Simple Summary
Morphological divergence between Siamese and other crocodiles has been identified by size, number of scales, and patterns of cervical squamation with post-occipital scutes (P.O.). However, a large variation of P.O. has been observed in captive Siamese crocodiles in Thailand, leading to questions about possible crocodile hybrids. The genetic diversity and population structure of Siamese crocodiles were studied using mitochondrial DNA D-loop and microsatellite genotyping. The STRUCTURE plot revealed numerous distinct gene pools, indicating that the crocodiles in each farm descended from distinct lineages. Researchers also discovered evidence of introgression in several individual crocodiles, implying that Siamese and saltwater crocodiles may have hybridized. A schematic protocol for screening hybrids was proposed based on patterns observed in phenotypic and molecular data.
Abstract
Populations of Siamese crocodiles (Crocodylus siamensis) have severely declined because of hunting and habitat fragmentation, necessitating a reintroduction plan involving commercial captive-bred populations. However, hybridization between Siamese and saltwater crocodiles (C. porosus) has occurred in captivity. Siamese crocodiles commonly have post-occipital scutes (P.O.) with 4–6 scales, but 2–6 P.O. scales were found in captives on Thai farms. Here, the genetic diversity and population structure of Siamese crocodiles with large P.O. variations and saltwater crocodiles were analyzed using mitochondrial DNA D-loop and microsatellite genotyping. Possible crocodile hybrids or phenotypic variations were ascertained by comparison with our previous library from the Siam Crocodile Bioresource Project. Siamese crocodiles with <4 P.O. scales in a row exhibit normal species-level phenotypic variation. This evidence encourages the revised description of Siamese crocodiles. Moreover, the STRUCTURE plot revealed large distinct gene pools, suggesting crocodiles in each farm were derived from distinct lineages. However, combining both genetic approaches provides evidence of introgression for several individual crocodiles, suggesting possible hybridization between Siamese and saltwater crocodiles. We proposed a schematic protocol with patterns observed in phenotypic and molecular data to screen hybrids. Identifying non-hybrid and hybrid individuals is important for long-term in situ/ex situ conservation.
1. Introduction
Siamese crocodile (Crocodylus siamensis, Schneider, 1801) [1] is a freshwater species found in a wide range of lowland freshwater habitats including slow-moving rivers, streams, lakes, seasonal oxbow lakes, marshes, and swamps in mainland Southeast Asia, including Cambodia, Lao PDR and Thailand and on some islands of Indonesia and Malaysia [2,3,4,5,6]. This medium-sized crocodylian has a total length of less than 3.5 m [7]. However, its historical population distribution has decreased by 20% globally, with only 11% of its habitat range in nationally protected areas [8]. The Siamese crocodile was listed as a critically endangered species on the International Union for Conservation of Nature (IUCN) Red List and in Appendix I of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) in 1996 to aid conservation efforts. In Thailand, Siamese crocodiles are widely distributed in lowland regions; however, most populations have been extirpated because of hunting, habitat loss, and collection to stock commercial crocodile farms over the last 40 years [8,9]. The severe decline of Siamese crocodile populations has led to there being fewer than 20 wild individuals in Khao Ang Rue Nai Wildlife Sanctuary (13°13′2.13″ N, 101°42′37″ E), Kaeng Krachan National Park (12°54′1.9″ N, 99°38′13.98″ E), Namnao National Park (16°57′57.65″ N, 101°30′28.08″ E), Yod Dom Wildlife Sanctuary (14°26′6.06″ N, 105°6′ 1.2″ E), and Bueng Boraphet (15°41′2.67″ N, 100°14′59.07″ E) [6,10]. To restore Siamese crocodile wild populations, it is essential to reintroduce captive-bred individuals and implement in situ/ex situ management practices. [11]. By contrast, 1.3 million Siamese crocodiles were present on 1400 farms in 2020, and crocodile farming now accounts for approximately 1% of Thailand’s agricultural income [12]. However, the occurrence of hybridization between Siamese and saltwater crocodiles (C. porosus, Schneider, 1801) [13] is bidirectional between males and females of parental species in captivity. Interspecific hybridization frequently occurs in Southeast Asia due to the keeping of both species together in captivity, rather than from the wild [11,14]. Both F1 hybrid and backcross crocodiles are fertile and reportedly grow faster than either parental species [13]. The genetic integrity of the species is at risk, which could harm conservation management. Alien outbreeding depression hybrids must be identified from the parental species before the reintroduction program or to improve genetic diversity in the wild. Using an effective genetic diagnosis approach, a genetically diverse captive population of pure Siamese crocodiles was identified while hybrids were differentiated from the parental species. Siamese crocodile sources serve as critical genetic resources for reintroduction efforts [11,15].
The morphological divergence between Siamese and other crocodiles (Crocodylus spp.) has been identified by size, the number of scales, and patterns of cervical squamation with post-occipital scutes (P.O.) [16]. The P.O.s of Siamese crocodiles show one row with 4–6 scales and several small scales, but no P.O. is seen in saltwater crocodiles. However, a large variation of P.O., ranging from two to six scales, has been observed in captive Siamese crocodiles in Thailand. This leads us to question whether possible crocodile hybrids remain in captivity, or whether these are actual phenotypic variations of Siamese crocodiles. A genetic approach to identifying Siamese and saltwater crocodiles was developed together with morphological observations in our previous study [11,15] but the P.O. pattern of each crocodile was not photo-recorded as evidence in our library report. In this study, to test these hypotheses, the genetic diversity and structure of Siamese and saltwater crocodile populations were assessed by screening the gene pool using 22 microsatellite markers and mitochondrial (mtDNA) D-loop sequences coupled with each crocodile photo record. MtDNA and nuclear DNA microsatellites are molecular genetic markers that can identify population diversity, origins of individuals, and hybrids along with their parents, especially in crocodiles [11,15]. Results were compared with those of the large gene pool library under “the Siam Crocodile Bioresource Project” from our previous study [11]. These findings provide pivotal information for prospective reintroduction programs and in situ/ex situ management.
2. Materials and Methods
2.1. Specimen Collection and DNA Extraction
A total of 136 Siamese and 29 saltwater crocodile specimens were collected from 4 captive locations under the auspices of the Thai Crocodile Farm Association (TCFA) and in accordance with CITES regulations for the leather and food industries. Table S1 provides detailed information on the sampled individuals. Scale samples were collected from the tail of captive crocodiles registered at four crocodile farms in Chonburi (CB) (13°09′06.57″ N, 101°28′36.01″ E), Nakhon Ratchasima (NR) (14°57′21.19″ N, 101°28′36.01″ E), Chainat (CN) (15°15′10.44″ N, 100°02′38.27″ E) and Nakhon Pathom (NP) (13°43′20.25″ N, 100°15′20.83″ E) between January and August 2022. A piece of scale clipped from the tail of each specimen was collected as a DNA source. Permission was granted by the farm owners and the TCFA and also from unnamed crocodile farms. Individuals were classified as Siamese or saltwater crocodiles based on external morphological observation [17,18] and photographed. The dataset comprised photo images of 165 individual crocodiles, each captured in 30–50 different postures to minimize testing redundancy and bias. The DNA extraction and quality assessment were performed using the same methods as in previous studies [19] (Supplementary Data S1). All experimental procedures and animal care were carried out in compliance with the Regulations on Animal Experiments at Kasetsart University and approved by the Animal Experiment Committee under Approval No. ACKU64-SCI-011.
