Genetic Diversity and Connectivity of Ocypode ceratophthalmus in the East and South China Seas and Its Implications for Conservation
Abstract
Simple Summary
Abstract
1. Introduction
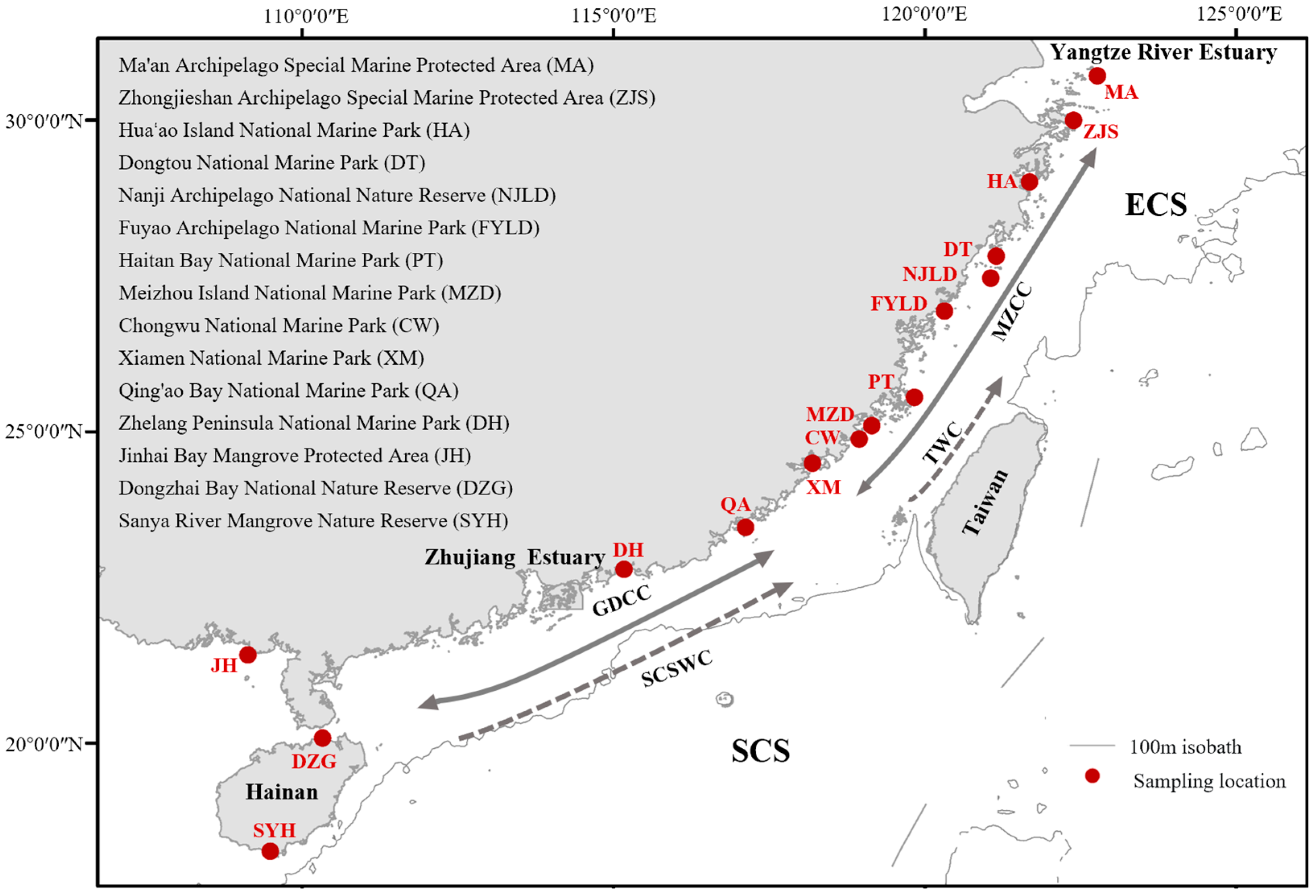
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Statistical Analyses of Genetic Data
3. Results
3.1. Population Genetic Diversity
3.2. Haplotype Analysis
3.3. Population Genetic Structure
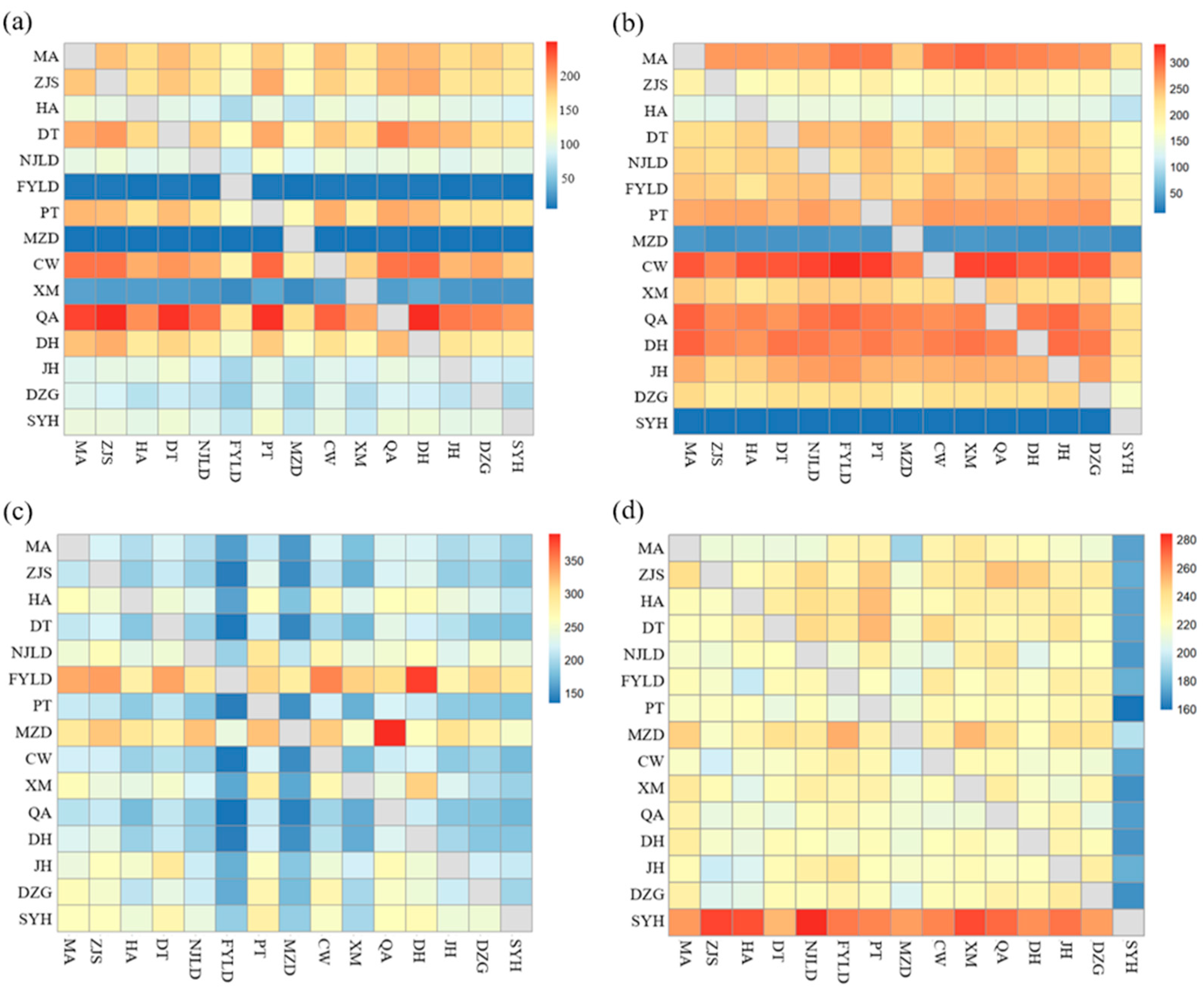
3.4. Migration and Connectivity
4. Discussion
4.1. Differences of Genetic Diversity in Populations
4.2. Connectivity Differences among Populations
4.3. Conservation and Management Implications for MPA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, H.; Xu, H.; Wu, J.; Le Quesne, W.; Sweeting, C.; Polunin, N. An overview of spatial management and marine protected areas in the East China Sea. Coast. Manag. 2008, 36, 443–457. [Google Scholar] [CrossRef]
- Teh, L.S.; Cashion, T.; Cheung, W.W.; Sumaila, U.R. Taking stock: A Large Marine Ecosystem perspective of socio-economic and ecological trends in East China Sea fisheries. Rev. Fish Biol. Fish. 2020, 30, 269–292. [Google Scholar] [CrossRef]
- Yao, Y. On the construction of South China Sea Mrine Protected Area from the perspective of a community of shared future of mankind: Realistic requirements, theoretical basisi and China’s countermeasures. J. Guangxi Univ. 2019, 41, 96–106. [Google Scholar]
- Asaad, I.; Lundquist, C.J.; Erdmann, M.V.; Costello, M.J. Delineating priority areas for marine biodiversity conservation in the Coral Triangle. Biol. Conserv. 2018, 222, 198–211. [Google Scholar] [CrossRef]
- He, J. A Legal Analysis of the Construction of Marine Protected Areas in the South China Sea and Its Impact. Master’s Thesis, Wuhan University, Wuhan, China, 2020. [Google Scholar]
- Zeng, X.; Chen, M.Y.; Zeng, C.; Cheng, S.; Wang, Z.H.; Liu, S.R.; Zou, C.X.; Ye, S.F.; Zhu, Z.G.; Cao, L. Assessing the management effectiveness of China’s marine protected areas: Challenges and recommendations. Ocean Coast. Manag. 2022, 224, 106172. [Google Scholar] [CrossRef]
- McLeod, E.; Salm, R.; Green, A.; Almany, J. Designing marine protected area networks to address the impacts of climate change. Front. Ecol. Environ. 2009, 7, 362–370. [Google Scholar] [CrossRef]
- Planes, S.; Jones, G.P.; Thorrold, S.R. Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl. Acad. Sci. USA 2009, 106, 5693–5697. [Google Scholar] [CrossRef] [PubMed]
