulators in the development of the mammary glands and modulating lactation hormones during the reproductive cycle and pregnancy in mammals [80,99,100,101].
Furthermore, our study revealed a notable cooperation between the TFs FOXA1 and FOXM1 concerning the MY trait, which is pivotal for regulating hormones [102] that may significantly influence both milk production and immunity.
Other interesting TFs in our analysis included HOXA6 and HOXB5, which are members of the HOX family. These TFs govern cell differentiation, cellular functions, cell proliferation, embryonic development, and tissue stability by regulating the promoter regions of a diverse array of target genes [79]. In mammals, HOX genes play a critical role in shaping body anatomy, the skeletal and central nervous systems, and regulating mammary glands and lactation [103]. Additionally, they relate to genetic susceptibility to mammary gland tumors [79] and to human breast cancer [104].
Similarly, we also identified another member of the PPARs family that plays a vital role as a nuclear receptor crucial for processes such as lipid metabolism and immune responses [105]. Within the context of mammary glands, they hold a pivotal role in the regulation of milk fat synthesis, secretion, and transportation [106]. Notably, as elucidated by Liu et al. [107], PPARG is expressed not only within the nucleus of bovine mammary epithelial cells but also upregulates the expression of key genes, such as fatty acid synthase (FASN) and acyl-CoA synthetase short-chain family member 2 (ACSS2) during the lactation phase in goat mammary gland epithelial cells [108]. Collectively, these studies reveal the positive regulatory role of the PPARs family members in the synthesis of milk fat in ruminants [106].
Suppressor of mothers against decapentaplegic (SMAD) family members SMAD3 and SMAD4, on the other hand, modulate transcriptional activities either independently or in combination with various co-regulators [109]. Their scope includes tissue development and homeostasis, pivotal for cellular fate in multicellular systems [110]. They also interface with the transforming growth factor beta (TGF) signaling pathway [111], although TGF shows a negative correlation with certain milk components like casein production and milk secretion [112].
Altogether, our findings on TFs provide critical insights into the regulatory mechanisms of candidate genes, which may help decipher the genetic programs governing specific biological processes related to the development of various milk traits in mammary gland tissue in MUGs. Particularly, milk synthesis results from complex interactions among several tissues and organs, including the immune system, adipose tissue, liver, muscles, and, most importantly, the mammary gland tissue [113], where TFs play a key role in regulating gene expression leading to the production of lactose, protein, fat, and other milk components. This process highly influences both the production and quality of milk. Consequently, the elucidation of TFs and their trait-specific partner choice, on the one hand, enriches our understanding regarding individual milk traits, and, on the other hand, could lead the way for future research to further decipher the genetic basis of these traits and their practical application in MUG breeding.
4. Discussion
Goat milk is consumed as a beverage globally and is a key ingredient in cheese production. The quality of cheese produced from goat milk depends on its fat and protein composition, which are known to be inversely related to milk yield [4,114]. In contrast, the MUG breed demonstrates a significant positive correlation between milk yield and its fat and protein content, along with low SCC, resulting in superior milk quality [4,114]. This breed also maintains consistent milk production during reproductive seasonality [1]. Recent studies [6,17,18,24] have actively explored the genetic and genomic basis of these distinct milk traits in MUGs, aiming to understand the inherent differences and to develop precise breeding strategies for sustainable dairy production. This focus on precision breeding and optimizing dairy production underscores the importance of genetic and genomic research in this field.
In this context, Guan et al. [24] conducted an association analysis using a linear mixed model to explore the relationship between SNP genotypes and dairy traits, identifying, in total, 26 quantitative trait loci linked to seven milk traits (DM, DPM, FP, LP, MY, PP, and SCC) in MUGs. While single-SNP analysis methods, like those used by Guan et al. [24], are effective in identifying key SNPs, they capture only a portion of the genetic variation and make a modest contribution to the variance of complex traits [115,116]. Given the polygenic nature of quantitative traits and the potential for epistatic interactions, focusing solely on individual SNP effects may overlook the broader genetic architecture of these traits. To address this, we employed two distinct approaches in our study: the machine learning-based RF and the information theory-based MIDESP algorithms. These methods were used to identify genotype–phenotype associations, accounting for both single-SNP associations and epistatic interactions. Although both methods aimed to unravel the complex genetic underpinnings of the seven milk traits in the MUGs, Table 2 demonstrates that they identified considerably different candidate genes for each trait, using the same dataset. A limited number of common genes were found for each of the seven milk traits, using both methods. To gain a more comprehensive understanding of the genomic architecture of these traits, it was crucial to consider the combined set of candidate genes identified by both the RF and MIDESP algorithms. This integrative approach has the potential to uncover genetic patterns with subtle individual SNP effects and their significant interactions, revealing the intricate genetic basis of these traits. Of particular interest, we further made a pairwise comparison using combined gene sets among the seven milk traits (Figure 2). Despite the known associations between mammary gland tissue and these traits, our comparative analysis revealed a striking pattern of disparity in the underlying genes for these traits, suggesting a more complex genetic interplay that requires further investigation.
Hence, another fundamental step in our study was the application of the PC-TraFF algorithm [42] for analyzing TFs and their cooperation, since TFs play a pivotal role in regulating gene expression, thereby strongly influencing the traits by directing biological processes and developmental pathways [117,118,119]. These are crucial for understanding the complex genetic programs and the transcriptional machinery of these traits. It is widely recognized today that TFs change their partners depending on their biological functions, and their cooperation is essential for the precise orchestration of specific genetic programs [61]. Previous studies have demonstrated that the preferential partner choice of TFs is achieved through a non-random process, which is strongly influenced by their specific roles in distinct biological functions and the cellular context [59,60,61,118,120]. In particular, within this context, we identified the TF DBP as a central hub for DPM and LP traits, consistently cooperating with TFs HAND1E47, SMAD4, E2F1, and THAP1 across various traits (Figure 3). The role of the DBP factor in immunity and milk production in sheep and cattle [121,122] has also been discovered, thereby strengthening our results. Similarly, Lemay et al. [68] uncovered the role of DBP in regulating mammary development by controlling circadian rhythms during lactation in mice. In contrast, Casey et al. [123] noted DBP’s downregulation for intracellular circadian rhythms in mammary gland tissue during the pregnancy-to-lactation transition. This difference in the role of DBP in the mammary gland tissue may be attributed to the transition from one phase of pregnancy to the next productive phase of lactation. Additionally, we frequently observed the cooperation of DBP with HAND1E47, possibly suggesting their potential involvement in regulating processes related to immunity and milk yield synthesis in the udder in response to circadian rhythms during the lactation period, as explained in Section 3.