2.2. Microsatellite Genotyping and Microsatellite Data Analysis
Twenty-two microsatellite primer sets, developed originally from saltwater crocodiles (Table S2) [20,21], were used for the genotyping of all crocodile individuals. The genotypic data resulting from this study were deposited in the Dryad Digital Repository Dataset (https://datadryad.org/stash/share/s4zREYQ1AUUXTsaIpk0r1HSdkYljvu8yvJOVR143K7Y, accessed on 18 February 2023). We used the same methods as previous studies for PCR amplification to analyze genetic diversity and population structure of the crocodile populations [11,22,23,24,25,26,27] (Supplementary Data S1).
2.3. Mitochondrial DNA D-Loop Sequencing and Data Analysis
The mtDNA D-loop sequences of DNA fragments were amplified using the primers mtCytbf2 (5′-TGCCATGTTCGCATCCATCC-3) and mt12srRNAr2 (5′-CCAGAGGCTA GGCGTCGTGG-3) [11]. We used the same methods as previous studies for PCR amplification and analyze genetic diversity of the crocodile populations [11,22] (Supplementary Data S1).
3. Results
Genetic Variability of Captive Crocodile Population Based on Microsatellite Data
All captive individuals were genotyped, and 459 alleles were found across all loci, with the mean number of alleles per locus being 20.864 ± 2.351 (Table 1). All allelic frequencies in the captive population significantly deviated from what would be expected under the Hardy–Weinberg equilibrium, indicating the presence of linkage disequilibrium (Tables S3–S7). Null alleles were frequently observed for 13 loci (CpP3001, CpP501, CpP214, CpP2206, CpP3313, CpP2504, CpP203, CpP1308, CpP4004, CpP3008, CpP2904, CpP3004, CpP1409), and all markers listed were treated similarly. Siamese crocodiles from CB and NR populations exhibited negative F values, but Siamese crocodiles from CN and NP populations exhibited positive F values, similar to saltwater crocodiles from NR. The PIC of all captive populations ranged from 0.057 to 0.932 and I ranged from 0.153 to 3.031 (Table S8). The Ho values ranged from 0.059 to 1.000 (mean ± standard error (SE): 0.629 ± 0.033) and the He values ranged from 0.058 to 0.936 (mean ± SE: 0.718 ± 0.037) (Table 1 and Table S8). Welch’s t-test showed that Ho was not significantly different from He in Table 2. By comparing pairwise Ho values between populations, there were statistical differences between six pairs, while pairwise He values were different between five pairs (Table 3). The AR value of the population was 20.787 ± 2.332. The standard genetic diversity indices are summarized in Table 1 and Supplementary Table S8.

Table 1.
Genetic diversity of 136 Siamese (Crocodylus siamensis, Schneider, 1801) [1] and 29 saltwater crocodiles (C. porosus, Schneider, 1801) [13] based on 22 microsatellite loci. Table S1 provides detailed information on the sampled individuals.

Table 2.
Welch’s t-test of observed heterozygosity (Ho) and expected heterozygosity (He) of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13] based on 22 microsatellite loci.

Table 3.
Comparison of genetic diversity parameters between Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13] based on 22 microsatellite loci. Table S1 provides detailed information on the sampled individuals.
We determined the degree of relatedness between individuals in the captive crocodile population by employing a pairwise test. The mean pairwise r values were calculated for a total of 13,530 combinations of crocodiles, which included all 165 sampled individuals, including Siamese and saltwater crocodiles, were −0.018 ± 0.033 (CB population = −0.018 ± 0.042, NR population = −0.019 ± 0.029, CN population = −0.017 ± 0.030, NP population = −0.019 ± 0.032 for Siamese crocodiles and NR population of saltwater crocodiles = −0.020 ± 0.032). No pairs showed r < −0.25. There were 13,525 pairs with −0.25 < r < 0.25 and 5 pairs with 0.25 > r (Table 2, Tables S9–S13). Distribution of r values for the crocodiles exhibited a left skew, indicating lower pairwise r values than what would be expected under a null hypothesis of unrelated individuals by chance. The distributions of pairwise r differed significantly between the CB and NP populations, and the mean pairwise r values were also significantly different across all populations. (Figure 1, Table S14). The mean FIS was −0.074 ± 0.076 (Table 3), with individual values of FIS ranging from −0.191 to 0.085 (Tables S15–S19). However, distributions of FIS from all populations differed significantly from each other (Figure 1, Table S14). The Ne of Siamese crocodiles for individuals that contributed genetically to the CB population was 41.3 (95% CI: 32.2.8–46.5), 202.2 (95% CI: 87.7–113.3) for the NR population, 115.5 (95% CI: 69.5–88.1) for the CN population, 45.2 (95% CI: 37.1–136.1) for the NP population, and 103.7 (95% CI: 66.7–74.8) for saltwater crocodiles in the NR population (Table 4).
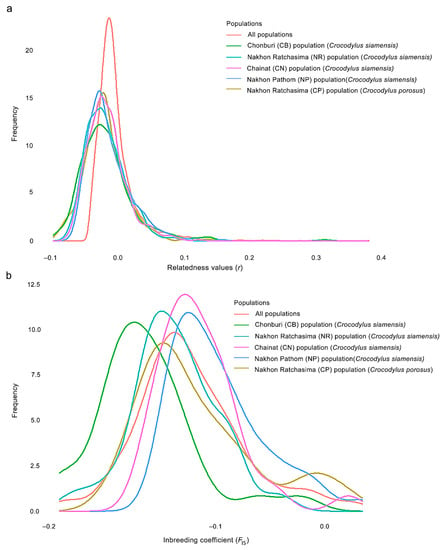
Figure 1.
Observed distribution of (a) pairwise relatedness (r) and (b) inbreeding coefficients (FIS) for 136 Siamese (Crocodylus siamensis, Schneider, 1801) [1] and 29 saltwater crocodiles (C. porosus, Schneider, 1801) [13] individuals, plotted against the expected distributions.

Table 4.
Inbreeding coefficients, relatedness, effective population size, and ratio of effective population size and census population (Ne/N) of 136 Siamese (Crocodylus siamensis, Schneider, 1801) [1] and 29 saltwater crocodiles (C. porosus, Schneider, 1801) [13].
Significant differences (p < 0.05) were observed in the estimates of FST between captive populations after 110 permutations. The AMOVA showed that genetic variation was 84% among individuals crocodiles within a population and 11% between populations (Table S21). Nei’s genetic distances and RST showed that the CN population was closer than the NP to the others (Tables S20 and S22). The distinction between the five crocodile groups and the three suspected individuals (CSI05, CSI06, and CPO09) from our previous study was supported by the first, second, and third principal components, which accounted for 10.48, 8.87, and 4.24% of the total variation, respectively, as revealed by PCoA [11] (Figure 2). Different population patterns were generated by the model-based Bayesian clustering algorithms implemented in STRUCTURE with increasing K values; however, the highest posterior probability with one peak (K = 3) based on Evanno’s ΔK, while the mean ln P(K) also revealed one peak (K = 16) (Figure 3 and Figure S1).