- Toonen, R.J.; Andrews, K.R.; Baums, I.B.; Bird, C.E.; Concepcion, G.T.; Daly-Engel, T.S.; Eble, J.A.; Faucci, A.; Gaither, M.R.; Iacchei, M. Defining boundaries for ecosystem-based management: A multispecies case study of marine connectivity across the Hawaiian Archipelago. J. Mar. Biol. 2011, 2011, 460173. [Google Scholar] [CrossRef] [PubMed]
- Spalding, M.D.; Fox, H.E.; Allen, G.R.; Davidson, N.; Ferdaña, Z.A.; Finlayson, M.; Halpern, B.S.; Jorge, M.A.; Lombana, A.; Lourie, S.A. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. BioScience 2007, 57, 573–583. [Google Scholar] [CrossRef]
- Hashimoto, K.; Yamada, K.; Sekino, M.; Kobayashi, M.; Sasaki, T.; Fujinami, Y.; Yamamoto, M.; Choi, K.S.; Henmi, Y. Population genetic structure of the pen shell Atrina pectinata sensu lato (Bivalvia: Pinnidae) throughout East Asia. Reg. Stud. Mar. Sci. 2021, 48, 102024. [Google Scholar] [CrossRef]
- Xu, D.D.; Lou, B.; Shi, H.L.; Geng, Z.; Li, S.L.; Zhang, Y.R. Genetic diversity and population structure of Nibea albiflora in the China Sea revealed by mitochondrial COI sequences. Biochem. Syst. Ecol. 2012, 45, 158–165. [Google Scholar] [CrossRef]
- Dou, C.F. Analysis of genetic diversity and structure of Octopus ovulum. China Water Transp. 2017, 17, 165–167. [Google Scholar]
- Xu, Y. Studies on Molecular Phylogeny of Sesarmid Crabs from the Coast of China and Molecular Phylogeography of Two Mangrove Crabs. Master’s Thesis, Guangxi University, Nanjing, China, 2020. [Google Scholar]
- He, L.J.; Mukai, T.; Chu, K.H.; Ma, Q.; Zhang, J. Biogeographical role of the Kuroshio Current in the amphibious mudskipper Periophthalmus modestus indicated by mitochondrial DNA data. Sci. Rep. 2015, 5, 15645. [Google Scholar] [CrossRef]
- Wang, J.; Tsang, L.M.; Dong, Y.W. Causations of phylogeographic barrier of some rocky shore species along the Chinese coastline. Bmc Evol. Biol. 2015, 15, 114. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zhang, Y.; Zhao, L.L.; Song, N.; Han, Z.Q.; Gao, T.X. Shallow mitochondrial phylogeographical pattern and high levels of genetic connectivity of Thamnaconus hypargyreus in the South China Sea and the East China Sea. Biochem. Syst. Ecol. 2016, 67, 110–118. [Google Scholar] [CrossRef]
- JinXian, L.I.U.; TianYiang, G.A.O.; ShiFang, W.U. Pleistocene Isolation in the Northwestern Pacific Marginal Seas and Limited Dispersal in a Marine Fish, Chelon haematocheilus (Temminck & Schlegel, 1845). J. Ocean Univ. China 2007, 37, 931–938. [Google Scholar]
- Wang, J.J.; Sun, P.; Yin, F. Low mtDNA Cytb diversity and shallow population structure of Eleutheronema tetradactylum in the East China Sea and the South China Sea. Biochem. Syst. Ecol. 2014, 55, 268–274. [Google Scholar] [CrossRef]
