Identification of Candidate Genes for Cold Tolerance at Seedling Stage by GWAS in Rice (Oryza sativa L.)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Evaluation
2.3. Genome-Wide Association Mapping
2.4. Haplotype Analysis for Candidate Genes
3. Results
3.1. Phenotypic Variations in the Cold Tolerance (CT) of Rice at the Seedling Stage
3.2. GWAS for CT
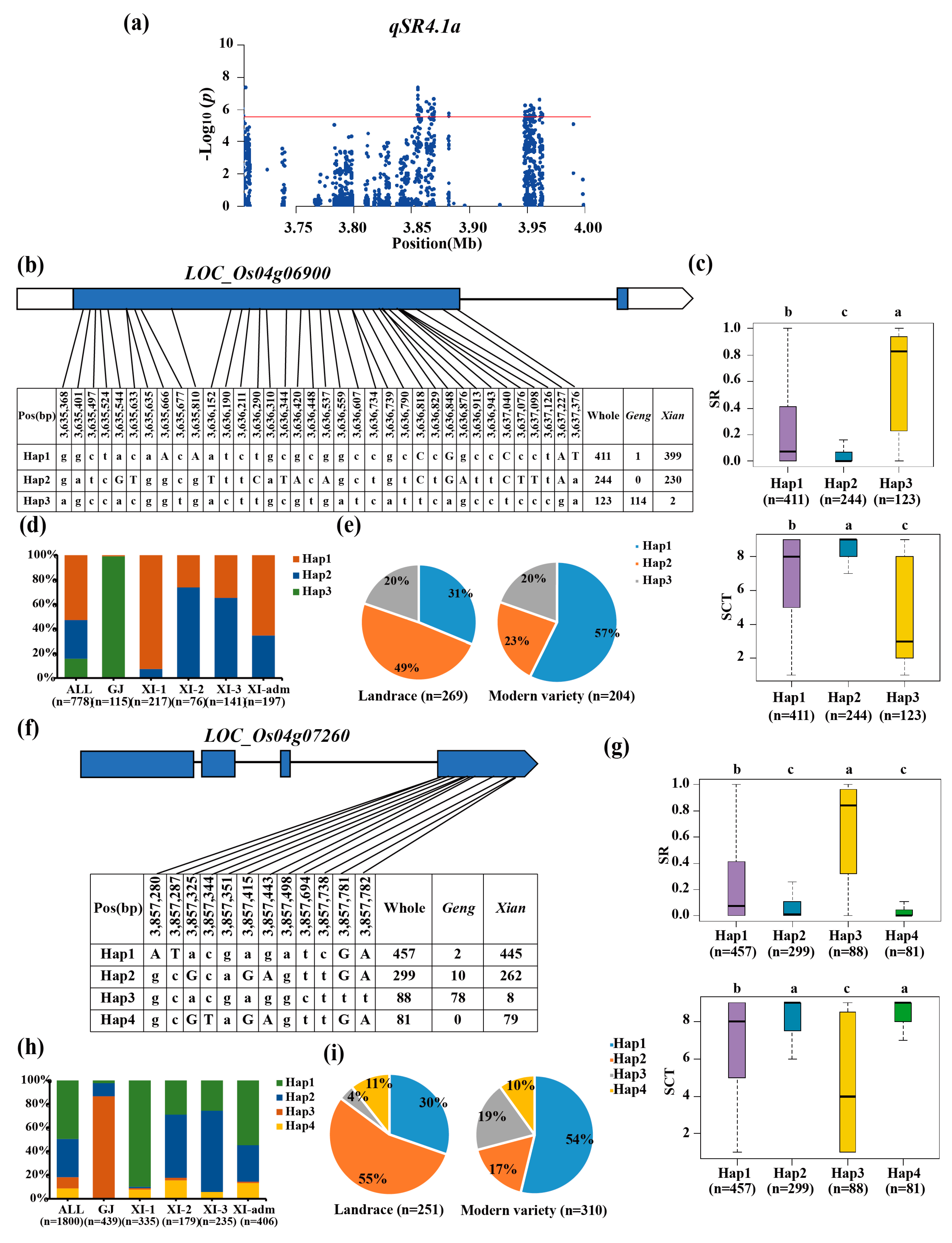
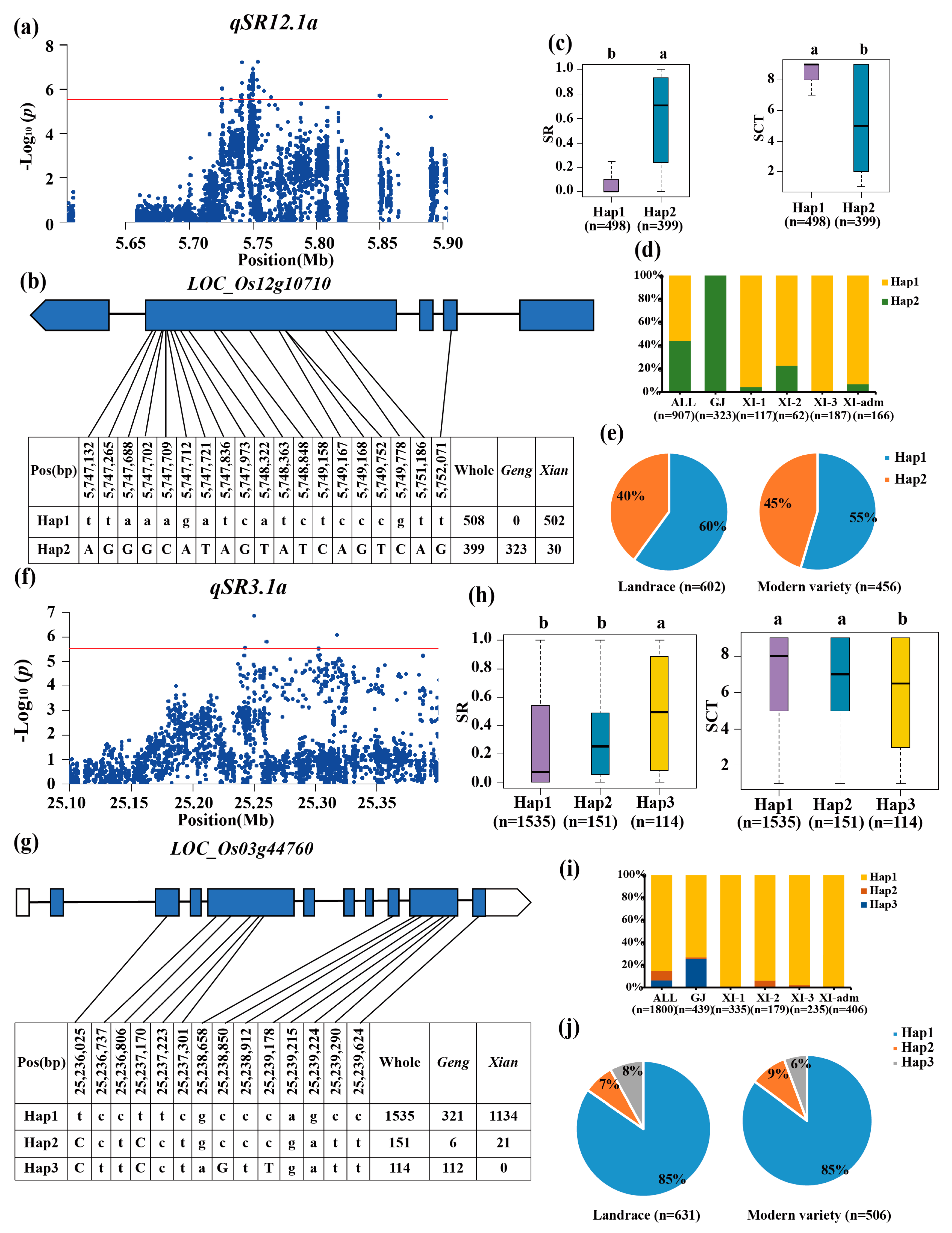
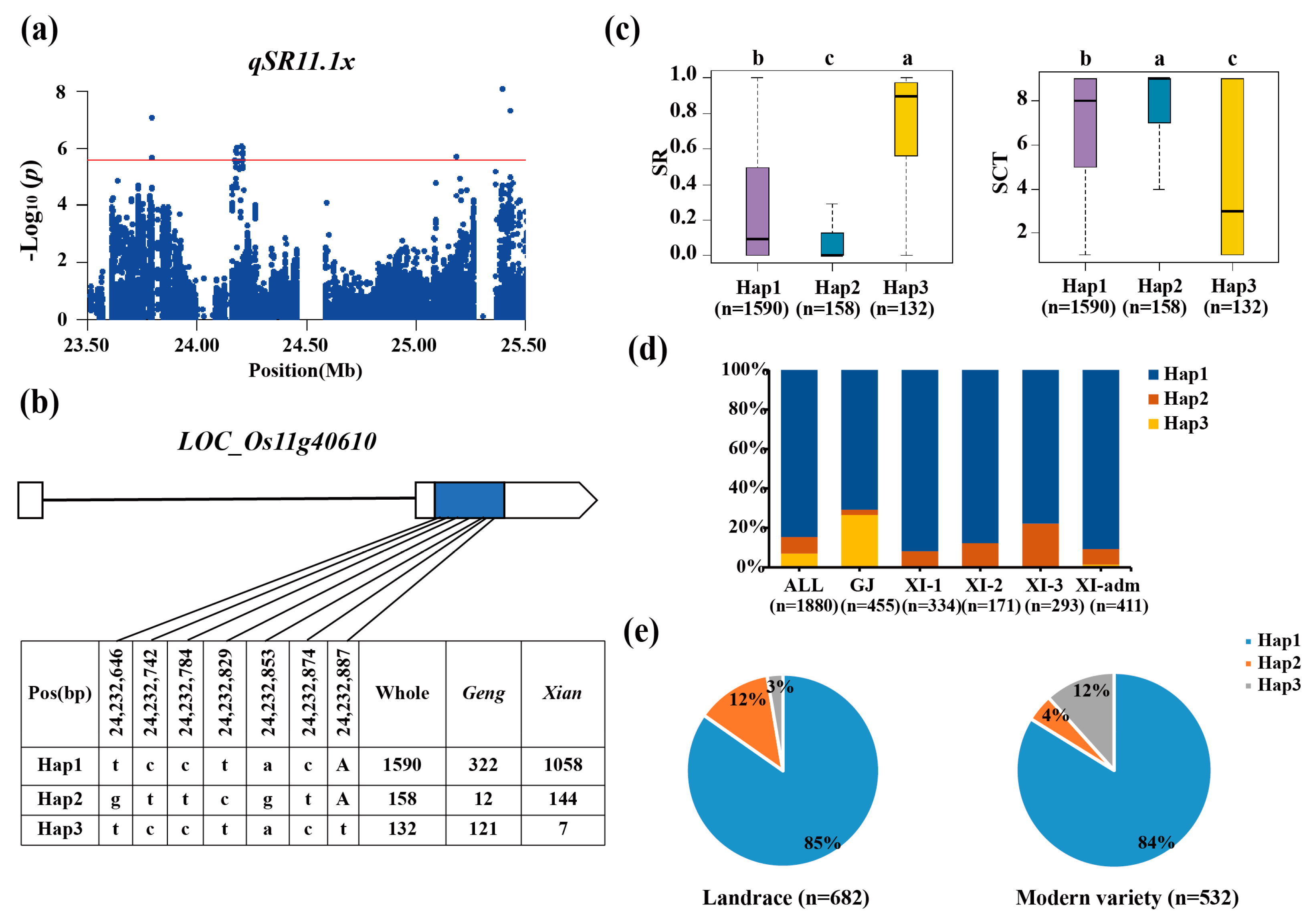
3.3. Haplotype Analyses of the Candidate Genes
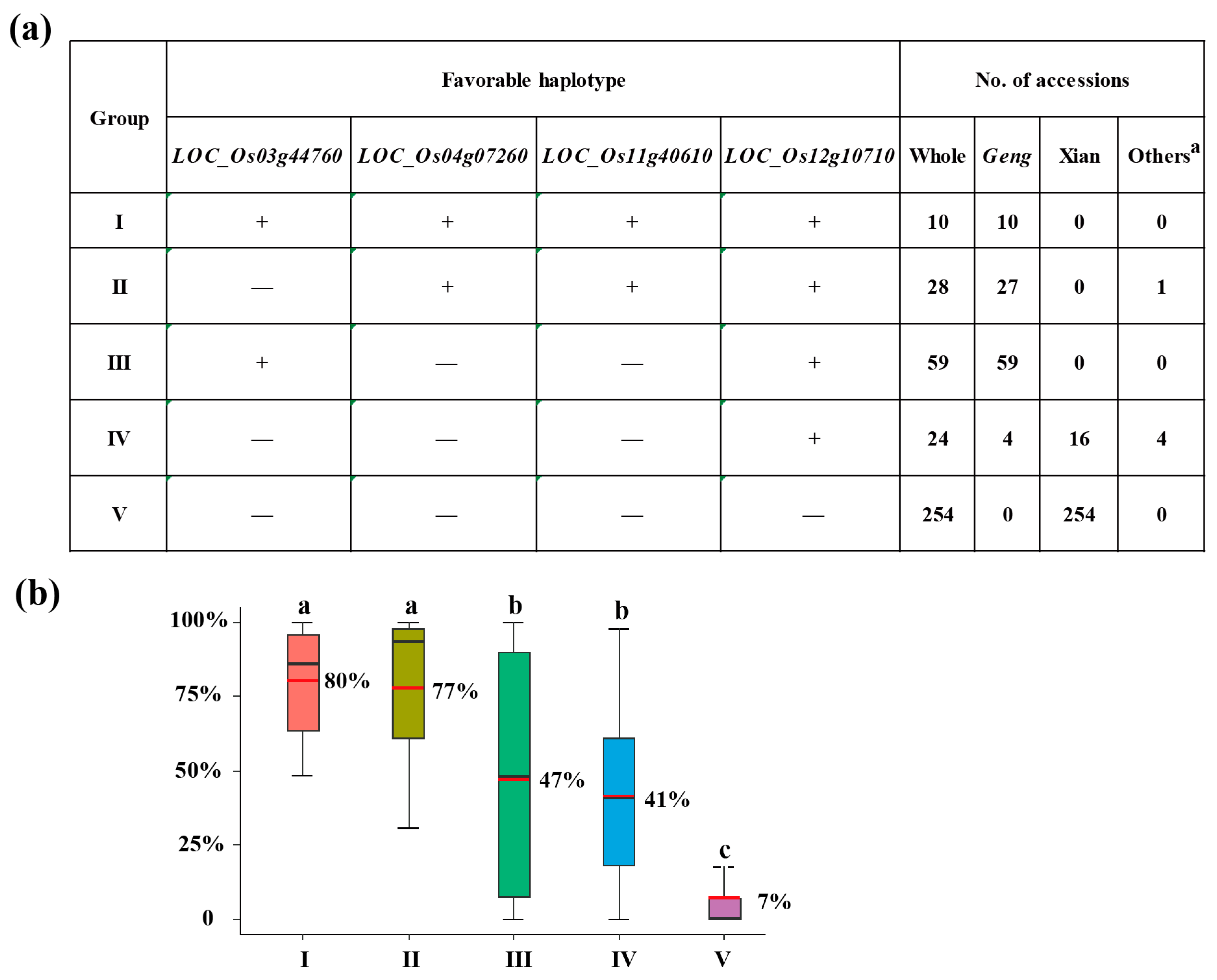
3.4. Optimal Combination of CT-Haplotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W.; et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 1. [Google Scholar] [CrossRef]
- Li, L.; Mao, D.; Prasad, M. Deployment of cold tolerance loci from Oryza sativa ssp. Japonica cv. ‘Nipponbare’ in a highyielding Indica rice cultivar ‘93-11’. Plant Breed. 2018, 137, 553–560. [Google Scholar]
- Fujino, K.; Matsuda, Y. Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Mol. Biol. 2009, 72, 137–152. [Google Scholar] [CrossRef]
- Mao, D.; Xin, Y.; Tan, Y.; Hu, X.; Bai, J.; Liu, Z.; Yu, Y.; Li, L.; Peng, C.; Fan, T.; et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. USA 2019, 116, 3494–3501. [Google Scholar] [CrossRef]