Similarly, SMAD3/4 were involved in regulating various milk traits in our study. They influence lactation performance through signaling pathways within the mammary gland, as found in mammals [124]. Our findings revealed the cooperation of E2F1 with SMAD3/4, particularly concerning protein percentage, dry matter percentage, and milk yield. This collaboration was also identified in mammary cancer, where it acts as a suppressor [125,126], and both factors are additionally implicated in the regulation of fat metabolism [127].
Another important TF identified in our findings, BATF, serves as a central node in regulating fat percentage along with other milk production traits. The role of BATF in immunity and fat metabolism, as identified in previous studies [128,129], aligns with our results. Remarkably, the cooperation between FOXO1 and BATF, identified for the DM trait, has a substantial impact on correct B cell proliferation [130]. Additionally, BATF and FOXA1 factors have been previously noted in various processes, including T cell development [131] and genomic regulatory regions in mammary glands epithelial cells [80]. Additionally, our findings indicate THAP1’s frequent cooperation with PPARA for different traits, suggesting its regulatory role in gene expression within the mammary glands. This contributes to lactation by assisting in the synthesis of milk constituents. Likewise, the positive regulatory role of PPARA in goat mammary glands epithelial cells for the synthesis and transportation of fat has been confirmed by various studies [106,132].
Overall, our findings revealed a notable divergence in the candidate genes among the seven milk traits in the MUG breed, while highlighting a surprising similarity in the TFs regulating them. This observation underscores a fundamental aspect of gene regulation: distinct gene profiles are governed by specific sets of TFs. Our analysis further unveiled a critical mechanism in the regulation of specific traits—most TFs are shared by all networks (Figure 3), yet they exhibit dynamic partner-switching behavior. This could suggest a highly evolved regulatory flexibility, wherein TFs modulate different phenotypic traits by changing their cooperation partners. These insights not only deepen our understanding of gene regulatory networks but also pave the way for further investigation into the intricate role of TFs in phenotype determination. These factors play direct or indirect roles in lactation through the synthesis of milk components. Specific regulatory factors such as FOXA1, FOXM1, BATF, DBP, and HAND1E47 are involved in regulating target genes crucial for both milk production and udder immunity, thereby influencing milk quality. However, it is crucial to note that our knowledge about the effects of these regulators is still evolving.
While this study focused on TFs and their preferred partner choices influencing milk traits in the MUG breed, similar regulatory mechanisms may be relevant across other dairy breeds, such as sheep. Many TFs involved in milk production might be conserved among ruminants [106], suggesting that consideration of TF cooperation could potentially be used for improved dairy performance. However, breed-specific factors, such as unique genetic backgrounds and environmental adaptations, could influence the extent and nature of TF interactions. Further studies across diverse dairy breeds would help validate the broader applicability of these regulatory mechanisms.
5. Conclusions
Milk production is a primary trait of the MUG breed, and understanding the genetic framework behind complex milk traits, which are regulated by multiple genes, is essential for optimizing milk yield and quality. In this study, we applied a machine learning approach and an information theory-based approach to analyze genotype–phenotype data. Our results reveal that, although distinct sets of genes contribute to different milk traits, their regulatory factors show strong similarities. Specifically, networks of TF pairs indicate that five key families dominate, comprising 13 of the 32 unique TFs identified. This underscores the importance of examining the cooperative interactions and partner-switching behavior of these TFs, based on their roles in regulating various traits, to gain valuable insights into the biological mechanisms underlying milk production in MUGs.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biology13110929/s1: Figure S1: Identification of the optimal number of SNPs associated with milk yield at 210 days through using incremental feature selection by considering 3000 SNPs.; Figure S2: Identification of the optimal number of SNPs associated with milk yield at 210 days through using incremental feature selection by considering 5000 SNPs; Figure S3: Expression values of TF genes (DBP, HAND1E47, HOXA4, PPARA, and THAP1) in mammary gland tissue of Murciano-Granadina goats at three different time points; Table S1: Number of SNPs and the corresponding genes identified using RF-based GWAS for each trait; Table S2: Number of SNP pairs and the corresponding genes identified in SNP–SNP interaction epistasis analysis for each trait; Table S3: Expression values of TF genes, measured in three different time points. The expression values are given as counts per million (CPM) values.
Author Contributions
M.G. designed and supervised the research; M.I.K. conducted computational analyses, prepared the data sets, and performed the literature survey; H.B. and M.I.K. developed the analysis pipeline and conducted the bioinformatics analyses; A.O.S. and F.R. were involved in the interpretation of the results, together with M.I.K.; M.G. and M.I.K. wrote the final version of the manuscript; M.G. conceived and managed the project. All authors have read and agreed to the published version of the manuscript.