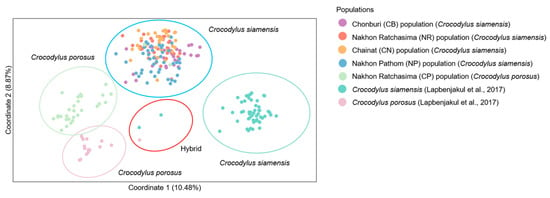
Figure 2.
Principal component analysis of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (C. porosus, Schneider, 1801) [13]. Table S1 provides detailed information on the sampled individuals and Lapbenjakul et al. [11].
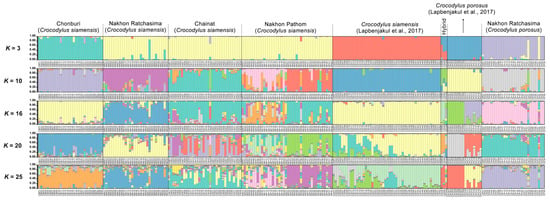
Figure 3.
Population structure of 136 Siamese (Crocodylus siamensis, Schneider, 1801) [1] individuals and 29 saltwater crocodile (C. porosus, Schneider, 1801) [13] individuals. Each vertical bar on the x-axis represents an individual, while the y-axis represents the proportion of membership (posterior probability) in each genetic cluster. Crocodiles are superimposed on the plot, with black vertical lines indicating the boundaries. Detailed information for all crocodile individuals is presented in Table S1 and Lapbenjakul et al. [11].
The amplicon length and alignment length of the mtDNA D-loop sequences were 1500 and 1350 bp, respectively. The numbers of haplotypes were 46 and 28 for Siamese and saltwater crocodiles, respectively. Overall haplotype and nucleotide diversities were 0.846 ± 0.022 and 0.015 ± 0.003 for Siamese and 0.998 ± 0.055 and 0.069 ± 0.025 for saltwater crocodiles (Table 5). A complex haplotype network was constructed from the many detected polymorphic sites and haplotypes (Figure 4). The most common haplotype of all Siamese crocodile populations was haplotype CS36. Seven haplotypes (CS01, CS16, CS20, CS21, CS27, CS30, and CS36) were shared in the CB, NR, CN, and NP populations. Furthermore, the most common haplotype of all populations of saltwater crocodiles was haplotype CD04 and CD05. Phylogenetic analysis of a combined data set for the mtDNA D-loop sequences from both Siamese and saltwater crocodiles, together with those for 21 crocodile species obtained from the public repositories (GenBank/DDBJ/European Nucleotide Archive (ENA)), indicated that most Siamese and saltwater crocodile sequences each formed a monophyletic clade. However, NP04, NP06, NP07, NP09, NP13, NP14, NP16, and NP20, first assigned to Siamese crocodile, were grouped with Cuban crocodile (C. rhombifer) (GenBank accession number: NC_024513); whereas CP01, CP05, CP06, CP11, CP12, CP13, CP15, CP20, CP23, CP25, CP27, CP28, CP29, and CP30, categorized with the saltwater crocodile, were placed as a sister clade to Siamese crocodile (Figure S2). These results agreed with BLAST results of sequence identity (Table S1). To examine the genetic differentiation among the five populations, we calculated FST, GST, ΦST, Dxy, Da, and Nm. The FST values ranged from −0.015 to 0.347, the GST values ranged from −0.008 to 0.071 and the ΦST values ranged from 0.013 to 0.330, The Nm values ranged from 0.947 to infinite, the Dxy values ranged from 0.002 to 0.065 and the Da values ranged from 0.000 to 0.026 (Table 6).

Table 5.
Mitochondrial DNA D-loop sequence diversity of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13].
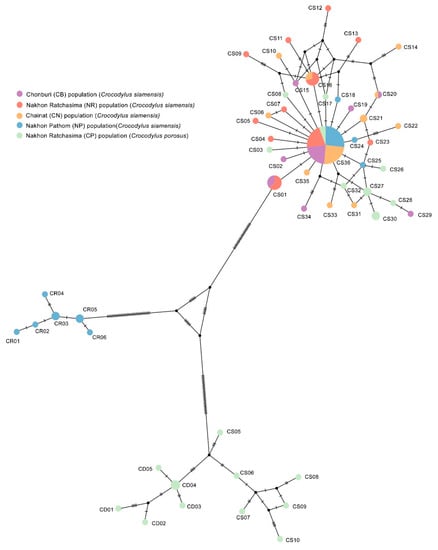
Figure 4.
Haplotype network based on sequence data for the mitochondrial DNA D-loop region of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13].

Table 6.
Genetic differentiation between the three populations of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13] for the mitochondrial DNA D-loop sequence. Genetic differentiation coefficient (GST), Wright’s F-statistic for the subpopulations within the total population (FST), ΦST, gene flow (Nm) from the sequence data and the haplotype data, the average number of nucleotide substitutions per site between populations (Dxy), and the net nucleotide substitutions per site between populations (Da).
4. Discussion
Siamese crocodile is well-represented in captivity, with possibly over 1.5 million individuals in farms in Thailand, Cambodia, and Vietnam [31,32,33,34], and smaller numbers in farms in China and zoos in Europe and North America. In Thailand, 1400 crocodile farms with 1,319,395 Siamese and 162,449 saltwater crocodiles were operating in 2020, while 47,367 skins were sold in international trade in 2020 [35,36,37]. However, the captive population in Thailand includes an unknown number of individuals hybridized with saltwater crocodiles [11,33,38,39,40], similar to captive crocodiles in Cambodia, Lao PDR, and Vietnam [33,41,42,43]. Observations of hybrids between Siamese and saltwater crocodiles have been reported as a consequence of anthropogenic impacts [11,39,44,45]. Most anthropogenic crocodile hybrids pose a serious problem for conservation management because hybrids a possess highly similar morphology to their parental species, which might lead to introgression if they are included in a reintroduction program [11].
Most crocodile farms are members of the TCFA, which aims to keep purely captive-bred individuals of both species to comply with the recommendations of CITES Appendix I captive breeding operation [11,15,46]. Most Thai crocodile farms have thus pledged not to produce hybrid offspring in the interest of conservation. However, we found discrepancies in two genetic markers, microsatellite genotyping and mtDNA D-loop in several crocodile individuals (both Siamese and saltwater crocodiles), suggesting the possibility of hybrids in the population examined. Crocodiles that were clearly identified as being pure specimens of a species were designated as either “identified Siamese crocodile” or “identified saltwater crocodile”. However, if the results of the two genetic markers were not consistent, the crocodiles were designated “unidentified crocodile”. To ensure compliance between genetic tools and phenotypic variation for Siamese and saltwater crocodile identification, we compared the results of genetic diversity and structure with phenotypic observation in each crocodile. The most frequently observed means of identifying hybrid characteristics between Siamese and saltwater crocodiles are the presence of P.O. This has led to the misunderstanding of many crocodile experts and non-governmental organizations (NGOs) who have visited crocodile farms in Southeast Asia as to whether Siamese crocodiles with fewer than four post-occipital scales in a row are hybrids [16,47].