- Saito, K.; Hayano, S.Y.; Kuroki, M.; Sato, Y. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 2010, 179, 97–102. [Google Scholar] [CrossRef]
- Saito, K.; Miura, K.; Nagano, K.; Hayano, S.Y.; Araki, H.; Kato, A. Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length. Theor. Appl. Genet. 2001, 103, 862–868. [Google Scholar] [CrossRef]
- Saito, K.; Hayano, S.Y.; Maruyama-Funatsuki, W.; Sato, Y.; Kato, A. Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor. Appl. Genet. 2004, 109, 515–522. [Google Scholar] [CrossRef]
- Jorde, L.B. Linkage Disequilibrium and the Search for Complex Disease Genes. Genome Res. 2000, 10, 1435–1444. [Google Scholar] [CrossRef]
- Wang, D.; Liu, J.; Li, C.; Kang, H.; Wang, Y.; Tan, X.; Liu, M.; Deng, Y.; Wang, Z.; Liu, Y.; et al. Genome-wide Association Mapping of Cold Tolerance Genes at the Seedling Stage in Rice. Rice 2016, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Guo, Z.; Li, X.; Ye, H.; Li, X.; Xiong, L. New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant Cell Environ. 2015, 39, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zou, B.; Shao, Q.; Cui, Y.; Lu, S.; Zhang, Y.; Huang, Q.; Huang, J.; Hua, J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 2018, 69, 413–421. [Google Scholar] [CrossRef]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic Structure and Diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Tanksley, S.D. Restriction fragment length polymorphism in Oryza sativa L. Genome 1989, 32, 1113–1118. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, L.; Chen, D.; Li, L.; Zhu, Y.; Xiao, Y.; Zhang, D.; Chen, C. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor. Appl. Genet. 2015, 128, 1359–1371. [Google Scholar] [CrossRef]
- Sales, M.A.; Burgos, N.R.; Shivrain, V.K.; Murphy, B.; Gbur, E.E. Morphological and Physiological Responses of Weedy Red Rice (Oryza sativa L.) and Cultivated Rice (O. sativa) to N Supply. Am. J. Plant Sci. 2011, 2, 569–577. [Google Scholar] [CrossRef]
- Alexandrov, N.; Tai, S.; Wang, W.; Mansueto, L.; Palis, K.; Fuentes, R.R.; Ulat, V.J.; Chebotarov, D.; Zhang, G.; Li, Z.; et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015, 43, 1023–1027. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Turner, S. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 25. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.; Xu, J.; He, W.; Yang, T. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Li, G.; Yu, Y.; Ou, Y. funRiceGenes dataset for comprehensive understanding and application of rice functional genes. GigaScience 2018, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ou, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, C.; Li, M.; Cui, Y.; Shi, Y.; Wu, Z.; Hu, Z.; Wang, W.; Xu, J.; Li, Z. The landscape of gene-CDS-haplotype diversity in rice: Properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol. Plant 2021, 14, 787–804. [Google Scholar] [CrossRef] [PubMed]
- Lou, Q.; Guo, H.; Li, J.; Han, S.; Khan, N.U.; Gu, Y.; Zhao, W.; Zhang, Z.; Zhang, H.; Li, Z.; et al. Cold-adaptive evolution at the reproductive stage in Geng/japonica subspecies reveals the role of OsMAPK3 and OsLEA9. Plant J. 2022, 111, 1032–1051. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, L.; Zheng, J.; Chen, F.; Wang, T.; Wang, M.; Tao, Y.; Wang, H.; Hong, Z.; Huang, Y.; et al. A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice. J. Exp. Bot. 2019, 70, 3851–3866. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.; Zhang, B.; Wang, H.; Wang, F.; Liu, M.; Gao, Y.; Zhang, W.; Deng, Z.; Sun, D.; Tang, W. Comparative Study of Early Cold-Regulated Proteins by Two-Dimensional Difference Gel Electrophoresis Reveals a Key Role for Phospholipase Dα1 in Mediating Cold Acclimation Signaling Pathway in Rice. Mol. Cell. Proteom. 2016, 15, 1397–1411. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; Wei, H.; Wang, L. A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ. 