Funding
This article was funded by the Open Access Publication Fund of South Westphalia University of Applied Sciences.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We acknowledge support by the Open Access Publication Fund of South Westphalia University of Applied Sciences. This work is part of MIK’s doctoral program, which is funded by the Higher Education Commission of Pakistan (HEC) in cooperation with Deutscher Akademischer Austauschdienst (DAAD). We would like to thank Akram Abdolmaleki, Muhammad Jawad, and Ghulam Asghar Sajid for proofreading the manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of commercial or financial relationships that could be construed as potential conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MUG | Murciano-Granadina goat |
| CAPRIGRAN | National Association of Murciano-Granadina Goat Breeders |
| SNPs | single-nucleotide polymorphisms |
| TFs | transcription factors |
| TFBSs | transcription factor binding sites |
| MIDESP | mutual information-based detection of epistatic SNP pairs |
| RF | random forest |
| PC-TraFF | potentially collaborating transcription factor finder |
| DM | dry matter percentage |
| DPM | milk production days |
| FP | fat percentage |
| PP | protein percentage |
| LP | lactose percentage |
| MY | milk yield at 210 days |
| SCC | somatic cell count |
| IFS | incremental feature selection |
| MI | mutual information |
| PMI | pointwise mutual information |
| PWM | position weight matrices |
| DBP | D-site binding protein |
| HOX | homeobox |
| PPARs | peroxisome proliferator activated receptors |
| THAP | Thanatos-associated proteins |
| DLX | distal-less homeobox |
| FOX | forkhead box |
| SMAD | suppressor of mothers against decapentaplegic |
References
- Delgado, J.V.; Landi, V.; Barba, C.J.; Fernández, J.; Gómez, M.M.; Camacho, M.E.; Martínez, M.A.; Navas, F.J.; León, J.M. Murciano-Granadina goat: A Spanish local breed ready for the challenges of the twenty-first century. In Sustainable Goat Production in Adverse Environments: Volume II: Local Goat Breeds; Springer: Cham, Switzerland, 2017; pp. 205–219. [Google Scholar]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.D.A. Software-automatized individual lactation model fitting, peak and persistence and Bayesian criteria comparison for milk yield genetic studies in Murciano-Granadina goats. Mathematics 2020, 8, 1505. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.V.; Delgado Bermejo, J.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Goat milk nutritional quality software-automatized individual curve model fitting, shape parameters calculation and Bayesian flexibility criteria comparison. Animals 2020, 10, 1693. [Google Scholar] [CrossRef] [PubMed]
- Magro, S.; Costa, A.; De Marchi, M.; Manuelian, C.L. Milk-related performances of Murciano-Granadina goats reared in Italy compared to cosmopolitan breeds. Ital. J. Anim. Sci. 2022, 21, 1170–1180. [Google Scholar] [CrossRef]
- Vacca, G.M.; Stocco, G.; Dettori, M.L.; Pira, E.; Bittante, G.; Pazzola, M. Milk yield, quality, and coagulation properties of 6 breeds of goats: Environmental and individual variability. J. Dairy Sci. 2018, 101, 7236–7247. [Google Scholar] [CrossRef]
- Guan, D.; Martínez, A.; Luigi-Sierra, M.G.; Delgado, J.V.; Landi, V.; Castelló, A.; Fernández Álvarez, J.; Such, X.; Jordana, J.; Amills, M. Detecting the footprint of selection on the genomes of Murciano-Granadina goats. Anim. Genet. 2021, 52, 683–693. [Google Scholar] [CrossRef]
- Martin, P.; Palhière, I.; Maroteau, C.; Bardou, P.; Canale-Tabet, K.; Sarry, J.; Woloszyn, F.; Bertrand-Michel, J.; Racke, I.; Besir, H.; et al. A genome scan for milk production traits in dairy goats reveals two new mutations in Dgat1 reducing milk fat content. Sci. Rep. 2017, 7, 1872. [Google Scholar] [CrossRef]
- Mucha, S.; Mrode, R.; Coffey, M.; Kizilaslan, M.; Desire, S.; Conington, J. Genome-wide association study of conformation and milk yield in mixed-breed dairy goats. J. Dairy Sci. 2018, 101, 2213–2225. [Google Scholar] [CrossRef]
- Scholtens, M.; Jiang, A.; Smith, A.; Littlejohn, M.; Lehnert, K.; Snell, R.; Lopez-Villalobos, N.; Garrick, D.; Blair, H. Genome-wide association studies of lactation yields of milk, fat, protein and somatic cell score in New Zealand dairy goats. J. Anim. Sci. Biotechnol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Massender, E.; Oliveira, H.R.; Brito, L.F.; Maignel, L.; Jafarikia, M.; Baes, C.F.; Sullivan, B.; Schenkel, F.S. Genome-wide association study for milk production and conformation traits in Canadian Alpine and Saanen dairy goats. J. Dairy Sci. 2023, 106, 1168–1189. [Google Scholar] [CrossRef]
- Roldán, D.; Rabasa, A.E.; Saldaño, S.; Holgado, F.; Poli, M.; Cantet, R.J.C. Qtl detection for milk production traits in goats using a longitudinal model. J. Anim. Breed. Genet. 2008, 125, 187–193. [Google Scholar] [CrossRef]
- Maroteau, C.; Palhière, I.P.; Larroque, H.H.; Clément, V.; Tosser-Klopp, G.; Rupp, R. QTL detection for traits of interest for the dairy goat industry. In Proceedings of the 64th Annual Meeting of the European Federation of Animal Science (EAAP), Nantes, France, 26–30 August 2013; Wageningen Academic Publishers: Wageningen, The Netherlands, 2013; Volume 19, p. 665. [Google Scholar]