4.1. Are Different Numbers of Post-Occipital Scutes Due to Phenotypic Variation within Siamese Crocodiles or the Consequence of Hybridization with Saltwater Crocodiles?
As shown in the STRUCTURE plot and PCoA, Siamese crocodiles from CB, CN, NR, and NP, and saltwater crocodiles from NR, were likely clustered into different groups. We analyzed the clustering order and gene pool pattern from K = 2–25. Identified pure Siamese crocodiles, which have two P.O. scales shared the same gene pool with Siamese crocodiles having three or four scales, whereas no P.O. scales were found in identified saltwater crocodiles (Figure 3, Figure 5 and Figure S3). For K = 3, Siamese crocodiles (both identified and unclear individuals) from CB, CN, NR, and NP were grouped in the same gene pool, while saltwater crocodiles were part of a new group, with a small part shared with Siamese crocodiles. At higher K levels, saltwater crocodiles (both identified and unclear individuals) became identifiable, whereas Siamese crocodiles from different farms were separated from each other. Gene pool structuring from both species or each farm showed admixture at higher K levels; however, P.O. scale number variation also appeared in Siamese crocodiles with mixtures of specific gene pools of Siamese crocodiles. This suggests that Siamese crocodiles with fewer than four scales in a row are part of normal phenotypic variations at the species level. Similarly, the first version of the species identification guideline was revised after DNA analysis and proved various characteristics under the same species such as in fighting fishes [48,49].
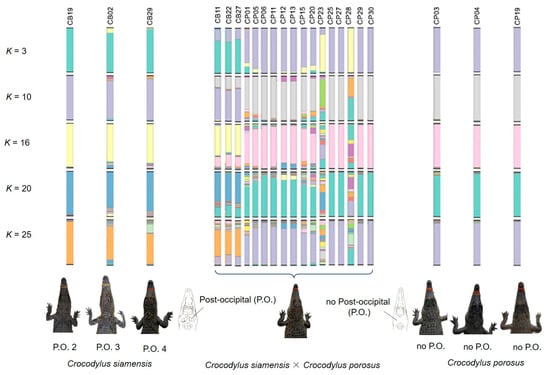
Figure 5.
Representation of structure plot and post-occipital scutes (P.O.) for hybrids between Siamese and saltwater crocodiles (Crocodylus siamensis × Crocodylus porosus) and pure Siamese and saltwater crocodiles. Detailed information for all hybrids between Siamese and saltwater crocodile individuals is presented in Figure 3 and Figure S3.
This misunderstanding of widespread hybridization in Thailand has probably resulted from personal communication among experts from IUCN/SSC/Crocodile Specialist Group and other NGOs who visited farms and followed the CITES Identification Guide based on morphological characteristics in Charette [16], leading to an erroneous judgment of crocodile hybridization events in Thai crocodile farms [46,47]. Revision of Siamese crocodile identification should be reconsidered for scientific taxonomic study, which is relevant to conservation management and economic value. However, the limited number of microsatellite markers located at regular intervals cannot cover species-specific genomic regions [50]. The 22 microsatellite marker loci in this study may have caused bias due to limited population history, the timing of selection, phasing error, and false LD resolution [51,52]. Therefore, larger sample sizes with higher numbers of microsatellite loci are required to extensively investigate the evidence. Genome-wide SNP are also needed to identify signature selection between species or specific phenotypic issues such as P.O.
4.2. Large Gene Pool Variation Reflects Different Historical Origins in the Wild Population
Using data generated from microsatellite loci derived from Siamese and saltwater crocodiles by Lapbenjakul et al. [11] and this study, we addressed the genetic structure and gene pool pattern between the two crocodile species. As shown by PCoA, Siamese and saltwater crocodiles were clustered into different groups. However, sharing of gene pools by Siamese crocodiles between the crocodile farms was observed at different K levels of the STRUCTURE plot. Although Siamese crocodile individuals from this study grouped in the same gene pool at K = 3, and Siamese and saltwater crocodile individuals from our previous study [11] were shown to be part of a new group with saltwater crocodiles, large differences in gene pools were observed among the four populations from K = 5–25. In the NP population, two subpopulations of different gene pools were found, consistent with the positive fixation index value. It can be inferred from this that Siamese crocodiles in each farm had different ancestral original lineages.
Historically, Siamese crocodiles were widespread in Central Thailand. The current captive Siamese crocodile populations might reflect differences in the original sources brought into farms in Thailand. This result agreed with the genetic diversity parameters showing high values of heterozygosity, AR, and Ne in Siamese crocodiles from the four captive sites in this study. The pairwise FST value was statistically significant among the four populations, implying genetic structure differentiation between the farms. The differentiation in genetic makeup reflects the accumulation of variations in allelic frequencies, providing crucial insights into the evolutionary history, genetic drift, and selection of distinct populations [53,54]. However, we have no evidence to identify gene pool associations and geographic origin as known ecotypes, as no capture records exist for Siamese crocodile individuals. Interestingly, many Siamese crocodiles from the four captivities showed a genetic admixture of different gene pools of Siamese crocodile from K = 5–25. This was also observed in the saltwater crocodile population, which might have resulted from the historical genetic exchange of parental stocks between farms. Regarding mtDNA D-loop sequences, positive Nm, low FST, and sharing haplotypes of crocodiles between farms also confirmed the presence of crocodile genetic exchange in the market farms. The exchange of Siamese crocodile parental stocks has been conducted to impede inbreeding within each captive site, which may provide a negative inbreeding level. These findings collectively suggest that large captive populations of Siamese crocodiles held on farms represent a good potential source for reintroduction programs. Crocodile farms under TCFA are willing to donate Siamese crocodiles for this purpose [55].
4.3. Siamese Crocodile Identification Protocol Based on Morphology and DNA Fingerprinting
Captive crocodiles must be genetically identified at the species level before release [9,56,57]. However, hybridization between Siamese and saltwater crocodiles is widespread among some captive populations in Southeast Asia [33,34,39,58]. Differentiating hybrids from parental species based on phenotype alone is very challenging; thus, genetic screening is necessary to confirm species identity [11,15,40,58]. To ensure the success of reintroduction programs, we must first address the complex hybrid issue. Cluster analysis using STRUCTURE can now determine the degree of hybridization and gene pool pattern by aggregating individuals into a single cluster relative to additional highly differentiated populations/species [11,59,60]. Our previous study indicated that three individuals (CPO09, CSI05, and CSI06) may have been the result of interspecific hybridization between Siamese and saltwater crocodiles [11]. In this study, after we added more Siamese and saltwater crocodiles to the library analysis, the three crocodile individuals were still identified as hybrids. However, high levels of genetic admixture were observed in many crocodile individuals, and this might result in misleading conclusions about the genetic admixture of gene pools under the STRUCTURE plot with the probability of identifying the state of hybrids alone. According to genetic diversity parameters and the STRUCTURE plot, great genetic diversity and large gene pools of both species likely remain in the population, while both species are very closely related lineages [15]. The two species may share some alleles of microsatellite repeats at the same genomic locus, which is often observed in many closely related species in vertebrates [61,62]. We, therefore, proposed criteria to screen hybrid crocodiles between Siamese and saltwater crocodiles as follows: (i) Consideration of genetic admixture at different K levels to examine the trend of clustering, separation of allelic signals and the majority of allelic pattern, although the best K level might be predicted from different algorithms; (ii) sharing a gene pool between the two species might be possible, but should not have more than a posterior probability of 0.05 at the K level, which shows the trend of separation between the two species; (iii) clustering by PCoA should be considered together with the STRUCTURE plot to test the group of crocodile specimens; (iv) determination of maternal lineage by mtDNA D-loop sequences may be added to confirm; and (v) external morphological observation with updated phenotypic variations in the P.O. should be scored together with genetic screening.