2021, 44, 3283–3301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Li, F.; Liu, H.; Yang, W.; Chong, K.; Xu, Y. OsMAPK3 Phosphorylates OsbHLH002/OsICE1 and Inhibits Its Ubiquitination to Activate OsTPP1 and Enhances Rice Chilling Tolerance. Dev. Cell 2017, 43, 731–743.e735. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zeng, Y.; Li, J.; Ma, X.; Zhang, Z.; Lou, Q.; Li, J.; Gu, Y.; Zhang, H.; Li, J.; et al. Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol. J. 2020, 18, 2491–2503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Hao, X.; Gao, Y.; Hua, Z.; Ma, X.; Chen, W.; Xu, Z.; Zhu, L.; Li, Z. Improving seedling cold tolerance of japonica rice by using the “Hidden Diversity” in indica rice germplasm in backcross breeding program. Acta Agron. Sin. 2007, 33, 1618–1624. [Google Scholar]
- Ali, J.; Pan, Y.; Zhang, H.; Zhang, D.; Li, J.; Xiong, H.; Yu, J.; Li, J.; Rashid, M.A.R.; Li, G.; et al. Genetic Analysis of Cold Tolerance at the Germination and Booting Stages in Rice by Association Mapping. PLoS ONE 2015, 10, 3. [Google Scholar]
- Cai, Z.; Zhang, Y.; Tang, W.; Chen, X.; Lin, C.; Liu, Y.; Ye, Y.; Wu, W.; Duan, Y. LUX ARRHYTHMO Interacts With ELF3a and ELF4a to Coordinate Vegetative Growth and Photoperiodic Flowering in Rice. Front. Plant Sci. 2022, 13, 853042. [Google Scholar] [CrossRef]