- Jiang, A.; Ankersmit-Udy, A.; Turner, S.A.; Scholtens, M.; Littlejohn, M.D.; Lopez-Villalobos, N.; Proser, C.G.; Snell, R.G.; Lehnert, K. A capra hircus chromosome 19 locus linked to milk production influences mammary conformation. J. Anim. Sci. Biotechnol. 2022, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Badaoui, B.; Serradilla, J.; Tomas, A.; Urrutia, B.; Ares, J.; Carrizosa, J.; Sanchez, A.; Jordana, J.; Amills, M. Goat acetyl-coenzyme A carboxylase α: Molecular characterization, polymorphism, and association with milk traits. J. Dairy Sci. 2007, 90, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Badaoui, B.; Serradilla, J.; Tomas, A.; Urrutia, B.; Ares, J.; Carrizosa, J.; Sànchez, A.; Jordana, J.; Amills, M. Identification of two polymorphisms in the goat lipoprotein lipase gene and their association with milk production traits. J. Dairy Sci. 2007, 90, 3012–3017. [Google Scholar] [CrossRef] [PubMed]
- Zidi, A.; Amills, M.; Tomás, A.; Vidal, O.; Ramírez, O.; Carrizosa, J.; Urrutia, B.; Serradilla, J.M.; Clop, A. Genetic variability in the predicted microRNA target sites of caprine casein genes. J. Dairy Sci. 2010, 93, 1749–1753. [Google Scholar] [CrossRef]
- Inostroza, M.G.P.; González, F.J.N.; Landi, V.; Jurado, J.M.L.; Bermejo, J.V.D.; Fernández Álvarez, J.; Martínez Martínez, M.D.A. Bayesian analysis of the association between casein complex haplotype variants and milk yield, composition, and curve shape parameters in Murciano-Granadina goats. Animals 2020, 10, 1845. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Landi, V.; Navas González, F.J.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.D.A. Integrating casein complex SNPs additive, dominance and epistatic effects on genetic parameters and breeding values estimation for murciano-granadina goat milk yield and components. Genes 2020, 11, 309. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Landi, V.; Guan, D.; Delgado, J.V.; Castelló, A.; Cabrera, B.; Mármol-Sánchez, E.; Alvarez, J.F.; Gómez-Carpio, M.; Martínez, A.; et al. A genome-wide association analysis for body, udder, and leg conformation traits recorded in Murciano-Granadina goats. J. Dairy Sci. 2020, 103, 11605–11617. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Fernández, A.; Martínez, A.; Guan, D.; Delgado, J.V.; Álvarez, J.F.; Landi, V.; Such, F.X.; Jordana, J.; Saura, M.; et al. Genomic patterns of homozygosity and inbreeding depression in Murciano-Granadina goats. J. Anim. Sci. Biotechnol. 2022, 13, 35. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Landi, V.; Guan, D.; Delgado, J.V.; Castelló, A.; Cabrera, B.; Mármol-Sánchez, E.; Fernández-Álvarez, J.; Martínez, A.; Such, X.; et al. Identification of genomic regions associated with morphological traits in Murciano-Granadina goats. In Proceedings of the 37th International Conference on Animal Genetics (ISAG), Lleida, Spain, 7–12 July 2019. [Google Scholar]
- Zidi, A.; Serradilla, J.M.; Jordana, J.; Carrizosa, J.; Urrutia, B.; Polvillo, O.; González-Redondo, P.; Gallardo, D.; Amills, M.; Fernández-Cabanás, V.M. Polymorphism of the caprine malic enzyme 1 (ME1) gene and its association with milk quality traits in Murciano-Granadina goats. Animal 2010, 4, 1953–1957. [Google Scholar] [CrossRef]
- Zidi, A.; Fernández-Cabanás, V.M.; Urrutia, B.; Carrizosa, J.; Polvillo, O.; González-Redondo, P.; Jordana, J.; Gallardo, D.; Amills, M.; Serradilla, J.M. Association between the polymorphism of the goat stearoyl-CoA desaturase 1 (SCD1) gene and milk fatty acid composition in Murciano-Granadina goats. J. Dairy Sci. 2010, 93, 4332–4339. [Google Scholar] [CrossRef]
- Guan, D.; Landi, V.; Luigi-Sierra, M.G.; Delgado, J.V.; Such, X.; Castelló, A.; Cabrera, B.; Mármol-Sánchez, E.; Fernández-Alvarez, J.; de la Torre Casañas, J.L.R.; et al. Analyzing the genomic and transcriptomic architecture of milk traits in Murciano-Granadina goats. J. Anim. Sci. Biotechnol. 2020, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Manunza, A.; Zidi, A.; Fernández-Cabanás, V.M.; Jordana, J.; Carrizosa, J.; Belaifa, E.; Urrutia, B.; Polvillo, O.; González-Redondo, P.; Amills, M.; et al. An association analysis between one missense polymorphism at the SREBF1 gene and milk yield and composition traits in goats. Can. J. Anim. Sci. 2012, 92, 167–173. [Google Scholar] [CrossRef]
- Mackay, T.F. Epistasis and quantitative traits: Using model organisms to study gene-gene interactions. Nat. Rev. Genet. 2014, 15, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, F.; Ramzan, F.; Rajavel, A.; Schmitt, A.O.; Gültas, M. MIDESP: Mutual Information-Based Detection of Epistatic SNP Pairs for Qualitative and Quantitative Phenotypes. Biology 2021, 10, 921. [Google Scholar] [CrossRef]
- Wang, H.; Bennett, D.A.; De Jager, P.L.; Zhang, Q.Y.; Zhang, H.Y. Genome-wide epistasis analysis for Alzheimer’s disease and implications for genetic risk prediction. Alzheimer’s Res. Ther. 2021, 13, 55. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, S.; Zou, F.; Wang, W. TEAM: Efficient two-locus epistasis tests in human genome-wide association study. Bioinformatics 2010, 26, i217–i227. [Google Scholar] [CrossRef]
- Li, X. A fast and exhaustive method for heterogeneity and epistasis analysis based on multi-objective optimization. Bioinformatics 2017, 33, 2829–2836. [Google Scholar] [CrossRef]
- D’Silva, S.; Chakraborty, S.; Kahali, B. Concurrent outcomes from multiple approaches of epistasis analysis for human body mass index associated loci provide insights into obesity biology. Sci. Rep. 2022, 12, 7306. [Google Scholar] [CrossRef]