- Chen, S.P.; Liu, T.; Li, Z.F.; Gao, T.X. Genetic population structuring and demographic history of red spotted grouper (Epinephelus akaara) in South and East China Sea. Afr. J. Biotechnol. 2008, 7, 3554–3562. [Google Scholar]
- Perez-Enriquez, R.; Taniguchi, N. Genetic structure of red sea bream (Pagrus major) population off Japan and the Southwest Pacific, using microsatellite DNA markers. Fish. Sci. 1999, 65, 23–30. [Google Scholar] [CrossRef]
- Zeng, L.Y.; Cheng, Q.Q.; Chen, X.Y. Microsatellite analysis reveals the population structure and migration patterns of Scomber japonicus (Scombridae) with continuous distribution in the East and South China Seas. Biochem. Syst. Ecol. 2012, 42, 83–93. [Google Scholar] [CrossRef]
- Ni, G. Phylogeography of Four Marine Bivalves Along China’ Coastline, with Views into the Evolutionary Processes and Mechanisms. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2013. [Google Scholar]
- Ma, K.Y.; Chow, L.H.; Wong, K.J.H.; Chen, H.N.; Ip, B.H.Y.; Schubart, C.D.; Tsang, L.M.; Chan, B.K.K.; Chu, K.H. Speciation pattern of the horned ghost crab Ocypode ceratophthalmus (Pallas, 1772): An evaluation of the drivers of Indo-Pacific marine biodiversity using a widely distributed species. J. Biogeogr. 2019, 46, 830. [Google Scholar] [CrossRef]
- Shen, R.J.; Dai, A.Y. Illustrated Fauna of China: Crustacea. In Volume 2 Crabs; Science Press: Beijing, China, 1964; 142p. [Google Scholar]
- Jenkins, T.L.; Stevens, J.R. Assessing connectivity between MPAs: Selecting taxa and translating genetic data to inform policy. Mar. Policy 2018, 94, 165–173. [Google Scholar] [CrossRef]
- Roberts, K.E.; Cook, C.N.; Beher, J.; Treml, E.A. Assessing the current state of ecological connectivity in a large marine protected area system. Conserv. Biol. 2021, 35, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Cassone, B.J.; Boulding, E.G. Genetic structure and phylogeography of the lined shore crab, Pachygrapsus crassipes, along the northeastern and western Pacific coasts. Mar. Biol. 2006, 149, 213–226. [Google Scholar] [CrossRef]
- Ren, G.; Miao, G.; Ma, C.; Lu, J.; Yang, X.; Ma, H. Genetic structure and historical demography of the blue swimming crab (Portunus pelagicus) from southeastern sea of China based on mitochondrial COI gene. Mitochondrial DNA Part A 2018, 29, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Canales-Aguirre, C.B.; Ferrada-Fuentes, S.; Galleguillos, R.; Oyarzun, F.X.; Hernandez, C.E. Population genetic structure of Patagonian toothfish (Dissostichus eleginoides) in the Southeast Pacific and Southwest Atlantic Ocean. PeerJ 2018, 6, e4173. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Y.; Xu, D.; Lou, B.; Guo, Y.; Sun, X.; Guo, B. Genetic population structure of miiuy croaker (Miichthys miiuy) in the Yellow and East China Seas base on mitochondrial COI sequences. Biochem. Syst. Ecol. 2014, 54, 240–246. [Google Scholar] [CrossRef]
- Ma, Z.F.; Pan, Q.Z.; An, M.; Yu, K.; Huang, S.; Li, S.; He, X.K. Analysis of genetic diversity of Siniperca scherzeri population in Qinshui river based on Mitochondrial Cytb and D-Loop Sequences. Mar. Fish. 2022, 44, 657–669. [Google Scholar]
- Ewers, R.M.; Didham, R.K. Confounding factors in the detection of species responses to habitat fragmentation. Biol. Rev. 2006, 81, 117–142. [Google Scholar] [CrossRef]
- Pacioni, C.; Hunt, H.; Allentoft, M.E.; Vaughan, T.G.; Wayne, A.F.; Baynes, A.; Haouchar, D.; Dortch, J.; Bunce, M. Genetic diversity loss in a biodiversity hotspot: Ancient DNA quantifies genetic decline and former connectivity in a critically endangered marsupial. Mol. Ecol. 2015, 24, 5813–5828. [Google Scholar] [CrossRef]
- Feng, B. Study on the Management Effectiveness of Nature Reserve System of Guangxi in the Context of Climate Change. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2020. [Google Scholar]
- Zhuang, Q. Study on the Gap Analysis and Establishment Management of Xiamen Marine Protect Area. Master’s Thesis, Third Institute of Oceanography, Xiamen, China, 2020. [Google Scholar]
- Chen, M.Y.; Zeng, C.; Zeng, X.; Liu, Y.; Wang, Z.H.; Shi, X.J.; Cao, L. Assessment of Marine Protected Areas in the East China Sea Using a Management Effectiveness Tracking Tool. Front. Mar. Sci. 2022, 10, 174. [Google Scholar] [CrossRef]
- Eastwood, E.K.; Lopez, E.H.; Drew, J.A. Population Connectivity Measures of Fishery-Targeted Coral Reef Species to Inform Marine Reserve Network Design in Fiji. Sci. Rep. 2016, 6, 19318. [Google Scholar] [CrossRef] [PubMed]
- Duran, S.; Palacin, C.; Becerro, M.A.; Turon, X.; Giribet, G. Genetic diversity and population structure of the commercially harvested sea urchin Paracentrotus lividus (Echinodermata, Echinoidea). Mol. Ecol. 2004, 13, 3317–3328. [Google Scholar] [CrossRef]
- Robalo, J.I.; Francisco, S.M.; Vendrell, C.; Lima, C.S.; Pereira, A.; Brunner, B.P.; Dia, M.; Gordo, L.; Castilho, R. Against all odds: A tale of marine range expansion with maintenance of extremely high genetic diversity. Sci. Rep. 2020, 10, 12707. [Google Scholar] [CrossRef] [PubMed]