- Che, L.; Tang, D.; Wang, K.; Wang, M.; Zhu, K.; Yu, H.; Gu, M.; Cheng, Z. OsAM1 is required for leptotene-zygotene transition in rice. Cell Res. 2011, 21, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Storme, N.; Geelen, D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. Plant Cell Environ. 2013, 37, 1–18. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, L.; Yang, D.I.; Zhao, C.; Zheng, Y. Cold stress contributes to aberrant cytokinesis during male meiosis I in a wheat thermosensitive genic male sterile line. Plant Cell Environ. 2010, 34, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Schläppi, M.R.; Mao, B.; Wang, W.; Wang, A.; Chu, C. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 2019, 17, 1834–1849. [Google Scholar] [CrossRef]
- Xiao, N.; Gao, Y.; Qian, H.; Gao, Q.; Wu, Y.; Zhang, D.; Zhang, X.; Yu, L.; Li, Y.; Pan, C.; et al. Identification of Genes Related to Cold Tolerance and a Functional Allele That Confers Cold Tolerance. Plant Physiol. 2018, 177, 1108–1123. [Google Scholar] [CrossRef] [PubMed]

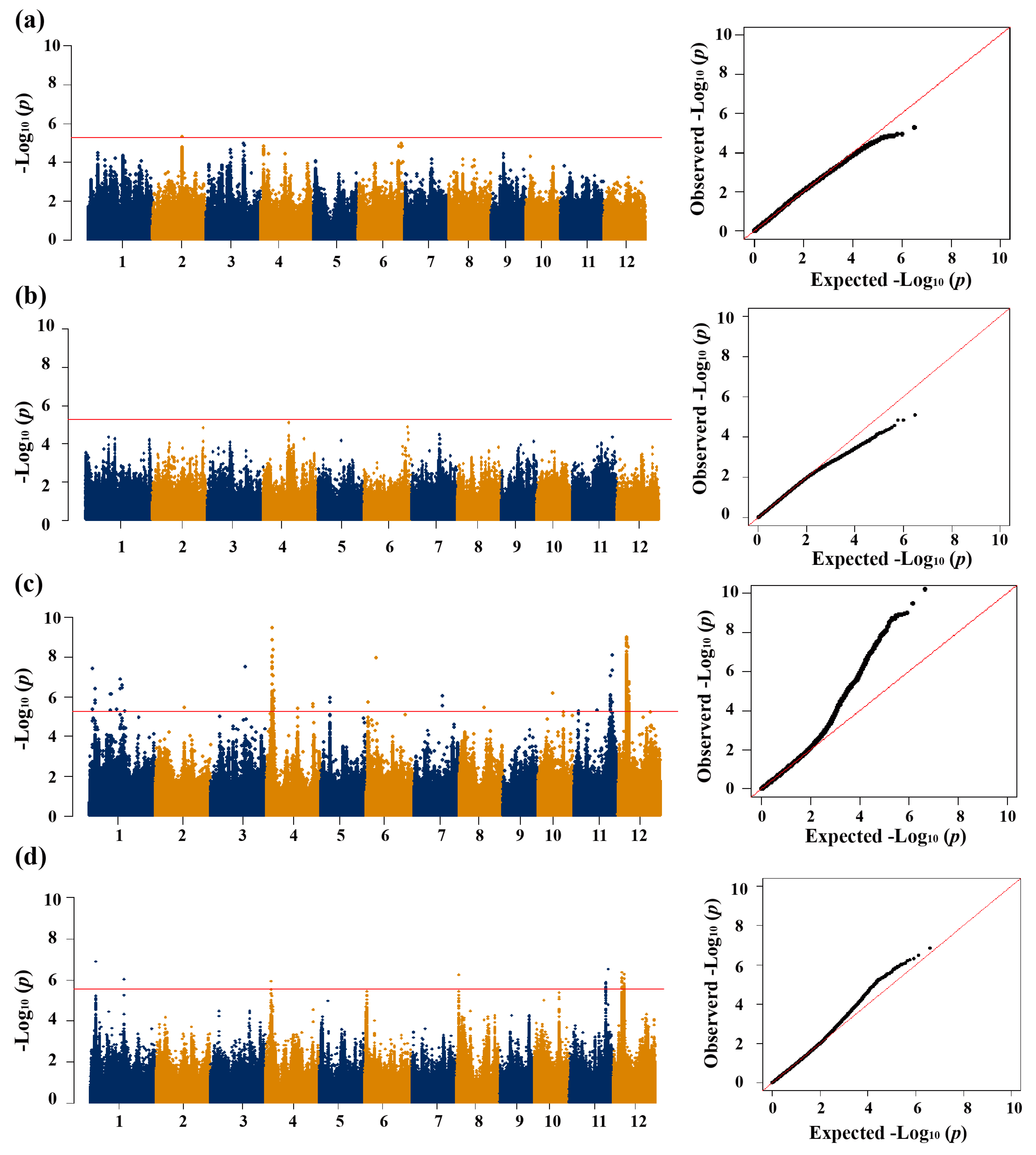
| Trait a | QTL | Chr. | QTL Region (Mb) | Lead SNP | p-Value | Cloned Gene |
|---|---|---|---|---|---|---|
| SR | qSR1.1a | 1 | 6.37–6.67 | 6,518,020 | 6.62 × 10−8 | |
| qSR1.2a | 1 | 11.71–12.01 | 11,864,873 | 6.05 × 10−7 | OsLEA9 [27] | |
| qSR1.3a | 1 | 19.98–20.27 | 20,125,327 | 6.39 × 10−7 | ||
| qSR2.1a | 2 | 6.26–6.56 | 6,416,777 | 3.57 × 10−7 | ||