- Liang, Z.; Prakapenka, D.; Parker Gaddis, K.L.; VandeHaar, M.J.; Weigel, K.A.; Tempelman, R.J.; Koltes, J.E.; Santos, J.E.P.; White, H.M.; Peñagaricano, F.; et al. Impact of epistasis effects on the accuracy of predicting phenotypic values of residual feed intake in U.S Holstein cows. Front. Genet. 2022, 13, 1017490. [Google Scholar] [CrossRef]
- Prakapenka, D.; Liang, Z.; Jiang, J.; Ma, L.; Da, Y. A Large-Scale Genome-Wide Association Study of Epistasis Effects of Production Traits and Daughter Pregnancy Rate in U.S. Holstein Cattle. Genes 2021, 12, 1089. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.H.; Xu, W.; Smith, C.W.; Murray, S.C.; Zhang, H.B. Analysis of the genes controlling three quantitative traits in three diverse plant species reveals the molecular basis of quantitative traits. Sci. Rep. 2020, 10, 10074. [Google Scholar] [CrossRef]
- Linder, R.A.; Seidl, F.; Ha, K.; Ehrenreich, I.M. The complex genetic and molecular basis of a model quantitative trait. Mol. Biol. Cell 2016, 27, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Yahyaoui, M.H. Genetic Polymorphism in Goat: Study of the Kappa Casein, Beta Lactoglobulin, and Stearoyl Coenzyme A Desaturase Genes. Ph.D. Dissertation, Facultad de Veterinaria, Universidad Autónoma de Barcelona, Barcelona, Spain, 2003. [Google Scholar]
- Ramzan, F.; Klees, S.; Schmitt, A.O.; Cavero, D.; Gültas, M. Identification of age-specific and common key regulatory mechanisms governing eggshell strength in chicken using random forests. Genes 2020, 11, 464. [Google Scholar] [CrossRef]
- Ramzan, F.; Gültas, M.; Bertram, H.; Cavero, D.; Schmitt, A.O. Combining random forests and a signal detection method leads to the robust detection of genotype-phenotype associations. Genes 2020, 11, 892. [Google Scholar] [CrossRef]
- Klees, S.; Lange, T.M.; Bertram, H.; Rajavel, A.; Schlüter, J.S.; Lu, K.; Schmitt, A.O.; Gültas, M. In Silico Identification of the Complex Interplay between Regulatory SNPs, Transcription Factors, and Their Related Genes in Brassica napus L. Using Multi-Omics Data. Int. J. Mol. Sci. 2021, 22, 789. [Google Scholar] [CrossRef]
- Haleem, A.; Klees, S.; Schmitt, A.O.; Gültas, M. Deciphering Pleiotropic Signatures of Regulatory SNPs in Zea mays L. Using Multi-Omics Data and Machine Learning Algorithms. Int. J. Mol. Sci. 2022, 23, 5121. [Google Scholar] [CrossRef]
- Meckbach, C.; Tacke, R.; Hua, X.; Waack, S.; Wingender, E.; Gültas, M. PC-TraFF: Identification of potentially collaborating transcription factors using pointwise mutual information. BMC Bioinform. 2015, 16, 400. [Google Scholar] [CrossRef]
- Meckbach, C.; Wingender, E.; Gültas, M. Removing background co-occurrences of transcription factor binding sites greatly improves the prediction of specific transcription factor cooperations. Front. Genet. 2018, 9, 189. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Mekonnen, Y.A.; Gültas, M.; Effa, K.; Hanotte, O.; Schmitt, A.O. Identification of candidate signature genes and key regulators associated with Trypanotolerance in the Sheko Breed. Front. Genet. 2019, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Bahbahani, H.; Salim, B.; Almathen, F.; Al Enezi, F.; Mwacharo, J.M.; Hanotte, O. Signatures of positive selection in African Butana and Kenana dairy zebu cattle. PLoS ONE 2018, 13, e0190446. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the Boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Heinrich, F.; Lange, T.M.; Kircher, M.; Ramzan, F.; Schmitt, A.O.; Gültas, M. Exploring the potential of incremental feature selection to improve genomic prediction accuracy. Genet. Sel. Evol. 2023, 55, 78. [Google Scholar] [CrossRef]
- Li, B.Q.; Hu, L.L.; Chen, L.; Feng, K.Y.; Cai, Y.D.; Chou, K.C. Prediction of protein domain with mRMR feature selection and analysis. PLoS ONE 2012, 7, e39308. [Google Scholar] [CrossRef]
- Li, B.Q.; Feng, K.Y.; Chen, L.; Huang, T.; Cai, Y.D. Prediction of Protein-Protein Interaction Sites by Random Forest Algorithm with mRMR and IFS. PLoS ONE 2012, 7, e43927. [Google Scholar] [CrossRef]
- Wei, W.H.; Hemani, G.; Haley, C.S. Detecting epistasis in human complex traits. Nat. Rev. Genet. 2014, 15, 722–733. [Google Scholar] [CrossRef]
- Phillips, P.C. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef]
- Huang, W.; Richards, S.; Carbone, M.A.; Zhu, D.; Anholt, R.R.; Ayroles, J.F.; Duncan, L.; Jordan, K.W.; Lawrence, F.; Magwire, M.M.; et al. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. USA 2012, 109, 15553–15559. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T.; et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017, 49, 643–650. [Google Scholar] [CrossRef]
- Wingender, E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Briefings Bioinform. 2008, 9, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.E.; Gossling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCHTM: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef] [PubMed]
- Taye, M.; Lee, W.; Jeon, S.; Yoon, J.; Dessie, T.; Hanotte, O.; Mwai, O.A.; Kemp, S.; Cho, S.; Oh, S.J.; et al. Exploring evidence of positive selection signatures in cattle breeds selected for different traits. Mamm. Genome 2017, 28, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Rajavel, A.; Klees, S.; Schlüter, J.S.; Bertram, H.; Lu, K.; Schmitt, A.O.; Gültas, M. Unravelling the Complex Interplay of Transcription Factors Orchestrating Seed Oil Content in Brassica napus L. Int. J. Mol. Sci. 2021, 22, 1033. [Google Scholar] [CrossRef]