These five steps would provide evidence that can prove the hybridization status of each Siamese crocodile individual under reasonable time before they are used in the reintroduction program (Figure 6). Crocodiles that pass the five tests of characteristics would be key sources for release to the wild. However, if the crocodile fails on some aspects with unclear determination, the individual should undergo more experimental tests such as karyotyping. Siamese and saltwater crocodiles have different chromosome numbers, whereas the F1 hybrid or backcross shows diverge chromosome constitution from the parental species [63,64]. However, karyotyping is time-consuming, expensive, and may not be practical to prove species purity for large numbers of individuals, whereas multiple types and generations of hybrid (both F2, F3 or backcross) might escape detection of chromosome number. More research utilizing genome-wide scans with single nucleotide polymorphisms (SNPs) is necessary to enhance our comprehension of selection signatures in diverse populations and species. However, genome-wide SNP analysis might not be reliable for multiple processes with small crocodile numbers in each reintroduction program. From this state, we found suspected hybrids with CB11 (P.O. 4), CB22 (P.O. 3), CB27 (P.O. 4), CP01 (P.O. 0), CP05 (P.O. 4), CP06 (P.O. 4), CP11 (P.O. 4), CP12 (P.O. 4), CP13 (P.O. 4), CP15 (P.O. 2), CP20 (P.O. 0), CP23 (P.O. 4), CP25 (P.O. 2), CP27 (P.O. 4), CP28 (P.O. 4), CP29 (P.O. 0), CP30 (P.O. 0), CSI05 (unidentified P.O.), CSI06 (unidentified P.O.), and CPO09 (unidentified P.O.) (Figure 5), which should be tested by karyotyping before release.
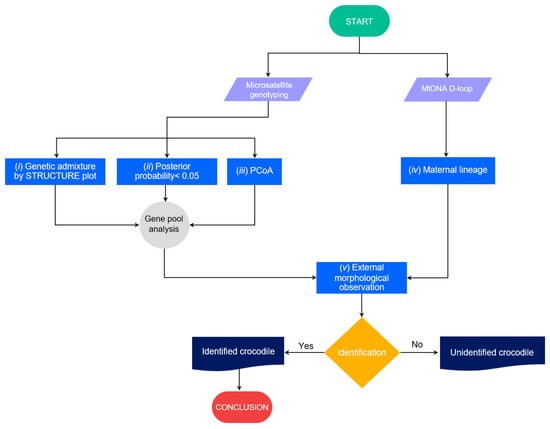
Figure 6.
Schematic representation of criteria to screen hybrid crocodiles between Siamese and saltwater crocodiles to prove the hybridization level status in each Siamese crocodile individual before they can be used in the reintroduction program.
The Thai government and TCFA have never promoted hybridization in crocodile farms under Appendix I captive breeding operation [46]; however, the incidence of contamination by hybrids was observed to be 5–10% here and in our previous study [11]. Hybrid contamination may result from a long history of crocodile trade from 30 years ago, which had no concrete plan of protection by the Thai Government. Hybridization between Siamese and Cuban crocodiles (C. rhombifer) as a result of human-mediated movement has also been observed in Southeast Asia [11,39,44,45]. Current results suggest that NP04, NP06, NP07, NP09, NP13, NP14, NP16, and NP20 are hybrid crocodiles derived from the Cuban crocodile lineage. We also found signs of a unique gene pool from unidentified crocodiles (suspected hybrid with Cuban crocodiles). However, we could not identify genetic admixture and introgression of Cuban crocodiles using microsatellite genotyping with our library, as there were no pure Cuban crocodiles in our experimental genetic stock [11] or in this study. Mitochondrial DNA analysis could allow us to infer the maternal lineage of Cuban crocodiles by comparing the DNA sequences with nucleotide data repositories such as GenBank, but this might not be enough for the cut-off determination of crocodile species. Thus, collaboration with crocodile research groups, governments, and NGOs such as the Crocodile Specialist Group (CSG) will be necessary for documenting and monitoring the introgression of several crocodile species in Southeast Asia.
Identifying purebred individuals from captive populations is a significant challenge for restocking and reintroduction efforts. The CSG has proposed various recommendations to support the conservation of Siamese crocodiles, including legislative and regulatory measures, compliance with CITES obligations, appropriate management of captive populations, conducting surveys and conservation initiatives, controlling illegal trade, promoting regional conservation initiatives, and exploring restocking options. These recommendations are aimed at supporting the current efforts of Thai national agencies to conserve the species, including the Department of Fisheries under the Ministry of Agriculture and Cooperatives and the Department of National Parks, Wildlife and Plant Conservation (DNP) under the Ministry of Natural Resources and Environment. To restore, protect, and create habitats for the Siamese crocodile, initiatives for public–private partnerships and sustainable financing will be launched. Prioritizing the management of key threats and conducting large-scale assessments of Siamese crocodile conservation status will be crucial for addressing challenges related to populations, protected areas, and other conservation initiatives.