- Fourdrilis, S.; Backeljau, T. Highly polymorphic mitochondrial DNA and deceiving haplotypic differentiation: Implications for assessing population genetic differentiation and connectivity. BMC Evol. Biol. 2019, 19, 92. [Google Scholar]
- Song, N.; Ma, G.; Zhang, X.; Gao, T.; Sun, D. Genetic structure and historical demography of Collichthys lucidus inferred from mtDNA sequence analysis. Environ. Biol. Fishes 2014, 97, 69–77. [Google Scholar] [CrossRef]
- Han, Z.; Xu, H.; Shui, B.; Zhou, Y.; Gao, T. Lack of genetic structure in endangered large yellow croaker Larimichthys crocea from China inferred from mitochondrial control region sequence data. Biochem. Syst. Ecol. 2015, 61, 1–7. [Google Scholar] [CrossRef]
- Sun, P.; Tang, B.; Yin, F. Population genetic structure and genetic diversity of Chinese pomfret at the coast of the East China Sea and the South China Sea. Mitochondrial DNA Part A 2018, 29, 643–649. [Google Scholar] [CrossRef]
- Hughes, R.N.; Hughes, D.J.; Smith, I.P. The ecology of ghost crabs. Oceanogr. Mar. Biol. Annu. Rev. 2014, 52, 201–256. [Google Scholar]
- Otwoma, L.M.; Reuter, H.; Timm, J.; Meyer, A. Genetic connectivity in a herbivorous coral reef fish (Acanthurus leucosternon Bennet, 1833) in the Eastern African region. Hydrobiologia 2018, 806, 237–250. [Google Scholar] [CrossRef]
- Zhang, Z.X. Observation and Analysis of the Coastal Current and Its Adjacent Current System in the China Offshore Waters. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2014. [Google Scholar]
- Geng, Q.F.; Wang, Z.S.; Tao, J.M.; Kimura, M.K.; Liu, H.; Hogetsu, T.; Lian, C.L. Ocean Currents Drove Genetic Structure of Seven Dominant Mangrove Species Along the Coastlines of Southern China. Front. Genet. 2021, 12, 615911. [Google Scholar] [CrossRef] [PubMed]
- Musilova, Z.; Kalous, L.; Petrtýl, M.; Chaloupková, P. Cichlid fishes in the Angolan headwaters region: Molecular evidence of the ichthyofaunal contact between the Cuanza and Okavango-Zambezi systems. PLoS ONE 2013, 8, e65047. [Google Scholar] [CrossRef]
- Gagnaire, P.A.; Broquet, T.; Aurelle, D.; Viard, F.; Souissi, A.; Bonhomme, F.; Arnaud-Haond, S.; Bierne, N. Using neutral, selected, and hitchhiker loci to assess connectivity of marine populations in the genomic era. Evol. Appl. 2015, 8, 769–786. [Google Scholar] [CrossRef] [PubMed]
- Munguia-Vega, A.; Green, A.L.; Suarez-Castillo, A.N.; Espinosa-Romero, M.J.; Aburto-Oropeza, O.; Cisneros-Montemayor, A.M.; Cruz-Pinon, G.; Danemann, G.; Giron-Nava, A.; Gonzalez-Cuellar, O.; et al. Ecological guidelines for designing networks of marine reserves in the unique biophysical environment of the Gulf of California. Rev. Fish Biol. Fish. 2018, 28, 749–776. [Google Scholar] [CrossRef]