- Rajavel, A.; Heinrich, F.; Schmitt, A.O.; Gültas, M. Identifying cattle breed-specific partner choice of transcription factors during the African trypanosomiasis disease progression using bioinformatics analysis. Vaccines 2020, 8, 246. [Google Scholar] [CrossRef]
- Steuernagel, L.; Meckbach, C.; Heinrich, F.; Zeidler, S.; Schmitt, A.O.; Gültas, M. Computational identification of tissue-specific transcription factor cooperation in ten cattle tissues. PLoS ONE 2019, 14, e0216475. [Google Scholar] [CrossRef]
- Zeidler, S.; Meckbach, C.; Tacke, R.; Raad, F.S.; Roa, A.; Uchida, S.; Zimmermann, W.H.; Wingender, E.; Gültas, M. Computational detection of stage-specific transcription factor clusters during heart development. Front. Genet. 2016, 7, 33. [Google Scholar] [CrossRef]
- Takahashi, S.; Inoue, I.; Nakajima, Y.; Seo, M.; Nakano, T.; Yang, F.; Kumagai, M.; Komoda, T.; Awata, T.; Ikeda, M.; et al. A promoter in the novel exon of hPPARγ. directs the circadian expression of PPARγ. J. Atheroscler. Thromb. 2010, 17, 73–83. [Google Scholar] [CrossRef]
- Haas, N.B.; Cantwell, C.A.; Johnson, P.F.; Burch, J. DNA-binding specificity of the PAR basic leucine zipper protein VBP partially overlaps those of the C/EBP and CREB/ATF families and is influenced by domains that flank the core basic region. Mol. Cell. Biol. 1995, 15, 1923–1932. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Mitsui, S.; Yan, L.; Yagita, K.; Miyake, S.; Okamura, H. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 2000, 20, 4773–4781. [Google Scholar] [CrossRef] [PubMed]
- Gachon, F.; Olela, F.F.; Schaad, O.; Descombes, P.; Schibler, U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006, 4, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Plaut, K.; Casey, T. Does the circadian system regulate lactation? Animal 2012, 6, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Lemay, D.G.; Neville, M.C.; Rudolph, M.C.; Pollard, K.S.; German, J.B. Gene regulatory networks in lactation: Identification of global principles using bioinformatics. BMC Syst. Biol. 2007, 1, 56. [Google Scholar] [CrossRef]
- Scott, I.C.; Anson-Cartwright, L.; Riley, P.; Reda, D.; Cross, J.C. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell. Biol. 2000, 20, 530–541. [Google Scholar] [CrossRef]
- Zhao, Y.; Johansson, C.; Tran, T.; Bettencourt, R.; Itahana, Y.; Desprez, P.Y.; Konieczny, S.F. Identification of a basic helix-loop-helix transcription factor expressed in mammary gland alveolar cells and required for maintenance of the differentiated state. Mol. Endocrinol. 2006, 20, 2187–2198. [Google Scholar] [CrossRef]
- Kumar, R.; Gupta, I.; Raja, K.; Kumar, V.; Saravanan, R.; Periasamy, K. Sequence characterization of S100A8 gene and its 5’flanking region in Indian Zebu (Bos indicus) and crossbred (Bos indicus X Bos Taurus) cattle. J. Entomol. Zool. Stud. 2020, 8, 1112–1117. [Google Scholar]
- Purba, F.; Nii, T.; Yoshimura, Y.; Isobe, N. Production of antimicrobial peptide S100A8 in the goat mammary gland and effect of intramammary infusion of lipopolysaccharide on S100A8 concentration in milk. J. Dairy Sci. 2019, 102, 4674–4681. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, Z.; Lian, X.; Cheng, X.; Liu, B.; Zhang, B.; Wang, H.; Wang, J.; Li, A.; Ren, Z.; et al. High expression of HOXA4 in patients with glioma indicates unfavorable clinical outcomes. Cell Cycle 2022, 21, 2387–2402. [Google Scholar] [CrossRef]
- Rezsohazy, R.; Saurin, A.J.; Maurel-Zaffran, C.; Graba, Y. Cellular and molecular insights into Hox protein action. Development 2015, 142, 1212–1227. [Google Scholar] [CrossRef]
- Ota, T.; Klausen, C.; Salamanca, M.C.; Woo, H.L.; Leung, P.C.; Auersperg, N. Expression and function of HOXA genes in normal and neoplastic ovarian epithelial cells. Differentiation 2009, 77, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Omatu, T. Overexpression of human homeobox gene in lung cancer A549 cells results in enhanced motile and invasive properties. [Hokkaido Igaku Zasshi] Hokkaido J. Med. Sci. 1999, 74, 367–376. [Google Scholar] [PubMed]
- Kim, H.J.; Roh, M.S.; Son, C.H.; Kim, A.J.; Jee, H.J.; Song, N.; Kim, M.; Seo, S.Y.; Yoo, Y.H.; Yun, J. Loss of Med1/TRAP220 promotes the invasion and metastasis of human non-small-cell lung cancer cells by modulating the expression of metastasis-related genes. Cancer Lett. 2012, 321, 195–202. [Google Scholar] [CrossRef]
- Cheng, S.; Qian, F.; Huang, Q.; Wei, L.; Fu, Y.; Du, Y. HOXA4, down-regulated in lung cancer, inhibits the growth, motility and invasion of lung cancer cells. Cell Death Dis. 2018, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Huang, Q.; Wei, G.H. The role of HOX transcription factors in cancer predisposition and progression. Cancers 2019, 11, 528. [Google Scholar] [CrossRef]
- Pellacani, D.; Bilenky, M.; Kannan, N.; Heravi-Moussavi, A.; Knapp, D.J.; Gakkhar, S.; Moksa, M.; Carles, A.; Moore, R.; Mungall, A.J.; et al. Analysis of normal human mammary epigenomes reveals cell-specific active enhancer states and associated transcription factor networks. Cell Rep. 2016, 17, 2060–2074. [Google Scholar] [CrossRef]
- Capuco, A.V.; Bickhart, D.; Li, C.; Evock-Clover, C.M.; Choudhary, R.K.; Grossi, P.; Bertoni, G.; Trevisi, E.; Aiken, G.E.; McLeod, K.R.; et al. Effect of consuming endophyte-infected fescue seed on transcript abundance in the mammary gland of lactating and dry cows, as assessed by RNA sequencing. J. Dairy Sci. 2018, 101, 10478–10494. [Google Scholar] [CrossRef]