5. Conclusions
These results indicate that a P.O. variation of 2–4 is within the species-level variation of Siamese crocodiles. In short, this is a phenotypic variation and not the result of hybridization with saltwater crocodiles. This baseline information on the association between genetic status and phenotypic variation of Siamese crocodiles in captive populations in Thailand is important for future conservation. Large differences in gene pools were observed in Siamese crocodiles, suggesting different historical origins of Siamese crocodiles in the wild population before massive capture and collection. Recently, a call was made to redefine the role of admixture in species conservation. It emphasized that crocodiles that have undergone gene flow and introgression during their evolutionary history or have been impacted by anthropogenic issues require protection measures. Ultimately, hybridization presents a management problem for Siamese crocodiles and complicates species identification based on morphology alone [65]. Adequate protocols to identify introgression and hybridization are urgently needed. Here, the genetic approach we followed proved that combining information from genetic and phenotypic approaches yielded more robust results. Accurate data on captive populations is critical for ensuring the long-term survival of the species through reintroduction programs and in situ/ex situ management, which helps to maintain sustainable genetic diversity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12040535/s1, Figure S1. Population structure of 136 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] and 29 saltwater crocodiles (C. porosus, Schneider, 1801) [13], (a) Plot of Evanno’s ΔK and (b) plot of ln P(K); Figure S2. Phylogenetic relationships among mitochondrial DNA D-loop region sequences were inferred using Bayesian inference analysis. Support values at each node denote the Bayesian posterior probability. Table S1 provides detailed information on the sampled individuals; Figure S3. Representation post-occipital scutes (P.O.) for hybrids between Siamese and saltwater crocodiles (Crocodylus siamensis × Crocodylus porosus). (a) CB11 (b) CB22 (c) CB27 (d) CP01 (e) CP05 (f) CP06 (g) CP11 (h) CP12 (i) CP13 (j) CP15 (k) CP20 (l) CP23 (m) CP25 (n) CP27 (o) CP28 (p) CP29 (q) CP30; Table S1. Specimen populations of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (C. porosus, Schneider, 1801) [13] All sequences were deposited in the DNA Data Bank of Japan (DDBJ) and BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 8 February 2023) of sequence identity; Table S2. Microsatellite primers and sequences; Table S3. Pairwise differentiation of linkage disequilibrium of Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] at Chonburi (CB) based on 22 microsatellite loci. Numbers indicate p-values with 110 permutations; Table S4. Pairwise differentiation of linkage disequilibrium of Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] at Nakhon Ratchasima (NR) based on 22 microsatellite loci. Numbers indicate p-values with 110 permutations; Table S5. Pairwise differentiation of linkage disequilibrium of Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] at Chainat (CN) based on 22 microsatellite loci. Numbers indicate p-values with 110 permutations; Table S6. Pairwise differentiation of linkage disequilibrium of Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] at Nakhon Pathom (NP) based on 22 microsatellite loci. Numbers indicate p-values with 110 permutations; Table S7. Pairwise differentiation of linkage disequilibrium of saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13] at Nakhon Ratchasima (CP) based on 22 microsatellite loci. Numbers indicate p-values with 110 permutations; Table S8. Genetic diversity of 136 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] and 29 saltwater crocodiles (C. porosus, Schneider, 1801) [13] based on 22 microsatellite loci. Table S1 provides detailed information on the sampled individuals; Table S9. Pairwise genetic relatedness (r) for all 30 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Chonburi (CB) Table S1 provides detailed information on the sampled individuals; Table S10. Pairwise genetic relatedness (r) for all 30 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Nakhon Ratchasima (NR) Table S1 provides detailed information on the sampled individuals; Table S11. Pairwise genetic relatedness (r) for all 34 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Chainat (CN). Table S1 provides detailed information on the sampled individuals; Table S12. Pairwise genetic relatedness (r) for all 42 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Nakhon Pathom (NP). Table S1 provides detailed information on the sampled individuals; Table S13. Pairwise genetic relatedness (r) for all 29 saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [3] in Nakhon Ratchasima (CP). Table S1 provides detailed information on the sampled individuals; Table S14. Distributions of r values and FIS values for Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [1]; Table S15. Pairwise inbreeding coefficients (FIS) for all 30 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Chonburi (CB). Detailed information on all Siamese crocodile individuals is presented in Table S1; Table S16. Pairwise inbreeding coefficients (FIS) for all 30 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Nakhon Ratchasima (NR). Detailed information on all Siamese crocodile individuals is presented in Table S1; Table S17. Pairwise inbreeding coefficients (FIS) for all 34 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Chainat (CN). Detailed information on all Siamese crocodile individuals is presented in Table S1; Table S18. Pairwise inbreeding coefficients (FIS) for all 42 Siamese crocodiles (Crocodylus siamensis, Schneider, 1801) [1] in Nakhon Pathom (NP). Detailed information on all Siamese crocodile individuals is presented in Table S1; Table S19. Pairwise inbreeding coefficients (FIS) for all 29 saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [1] in Nakhon Ratchasima (CP). Detailed information on all saltwater crocodile individuals is presented in Table S1; Table S20. Pairwise genetic differentiation (FST), pairwise FSTENA values with ENA correction for null alleles and RST values using FSTAT version 2.9.3 [66] and of Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13] between captive-bred individuals based on 22 microsatellite loci. The numbers indicate p values, with 110 permutations. Detailed information on all Siamese and saltwater crocodile individuals is presented in Table S1; Table S21. Analysis of molecular variance (AMOVA) results for Siamese crocodile (Crocodylus siamensis, Schneider, 1801) [1] and crocodiles (Crocodylus porosus, Schneider, 1801) [1] based on 22 microsatellite loci using Arlequin version 3.5.2.2 [67]. Detailed information on all Siamese and saltwater crocodiles is presented in Table S1; Table S22. Pairwise population Nei’s genetic distance (D) values using GenAlEx version 6.5 [28] of 166 Siamese (Crocodylus siamensis, Schneider, 1801) [1] and saltwater crocodiles (Crocodylus porosus, Schneider, 1801) [13] based on 22 microsatellite loci. Detailed information on all Siamese and saltwater crocodiles is presented in Table S1; Supplementary Data S1 Materials and Methods [66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85].
Author Contributions
Conceptualization, N.A., Y.T. and K.S.; funding acquisition, K.S.; formal analysis, N.A., W.W., P.W., N.M. and K.S.; investigation, N.A., W.W., P.W., T.P., W.S., T.T., A.L., S.F.A., N.M., K.H., P.D., Y.T. and K.S.; methodology, N.A., W.W., P.W., T.P., W.S., T.T. and K.S.; project administration, K.S.; resources, N.A., W.W., P.W., W.S. and Y.T.; software, N.A., W.W., P.W., T.P., W.S. and N.M.; supervision, P.D. and K.S.; validation, N.A., W.S. and K.S.; visualization, N.A. and K.S.; writing—original draft, N.A. and K.S.; writing—review and editing, N.A., W.W., P.W., T.P., W.S., T.T., A.L., S.F.A., N.M., K.H., P.D., Y.T. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported in part by the High-Quality Research Graduate Development Cooperation Project between Kasetsart University and the National Science and Technology Development Agency (NSTDA) (6417400247) awarded to T.P. and K.S.; the Thailand Science Research and Innovation through the Kasetsart University Reinventing University Program 2021 (3/2564) awarded to T.P., A.L. and K.S.; the Higher Education for Industry Consortium (Hi-FI) (6414400777) awarded to N.A.; the e-ASIA Joint Research Program (no. P1851131) awarded to W.S. and K.S.; a grant from the National Science and Technology Development Agency (NSTDA) (NSTDA P-19-52238 and JRA-CO-2564-14003-TH) awarded to W.S. and K.S.; a grant from Betagro Group (no. 6501.0901.1/68) awarded to K.S.; a grant from Kasetsart University Research and Development Institute (FF(KU)25.64) awarded to W.S., S.F.A. and K.S.; and support from the Office of the Ministry of Higher Education, Science, Research, and Innovation. International SciKU Branding (ISB), Faculty of Science, Kasetsart University awarded funds to K.S.
Institutional Review Board Statement
The animal care and experimental procedures were conducted according to the Regulations on Animal Experiments at Kasetsart University and were approved by the Animal Experiment Committee under the code ACKU64-SCI-011.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences were deposited in the DNA Data Bank of Japan (DDBJ). The online version contains Supplementary Materials available at https://datadryad.org/stash/share/s4zREYQ1AUUXTsaIpk0r1HSdkYljvu8yvJOVR143K7Y (accessed on 18 February 2018).