- Lu, J.Y.; Chen, Y.J.; Wang, Z.H.; Zhao, F.; Zhong, Y.S.; Zeng, C.; Cao, L. Larval dispersal modeling reveals low connectivity among national marine protected areas in the Yellow and East China. Biology 2023, 12, 396. [Google Scholar] [CrossRef]
- Chen, L.L.; Zhu, X.L.; Ni, S.F.; Yan, Z.Q.; Xu, F.C. Research on the policy instrument of the fishing ban in the autumn season in the East China Sea of China- based on the content analysis of policy texts since 2011. China Fish. 2022, 12, 60–63. [Google Scholar]
- Huang, C.D. A study on the administrative law of Fishing Ban in the summer in the South China Sea: A case of Naozhou island. Leg. Syst. Soc. 2020, 13, 125–126. [Google Scholar]
- Chen, R.L.; Wu, X.Q.; Liu, B.J.; Wang, Y.Q.; Gao, Z.Q. Mapping coastal fishing grounds and assessing the effectiveness of fishery regulation measures with AIS data: A case study of the sea area around the Bohai Strait, China. Ocean Coast. Manag. 2022, 223, 106136. [Google Scholar] [CrossRef]
- Yu, J.; Hu, Q.; Yuan, H.; Tong, F.; Chen, P.; Mao, J. Effects assessment of summer fishing moratorium in Daya Bay in the Northern South China Sea. J. Geosci. Environ. Prot. 2017, 5, 96. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.N.; Jiao, H.F.; Chen, C.; Liu, D.; Shi, H.X.; You, Z.J. Temporal Dynamics of Fishing Affect the Biodiversity of Macrobenthic Epifaunal Communities in the Coastal Waters of Ningbo, East China Sea. Thalassas 2021, 37, 39–49. [Google Scholar] [CrossRef]
- Meyer, C.G.; Holland, K.N.; Papastamatiou, Y.P. Seasonal and diel movements of giant trevally Caranx ignobilis at remote Hawaiian atolls: Implications for the design of Marine Protected Areas. Mar. Ecol. Prog. Ser. 2007, 333, 13–25. [Google Scholar] [CrossRef]

| Region | Ecoregion | MPA * | Population | Number of Samples | Weight (g) | Sampling Time |
|---|---|---|---|---|---|---|
| East China Sea | East China Sea Ecoregion | Dongtou National Marine Park (in) | DT | 20 | 18.06 ± 6.17 | 27 April 2022 |
| Fuyao Archipelago National Marine Park (in) | FYLD | 15 | 18.63 ± 5.70 | 30 March 2022 | ||
| Haitan Bay National Marine Park (in) | PT | 18 | 13.68 ± 4.54 | 6 March 2022 | ||
| Hua’ao Island National Marine Park (near) | HA | 15 | 15.89 ± 4.15 | 8 April 2022 | ||
| Ma’an Archipelago Special Marine Protected Area (in) | MA | 15 | 5.41 ± 2.25 | 10 December 2021 | ||