- Qian, Z.; Chen, L.; Liu, J.; Jiang, Y.; Zhang, Y. The emerging role of PPAR-alpha in breast cancer. Biomed. Pharmacother. 2023, 161, 114420. [Google Scholar] [CrossRef]
- Gimble, J.M.; Pighetti, G.M.; Lerner, M.R.; Wu, X.; Lightfoot, S.A.; Brackett, D.J.; Darcy, K.; Hollingsworth, A.B. Expression of peroxisome proliferator activated receptor mRNA in normal and tumorigenic rodent mammary glands. Biochem. Biophys. Res. Commun. 1998, 253, 813–817. [Google Scholar] [CrossRef]
- Richter, A.; Hollstein, R.; Hebert, E.; Vulinovic, F.; Eckhold, J.; Osmanovic, A.; Depping, R.; Kaiser, F.J.; Lohmann, K. In-depth characterization of the homodimerization domain of the transcription factor THAP1 and dystonia-causing mutations therein. J. Mol. Neurosci. 2017, 62, 11–16. [Google Scholar] [CrossRef]
- Fuchs, T.; Gavarini, S.; Saunders-Pullman, R.; Raymond, D.; Ehrlich, M.E.; Bressman, S.B.; Ozelius, L.J. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat. Genet. 2009, 41, 286–288. [Google Scholar] [CrossRef] [PubMed]
- LeDoux, M.S. Dystonia: Phenotypes and Genetics. In Movement Disorders; Academic Press: Cambridge, MA, USA, 2015; pp. 415–438. [Google Scholar]
- Zhao, Y.; Xiao, J.; Gong, S.; Clara, J.A.; LeDoux, M.S. Neural expression of the transcription factor THAP1 during development in rat. Neuroscience 2013, 231, 282–295. [Google Scholar] [CrossRef]
- Bozek, K.; Relógio, A.; Kielbasa, S.M.; Heine, M.; Dame, C.; Kramer, A.; Herzel, H. Regulation of clock-controlled genes in mammals. PLoS ONE 2009, 4, e4882. [Google Scholar] [CrossRef]
- Casey, T.; Crodian, J.; Suárez-Trujillo, A.; Erickson, E.; Weldon, B.; Crow, K.; Cummings, S.; Chen, Y.; Shamay, A.; Mabjeesh, S.J.; et al. CLOCK regulates mammary epithelial cell growth and differentiation. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2016, 311, R1125–R1134. [Google Scholar] [CrossRef]
- Morasso, M.I.; Radoja, N. Dlx genes, p63, and ectodermal dysplasias. Birth Defects Res. Part C Embryo Today Rev. 2005, 75, 163–171. [Google Scholar] [CrossRef]
- Lindtner, S.; Catta-Preta, R.; Tian, H.; Su-Feher, L.; Price, J.D.; Dickel, D.E.; Greiner, V.; Silberberg, S.N.; McKinsey, G.L.; McManus, M.T.; et al. Genomic resolution of DLX-orchestrated transcriptional circuits driving development of forebrain GABAergic neurons. Cell Rep. 2019, 28, 2048–2063. [Google Scholar] [CrossRef]
- Kouros-Mehr, H.; Werb, Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev. Dyn. 2006, 235, 3404–3412. [Google Scholar] [CrossRef]
- Durán Aguilar, M.; Román Ponce, S.; Ruiz López, F.; González Padilla, E.; Vásquez Peláez, C.; Bagnato, A.; Strillacci, M.G. Genome-wide association study for milk somatic cell score in holstein cattle using copy number variation as markers. J. Anim. Breed. Genet. 2017, 134, 49–59. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.L.; et al. Zebrafish HOX clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Stock, D.W.; Ellies, D.L.; Zhao, Z.; Ekker, M.; Ruddle, F.H.; Weiss, K.M. The evolution of the vertebrate DLX gene family. Proc. Natl. Acad. Sci. USA 1996, 93, 10858–10863. [Google Scholar] [CrossRef]
- Ghanem, N.; Jarinova, O.; Amores, A.; Long, Q.; Hatch, G.; Park, B.K.; Rubenstein, J.L.; Ekker, M. Regulatory roles of conserved intergenic domains in vertebrate DLX bigene clusters. Genome Res. 2003, 13, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Chen, H.; Cai, Z.; Yang, Y.; Feng, Z.; Zeng, M.; Chen, L.; Qin, Y.; Cai, B.; Zhu, P.; et al. Forkhead box family transcription factors as versatile regulators for cellular reprogramming to pluripotency. Cell Regen. 2021, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, M.; Sun, T.; Zhang, Z.; Liu, C. FOXM1: Functional Roles of FOXM1 in Non-Malignant Diseases. Biomolecules 2023, 13, 857. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, G.M.; Keri, R.A. FOXA1: A transcription factor with parallel functions in development and cancer. Biosci. Rep. 2012, 32, 113–130. [Google Scholar] [CrossRef]
- Sreekumar, A.; Toneff, M.J.; Toh, E.; Roarty, K.; Creighton, C.J.; Belka, G.K.; Lee, D.K.; Xu, J.; Chodosh, L.A.; Richards, J.S.; et al. WNT-mediated regulation of FOXO1 constitutes a critical axis maintaining pubertal mammary stem cell homeostasis. Dev. Cell 2017, 43, 436–448. [Google Scholar] [CrossRef]
- Carr, J.R.; Kiefer, M.M.; Park, H.J.; Li, J.; Wang, Z.; Fontanarosa, J.; DeWaal, D.; Kopanja, D.; Benevolenskaya, E.V.; Guzman, G.; et al. FoxM1 regulates mammary luminal cell fate. Cell Rep. 2012, 1, 715–729. [Google Scholar] [CrossRef]
- Castaneda, M.; Hollander, P.; Mani, S.A. Forkhead box transcription factors: Double-edged swords in cancer. Cancer Res. 2022, 82, 2057–2065. [Google Scholar] [CrossRef]
- Lewis, M.T. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2000, 2, 158–169. [Google Scholar] [CrossRef]
- Cantile, M.; Pettinato, G.; Procino, A.; Feliciello, I.; Cindolo, L.; Cillo, C. In vivo expression of the whole HOX gene network in human breast cancer. Eur. J. Cancer 2003, 39, 257–264. [Google Scholar] [CrossRef]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236. [Google Scholar] [CrossRef]