| qSR3.1a | 3 | 25.10–25.55 | 25,249,852 | 1.36 × 10−7 | ||
| qSR4.1a | 4 | 3.58–3.96 | 3,633,378 | 2.10 × 10−10 | ||
| qSR5.1a | 5 | 19.63–19.93 | 19,779,633 | 5.72 × 10−9 | ||
| qSR12.1a | 12 | 5.60–5.90 | 5,753,724 | 5.54 × 10−8 | ||
| SCT | qSCT1.1a | 1 | 11.53–11.97 | 11,956,876 | 4.74 × 10−8 | OsLEA9 [27] |
| qSCT1.2a | 1 | 28.47–28.77 | 28,623,028 | 1.88 × 10−7 | ||
| qSCT2.1a | 2 | 28.66–28.96 | 28,812,677 | 4.12 × 10−7 | LGS1 [28] | |
| qSCT3.1a | 3 | 25.11–25.41 | 25,260,301 | 8.78 × 10−7 | ||
| qSCT4.1a | 4 | 3.48–3.78 | 3,633,378 | 5.60 × 10−8 | ||
| qSCT8.1a | 8 | 9.18–9.48 | 9,334,380 | 8.64 × 10−7 | ||
| qSCT8.2a | 8 | 12.60–12.90 | 12,755,109 | 1.24 × 10−7 | ||
| qSCT9.1a | 9 | 14.24–14.54 | 14,393,189 | 5.25 × 10−7 |
| Population | Trait a | QTL | Chr. | QTL Region (Mb) | Lead SNP | p-Value | Cloned Gene |
|---|---|---|---|---|---|---|---|
| GJ | SR | qSR2.1g | 2 | 19.64–19.94 | 19,785,654 | 4.94 × 10−6 | |
| XI | SR | qSR1.1x | 1 | 3.45–3.75 | 3,604,713 | 4.19 × 10−7 | OsPLDα1 [29] |
| qSR1.2x | 1 | 20.23–21.03 | 20,231,449 | 1.39 × 10−7 | |||
| qSR4.1x | 4 | 3.63–4.80 | 3,855,187 | 3.33 × 10−10 | |||
| qSR5.1x | 5 | 5.71–6.01 | 5,861,711 | 1.21 × 10−6 | |||
| qSR11.1x | 11 | 23.79–25.42 | 25,393,345 | 8.00 × 10−9 | |||
| qSR12.1x | 12 | 5.71–7.25 | 5,754,489 | 6.05 × 10−11 | |||
| XI | SCT | qSCT1.1x | 1 | 3.45–3.75 | 3,604,730 | 1.34 × 10−7 | OsPLDα1 [29] |