| Meizhou Island National Marine Park (in) | MZD | 16 | 13.38 ± 2.66 | 5 March 2022 | ||
| Nanji Archipelago National Nature Reserve (in) | NJLD | 10 | 16.15 ± 6.95 | 18 April 2022 | ||
| Zhongjieshan Archipelago Special Marine Protected Area (near) | ZJS | 14 | 22.82 ± 7.11 | 18 April 2022 | ||
| Southern China Ecoregion | Chongwu National Marine Park (in) | CW | 24 | 16.59 ± 5.40 | 8 January 2022 | |
| Xiamen National Marine Park (in) | XM | 18 | 16.65 ± 5.72 | 2 January 2022 | ||
| South China Sea | Southern China Ecoregion | Qing’ao Bay National Marine Park (near) | QA | 22 | 15.01 ± 3.54 | 8 January 2022 |
| Zhelang Peninsula National Marine Park (near) | DH | 15 | 14.43 ± 4.56 | 22 March 2022 | ||
| Gulf of Tonkin Ecoregion | Dongzhai Bay National Nature Reserve (near) | DZG | 21 | 5.28 ± 2.51 | 18 September 2021 | |
| Jinhai Bay Mangrove Protected Area (near) | JH | 15 | 18.89 ± 6.09 | 1 April 2022 | ||
| Sanya River Mangrove Nature Reserve (near) | SYH | 15 | 5.54 ± 2.50 | 18 September 2021 |
| Region | Population | COI | D-Loop | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | h | Hd | π | N | h | Hd | π | ||
| The East China Sea | CW | 24 | 12 | 0.87 ± 0.05 | 0.0034 ± 0.0006 | 9 | 9 | 1.00 ± 0.05 | 0.0279 ± 0.0033 |
| XM | 18 | 12 | 0.95 ± 0.03 | 0.0054 ± 0.0007 | 9 | 9 | 1.00 ± 0.05 | 0.0281 ± 0.0028 | |
| MA | 15 | 9 | 0.88 ± 0.07 | 0.0037 ± 0.0007 | 5 | 5 | 1.00 ± 0.13 | 0.0314 ± 0.0052 | |
| PT | 18 | 10 | 0.88 ± 0.06 | 0.0050 ± 0.0010 | 5 | 4 | 0.90 ± 0.16 | 0.0271 ± 0.0050 | |
| MZD | 16 | 8 | 0.88 ± 0.05 | 0.0030 ± 0.0005 | 6 | 6 | 1.00 ± 0.10 | 0.0230 ± 0.0033 | |
| HA | 15 | 9 | 0.88 ± 0.07 | 0.0034 ± 0.0006 | 6 | 6 | 1.00 ± 0.10 | 0.0192 ± 0.0035 | |
| NJLD | 10 | 9 | 0.98 ± 0.05 | 0.0043 ± 0.0008 | 5 | 5 | 1.00 ± 0.13 | 0.0298 ± 0.0051 | |
| ZJS | 14 | 9 | 0.90 ± 0.06 | 0.0036 ± 0.0007 | 10 | 10 | 1.00 ± 0.05 | 0.0306 ± 0.0028 | |
| FYLD | 15 | 7 | 0.82 ± 0.08 | 0.0022 ± 0.0004 | 7 | 6 | 0.95 ± 0.10 | 0.0233 ± 0.0035 | |
| DT | 20 | 11 | 0.86 ± 0.07 | 0.0037 ± 0.0007 | 5 | 5 | 1.00 ± 0.13 | 0.0257 ± 0.0048 | |
| The South China Sea | QA | 22 | 14 | 0.93 ± 0.04 | 0.0036 ± 0.0005 | 5 | 5 | 1.00 ± 0.13 | 0.0331 ± 0.0063 |
| JH | 15 | 7 | 0.81 ± 0.08 | 0.0027 ± 0.0006 | 5 | 5 | 1.00 ± 0.13 | 0.0379 ± 0.0061 | |
| DZG | 21 | 9 | 0.86 ± 0.05 | 0.0027 ± 0.0004 | 5 | 5 | 1.00 ± 0.13 | 0.0268 ± 0.0064 | |
| SYH | 15 | 7 | 0.80 ± 0.08 | 0.0032 ± 0.0009 | 7 | 6 | 0.95 ± 0.10 | 0.0278 ± 0.0038 | |
| DH | 15 | 12 | 0.94 ± 0.05 | 0.0052 ± 0.0010 | 6 | 6 | 1.00 ± 0.10 | 0.0313 ± 0.0052 | |