- Mu, T.; Hu, H.; Ma, Y.; Feng, X.; Zhang, J.; Gu, Y. Regulation of key genes for milk fat synthesis in ruminants. Front. Nutr. 2021, 8, 765147. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lin, Y.; Liu, L.; Wang, L.; Bian, Y.; Gao, X.; Li, Q. Regulation of peroxisome proliferator-activated receptor gamma on milk fat synthesis in dairy cow mammary epithelial cells. Vitr. Cell. Dev. Biol.-Anim. 2016, 52, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhao, W.; Luo, J.; Yao, D.; Sun, Y.; Li, J.; Shi, H.P.; Loor, J.J. Peroxisome proliferator-activated receptor γ.1 and γ.2 isoforms alter lipogenic gene networks in goat mammary epithelial cells to different extents. J. Dairy Sci. 2014, 97, 5437–5447. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Le, H.V.; Shen, L.; Anderson, S.A.; Massagué, J. Integration of SMAD and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 2004, 117, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, A.; Ten Dijke, P.; van Dam, H. Key signaling nodes in mammary gland development and cancer: SMAD signal integration in epithelial cell plasticity. Breast Cancer Res. 2012, 14, 204. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, X.; Ren, X.; Tian, Y.; Chen, Z.; Xu, X.; Du, Y.; Jiang, C.; Fang, Y.; Liu, Z.; et al. SMAD2 and SMAD3 have differential sensitivity in relaying TGFβ. signaling and inversely regulate early lineage specification. Sci. Rep. 2016, 6, 21602. [Google Scholar] [CrossRef]
- Weaver, S.; Hernandez, L. Autocrine-paracrine regulation of the mammary gland. J. Dairy Sci. 2016, 99, 842–853. [Google Scholar] [CrossRef]
- Osorio, J.S.; Lohakare, J.; Bionaz, M. Biosynthesis of milk fat, protein, and lactose: Roles of transcriptional and posttranscriptional regulation. Physiol. Genom. 2016, 48, 231–256. [Google Scholar] [CrossRef]
- Pulina, G.; Nudda, A.; Battacone, G.; Fancellu, S.; Francesconi, A. Nutrition and quality of goat’s milk. In Dairy Goats Feeding and Nutrition; CAB International: Wallingford, UK, 2008; pp. 1–30. [Google Scholar]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- He, T.; Hill, C.B.; Angessa, T.T.; Zhang, X.Q.; Chen, K.; Moody, D.; Telfer, P.; Westcott, S.; Li, C. Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection. J. Exp. Bot. 2019, 70, 5603–5616. [Google Scholar] [CrossRef]
- Yang, S.; Gao, Y.; Zhang, S.; Zhang, Q.; Sun, D. Identification of Genetic Associations and Functional Polymorphisms of SAA1 Gene Affecting Milk Production Traits in Dairy Cattle. PLoS ONE 2016, 11, e0162195. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.; de León, N.; Grotewold, E. Challenges of translating gene regulatory information into agronomic improvements. Trends Plant Sci. 2019, 24, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Pajoro, A.; Angenent, G.C. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat. Rev. Genet. 2010, 11, 830–842. [Google Scholar] [CrossRef]
- Oget, C.; Tosser-Klopp, G.; Rupp, R. Genetic and genomic studies in ovine mastitis. Small Rumin. Res. 2019, 176, 55–64. [Google Scholar] [CrossRef]
- Olsen, H.G.; Knutsen, T.M.; Lewandowska-Sabat, A.M.; Grove, H.; Nome, T.; Svendsen, M.; Arnyasi, M.; Sodeland, M.; Sundsaasen, K.K.; Dahl, S.R.; et al. Fine mapping of a QTL on bovine chromosome 6 using imputed full sequence data suggests a key role for the group-specific component (GC) gene in clinical mastitis and milk production. Genet. Sel. Evol. 2016, 48, 79. [Google Scholar] [CrossRef]
- Casey, T.; Patel, O.; Dykema, K.; Dover, H.; Furge, K.; Plaut, K. Molecular signatures reveal circadian clocks may orchestrate the homeorhetic response to lactation. PLoS ONE 2009, 4, e7395. [Google Scholar] [CrossRef]
- Cocolakis, E.; Dai, M.; Drevet, L.; Ho, J.; Haines, E.; Ali, S.; Lebrun, J.J. SMAD signaling antagonizes STAT5-mediated gene transcription and mammary epithelial cell differentiation. J. Biol. Chem. 2008, 283, 1293–1307. [Google Scholar] [CrossRef]
- Zhang, S. Investigation of TGF-β. Associated Master-like Transcription Factors in Breast Cancer. Ph.D. Thesis, National University of Singapore, Singapore, 2016. [Google Scholar]
- Qi, J.C.; Yang, Z.; Zhang, Y.P.; Lu, B.S.; Yin, Y.W.; Liu, K.L.; Xue, W.Y.; Qu, C.B.; Li, W. miR-20b-5p, TGFBR2, and E2F1 form a regulatory loop to participate in epithelial to mesenchymal transition in prostate cancer. Front. Oncol. 2020, 9, 1535. [Google Scholar] [CrossRef]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef]
- Ji, L.S.; Sun, X.H.; Zhang, X.; Zhou, Z.H.; Yu, Z.; Zhu, X.J.; Huang, L.Y.; Fang, M.; Gao, Y.T.; Li, M.; et al. Mechanism of follicular helper T cell differentiation regulated by transcription factors. J. Immunol. Res. 2020, 2020, 1826587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liao, Q.; Pan, T.; Yu, L.; Luo, Z.; Su, S.; Liu, S.; Hou, M.; Li, Y.; Damba, T.; et al. BATF relieves hepatic steatosis by inhibiting PD1 and promoting energy metabolism. eLife 2023, 12, RP88521. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.; Hay, J.; Moles, M.W.; Michie, A.M. The discrete roles of individual FOXO transcription factor family members in B-cell malignancies. Front. Immunol. 2023, 14, 2557. [Google Scholar] [CrossRef]
- Pham, D.; Moseley, C.E.; Gao, M.; Savic, D.; Winstead, C.J.; Sun, M.; Kee, B.L.; Myers, R.M.; Weaver, C.T.; Hatton, R.D. Batf pioneers the reorganization of chromatin in developing effector T cells via Ets1-dependent recruitment of Ctcf. Cell Rep. 2019, 29, 1203–1220. [Google Scholar] [CrossRef]
- Tian, H.; Luo, J.; Shi, H.; Chen, X.; Wu, J.; Liang, Y.; Li, C.; Loor, J.J. Role of peroxisome proliferator-activated receptor-α on the synthesis of monounsaturated fatty acids in goat mammary epithelial cells. J. Anim. Sci. 2020, 98, skaa062. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).