| qSCT1.2x | 1 | 21.88–22.18 | 22,038,470 | 9.39 × 10−7 | |||
| qSCT4.1x | 4 | 3.50–3.80 | 3,653,716 | 1.19 × 10−7 | |||
| qSCT8.1x | 8 | 1.42–1.72 | 1,578,048 | 5.94 × 10−7 | |||
| qSCT11.1x | 11 | 24.03–24.33 | 25,393,345 | 3.17 × 10−7 | |||
| qSCT12.1x | 12 | 5.71–7.33 | 5,754,489 | 4.48 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Zhang, W.; Cao, H.; Zhai, L.; Song, Q.; Xu, J. Identification of Candidate Genes for Cold Tolerance at Seedling Stage by GWAS in Rice (Oryza sativa L.). Biology 2024, 13, 784. https://doi.org/10.3390/biology13100784
Shi H, Zhang W, Cao H, Zhai L, Song Q, Xu J. Identification of Candidate Genes for Cold Tolerance at Seedling Stage by GWAS in Rice (Oryza sativa L.). Biology. 2024; 13(10):784. https://doi.org/10.3390/biology13100784
Chicago/Turabian StyleShi, Huimin, Wenyu Zhang, Huimin Cao, Laiyuan Zhai, Qingxin Song, and Jianlong Xu. 2024. "Identification of Candidate Genes for Cold Tolerance at Seedling Stage by GWAS in Rice (Oryza sativa L.)" Biology 13, no. 10: 784. https://doi.org/10.3390/biology13100784
APA StyleShi, H., Zhang, W., Cao, H., Zhai, L., Song, Q., & Xu, J. (2024). Identification of Candidate Genes for Cold Tolerance at Seedling Stage by GWAS in Rice (Oryza sativa L.). Biology, 13(10), 784. https://doi.org/10.3390/biology13100784