Acknowledgments
We would like to thank the Thai Crocodile Farm Association for assistance with specimen collection and the provision of useful information. This project was conducted under a Memorandum of Agreement between the Faculty of Science at Kasetsart University and the Thai Crocodile Farm Association. The authors are also grateful to Ekaphan Kraichak (Department of Botany, Faculty of Science, Kasetsart University, Thailand), Chalitra Saysuk, and Rasoarahona Rivo Niaina Hajatiana Kevin Ryan (AGB Research Unit, Kasetsart University, Thailand) for helpful discussion. We thank the Center for Agricultural Biotechnology (CAB) at the Kasetsart University Kamphaeng Saen Campus and the NSTDA Supercomputer Center (ThaiSC) for support with server 561 analysis services. We also thank the Faculty of Science for providing research facilities.
Conflicts of Interest
The authors declare no conflict of interest. No funding source was involved in the study design, collection, analysis, and interpretation of the data, nor in writing the report or the decision to submit the article for publication.
References
- Schneider, J.G. Crocodylus siamensis. 1801. Available online: https://www.gbif.org/species/185108439 (accessed on 16 January 2023).
- Smith, M.A. The Fauna of British India including Ceylon and Burma; Reptilia and Amphibia V. 1. Loricata, Testudines; Taylor and Francis: London, UK, 1931. [Google Scholar]
- Daltry, J.C.; Chheang, D.; Em, P.; Poeung, M.; Sam, H.; Tan, T.; Simpson, B.K. Status of the Siamese crocodile in the central cardamom mountains, Cambodia. In Fauna & Flora International: Cambodia Programme and Department of Forestry and Wildlife: Phnom Penh; Fauna & Flora International: Phnom Penh, Cambodia, 2003. [Google Scholar]
- Platt, S.G.; Lynam, A.J.; Temsiripong, Y.; Kampanakngarn, M. Occurrence of the Siamese crocodile (Crocodylus siamensis) in Kaeng Krachan National Park, Thailand. Nat. Hist. Bull. Siam Soc. 2002, 50, 7–14. [Google Scholar]
- Platt, S.G.; Rainwater, T.R.; Finger, A.G.; Thorbjarnarson, J.B.; Anderson, T.A.; McMurry, S.T. Food habits, ontogenetic dietary partitioning and observations of foraging behaviour of Morelet’s crocodile (Crocodylus moreletii) in northern Belize. Herpetol. J. 2006, 16, 281–290. [Google Scholar]
- Simpson, B.K.; Bezuijen, M.R. Siamese crocodile Crocodylus siamensis. In Crocodiles Status Survey and Conservation Action Plan, 2nd ed.; Manolis, S.C., Stevenson, C., Eds.; Crocodile Specialist Group: Darwin, NT, Australia, 2010; pp. 120–126. [Google Scholar]
- Smith, M.A. Crocodylus siamensis. J. Nat. Hist. Soc. Siam 1919, 3, 217–222. [Google Scholar]
- Ilhow, F.; Bonke, R.; Hartman, T.; Geissler, P.; Behler, N.; Rödder, D. Habitat suitability, coverage by protected areas and population connectivity for the Siamese crocodile Crocodylus siamensis Schnider, 1801. Aquatic. Conserv. Mar. Fresh. Ecosyst. 2015, 25, 544–554. [Google Scholar] [CrossRef]
- Platt, S.G.; Rainwater, T.R.; Nichols, S. A recent population assessment of the American crocodile (Crocodylus acutus) in Turneffe Atoll, Belize. Herpetol. Bull. 2004, 89, 26–32. [Google Scholar]
- Thorbjarnarson, J.B.; Messel, H.; King, F.W.; Ross, J.P. Crocodiles: An Action Plan for their Conservation; IUCN: Gland, Switzerland, 1992. [Google Scholar]
- Lapbenjakul, S.; Thanapa, W.; Twilprawat, P.; Muangmai, N.; Khancanaketu, T.; Temsiripong, Y.; Unajak, S.; Peyachoknagul, S.; Srikulnath, K. High genetic diversity and demographic history of captive Siamese and saltwater crocodiles suggest the first step toward the establishment of a breeding and reintroduction program in Thailand. PLoS ONE 2017, 12, e0184526. [Google Scholar] [CrossRef]
- Suttimeechaikul, N.; Pakdeekong, M.; Tankitjanukit, S.; Keawjantavee, P.; Temsiripong, Y.; Phuangcharoen, W.; Kittiwanich, J.; Aiamsaart, P. Croccodile aquaculture. Available online: https://online.fliphtml5.com/vqklc/ljrd/?fbclid=IwAR0ZDRISD47Gh8t3#p=1 (accessed on 16 January 2023).
- Schneider. Crocodylus porosus. 1801. Available online: https://www.gbif.org/species/144104483 (accessed on 16 January 2023).
- Suvanakorn, P.; Youngprapakorn, C. Crocodile farming in Thailand. In Wildlife Management: Crocodiles and Alligators; Webb, G.J.W., Manolis, S.C., Whitehead, P.J., Eds.; Surrey Beatty and Sons, Pty. Ltd.: Chipping Norton, NSW, Australia, 1987; pp. 341–343. [Google Scholar]
- Srikulnath, K.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S. New haplotype of the complete mitochondrial genome of Crocodylus siamensis and its species-specific DNA markers: Distinguishing C. siamensis from C. porosus in Thailand. Mol. Biol. Rep. 2012, 39, 4709–4717. [Google Scholar] [CrossRef]
- Charette, R. CITES Identification Guide: Crocodilians; Environment Canada: Ottawa, ON, Canada, 1995.