| Different Classifications | Genetic Markers | Source of Variation | df | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|---|---|
| Region level (ECS and SCS) | COI | Between regions | 1 | 0.708 | −0.002 | −0.22 |
| Within regions among populations | 13 | 12.383 | −0.002 | −0.18 | ||
| within populations | 238 | 233.873 | 0.983 | 100.40 | ||
| Total | 252 | 246.964 | 0.979 | |||
| D-Loop | Between regions | 1 | 5.481 | −0.072 | −0.86 | |
| Within regions among populations | 13 | 108.307 | −0.037 | −0.44 | ||
| within populations | 80 | 685.181 | 8.565 | 101.29 | ||
| Total | 94 | 798.968 | 8.455 | |||
| Ecoregion level (East China Sea Ecoregion, Southern China Ecoregion, Gulf of Tonkin Ecoregion) | COI | Among ecoregions | 2 | 2.857 | 0.007 | 0.76 |
| Within ecoregions among populations | 12 | 10.234 | −0.008 | −0.80 | ||
| within populations | 238 | 233.873 | 0.983 | 100.04 | ||
| Total | 252 | 246.964 | 0.982 | |||
| D-Loop | Among ecoregions | 2 | 10.766 | −0.111 | −1.31 | |
| Within ecoregions among populations | 12 | 103.021 | 0.003 | 0.04 | ||
| within populations | 80 | 685.181 | 8.565 | 101.27 | ||
| Total | 94 | 798.968 | 8.457 | |||
| Population level | COI | Among populations | 14 | 13.091 | −0.003 | −0.29 |
| within populations | 238 | 233.873 | 0.983 | 100.29 | ||
| Total | 252 | 246.964 | 0.980 | |||
| D-Loop | Among populations | 14 | 113.787 | −0.070 | −0.82 | |
| within populations | 80 | 685.181 | 8.565 | 100.82 | ||
| Total | 94 | 798.968 | 8.495 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, F.; Liu, Y.; Wang, Z.; Lu, J.; Cao, L.; Zeng, C. Genetic Diversity and Connectivity of Ocypode ceratophthalmus in the East and South China Seas and Its Implications for Conservation. Biology 2023, 12, 437. https://doi.org/10.3390/biology12030437
Zhao F, Liu Y, Wang Z, Lu J, Cao L, Zeng C. Genetic Diversity and Connectivity of Ocypode ceratophthalmus in the East and South China Seas and Its Implications for Conservation. Biology. 2023; 12(3):437. https://doi.org/10.3390/biology12030437
Chicago/Turabian StyleZhao, Feng, Yue Liu, Zihan Wang, Jiaying Lu, Ling Cao, and Cong Zeng. 2023. "Genetic Diversity and Connectivity of Ocypode ceratophthalmus in the East and South China Seas and Its Implications for Conservation" Biology 12, no. 3: 437. https://doi.org/10.3390/biology12030437
APA StyleZhao, F., Liu, Y., Wang, Z., Lu, J., Cao, L., & Zeng, C. (2023). Genetic Diversity and Connectivity of Ocypode ceratophthalmus in the East and South China Seas and Its Implications for Conservation. Biology, 12(3), 437. https://doi.org/10.3390/biology12030437