- Ross, F.D.; Mayer, G.C. On the dorsal armor of the Crocodilia. In Advances in Herpetology and Evolutionary Biology; Museum of Comparative Zoology: Cambridge, MA, USA, 1983; pp. 305–331. [Google Scholar]
- Dinets, V. Long-distance signaling in Crocodylia. Copeia 2013, 2013, 517–526. [Google Scholar] [CrossRef]
- Supikamolseni, A.; Ngaoburanawit, N.; Sumontha, M.; Chanhome, L.; Suntrarachun, S.; Peyachoknagul, S.; Srikulnath, K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 2015, 14, 13981–13997. [Google Scholar] [CrossRef]
- Miles, L.G.; Isberg, S.R.; Glenn, T.C.; Lance, S.L.; Dalzell, P.; Thomson, P.C.; Moran, C. A genetic linkage map for the saltwater crocodile (Crocodylus porosus). BMC Genom. 2009, 10, 339. [Google Scholar] [CrossRef]
- Miles, L.G.; Isberg, S.R.; Moran, C.; Hagen, C.; Glenn, T.C. 253 Novel polymorphic microsatellites for the saltwater crocodile (Crocodylus porosus). Conserv. Genet. 2009, 10, 963–980. [Google Scholar] [CrossRef]
- Ariyaraphong, N.; Pansrikaew, T.; Jangtarwan, K.; Thintip, J.; Singchat, W.; Laopichienpong, N.; Pongsanarm, T.; Panthum, T.; Suntronpong, A.; Ahmad, S.F.; et al. Introduction of wild Chinese gorals into a captive population requires careful genetic breeding plan monitoring for successful long-term conservation. Glob. Ecol. Conserv. 2021, 28, e01675. [Google Scholar] [CrossRef]
- Chailertrit, V.; Swatdipong, A.; Peyachoknagul, S.; Salaenoi, J.; Srikulnath, K. Isolation and characterization of novel microsatellite markers from Siamese fighting fish (Betta splendens, Osphronemidae, Anabantoidei) and their transferability to related species, B. smaragdina and B. imbellis. Genet. Mol. Res. 2014, 13, 7157–7162. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Koomgun, T.; Prasongmaneerut, T.; Thongchum, R.; Singchat, W.; Tawichasri, P.; Fukayama, T.; Sillapaprayoon, S.; Kraichak, E.; Muangmai, N.; et al. Take one step backward to move forward: Assessment of genetic diversity and population structure of captive Asian woolly-necked storks (Ciconia episcopus). PLoS ONE 2019, 14, e0223726. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Kamsongkram, P.; Subpayakom, N.; Sillapaprayoon, S.; Muangmai, N.; Kongphoemph, A.; Wongsodchuen, A.; Intapan, S.; Chamchumroon, W.; Safoowong, M.; et al. Predictive genetic plan for a captive population of the Chinese goral (Naemorhedus griseus) and prescriptive action for ex situ and in situ conservation management in Thailand. PLoS ONE 2020, 15, e0234064. [Google Scholar] [CrossRef]
- Thintip, J.; Singchat, W.; Ahmad, S.F.; Ariyaraphong, N.; Muangmai, N.; Chamchumroon, W.; Pitiwong, K.; Suksavate, W.; Duangjai, S.; Duengkae, P.; et al. Reduced genetic variability in a captive-bred population of the endangered Hume’s pheasant (Syrmaticus humiae, Hume 1881) revealed by microsatellite genotyping and D-loop sequencing. PLoS ONE 2021, 16, e0256573. [Google Scholar] [CrossRef]
- Wongtienchai, P.; Lapbenjakul, S.; Jangtarwan, K.; Areesirisuk, P.; Mahaprom, R.; Subpayakom, N.; Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Songchan, R.; et al. Genetic management of a water monitor lizard (Varanus salvator macromaculatus) population at Bang Kachao Peninsula as a consequence of urbanization with Varanus Farm Kamphaeng Saen as the first captive research establishment. J. Zool. Syst. Evol. Res. 2021, 59, 484–497. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Wang, J. Coancestry: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 2011, 11, 141–145. [Google Scholar] [CrossRef]
- Temsiripong, Y.; Ratanakorn, P.; Kullavanijaya, B. Management of the Siamese crocodile in Thailand. In Proceedings of the 17th Working Meeting of the IUCN-SSC Crocodile Specialist Group, Darwin, NT, Australia, 24–29 May 2004; pp. 141–142. [Google Scholar]
- Jelden, D.C.; Manolis, C.; Giam, H.; Thomson, J.; Lopez, A. Crocodile Conservation and Management in Cambodia: A Review with Recommendations; IUCN Crocodile Specialist Group: Darwin, NT, Australia, 2005. [Google Scholar]
- Jelden, D.C.; Manolis, C.; Tsubouchi, T.; Nguyen Dao, N.V. Crocodile Conservation and Farming in the Socialist Republic of Viet Nam: A Review with Recommendations; Crocodile Specialist Group: Darwin, NT, Australia, 2008. [Google Scholar]
- Manolis, C. 2nd Siamese crocodile meeting on husbandry and conservation and Siamese crocodile task force meeting (1–2 June 2017). Crocodile Spec. Group Newsl. 2017, 36, 5–9. [Google Scholar]
- Caldwell, J. World Trade in Crocodilian Skins 2008–2010; UNEP-WCMC: Cambridge, CA, USA, 2010. [Google Scholar]
- Getpech, Y. Crocodile conservation and captive breeding in Thailand. In Proceedings of the 1st Regional Species Meeting of the IUCN-SSC Crocodile Specialist Group, Bangkok, Thailand, 4–6 April 2011; pp. 27–28. [Google Scholar]
- Caldwell, J. World Trade in Crocodilian Skins 2018–2020; UNEP-WCMC: Cambridge, CA, USA, 2022. [Google Scholar]
- Chavananikul, V.; Wattanodorn, S.; Youngprapakorn, P. Karyotypes of 5 species of crocodile kept in Samutprakan Crocodile Farm and Zoo. In Proceedings of the 12th Working Meeting of the IUCN-SSC Crocodile Specialist Group, Pattaya, Thailand, 2–6 May 1994; pp. 58–62. [Google Scholar]
- Thang, N.Q. The status of Crocodylus rhombifer in the Socialist Republic of Vietnam. In Proceedings of the 12th Working Meeting of the IUCN-SSC Crocodile Specialist Group, Pattaya, Thailand, 2–6 May 1994; pp. 141–142. [Google Scholar]
- Fitzsimmons, N.N.; Buchan, J.C.; Lam, P.V.; Polet, G.; Hung, T.T.; Thang, N.Q.; Gratten, J. Identification of purebred Crocodylus siamensis for reintroduction in Vietnam. J. Exp. Zool. A Ecol. Genet. Physiol. 2002, 294, 373–381. [Google Scholar] [CrossRef]
- Truyen, T. Country—Vietnam. In Proceedings of the 1st Regional Meeting of the IUCN-SSC Crocodile Specialist Group, Bangkok, Thailand, 4–6 April 2011; pp. 33–35. [Google Scholar]
- Phothitay, C.; Phommachanh, B.; Bezuijen, M.R. Siamese crocodiles at Ban Kuen Zoo, Lao PDR. Crocodile Spec. Group Newsl. 2005, 24, 11–12. [Google Scholar]
- Cox, J.H., Jr.; Phothitay, C. Surveys of the Siamese Crocodile Crocodylus Siamensis in Savannakhet Province, Lao PDR, 6 May–4 June 2008; OZ Minerals Ltd. & Wildlife Conservation Society: Vientiane, Laos, 2008. [Google Scholar]
- Cohen, M.M.; Gans, C. The chromosomes of the order Crocodilia. Cytogenet. Genome Res. 1970, 9, 81–105. [Google Scholar] [CrossRef]
- Chavananikul, V.; Suwattana, D.; Koykul, W.; Wattanodorn, S.; Sukkai, J. A Research Report on Karyotypes of Freshwater Crocodiles (Crocodylus siamensis), Saltwater Crocodiles (Crocodylus porosus) and the Inter-Specific Hybrids by Conventional and Banding Techniques; Chulalongkorn University: Bangkok, Thailand, 1998. [Google Scholar]
- Doc, C. Convention on international trade in endangered species of wild fauna and flora. In Proceedings of the Nineteenth meeting of the Conference of the Parties, Panama City, Panama, 14–25 November 2022. [Google Scholar]
- Wildlife Conservation Society. Recommendations for CITES CoP19; Wildlife Conservation Society: New York, NY, USA, 2022. [Google Scholar]
- Kowasupat, C.; Panijpan, B.; Ruenwongsa, P.; Sriwattanarothai, N. Betta mahachaiensis, a new species of bubble-nesting fighting fish (Teleostei: Osphronemidae) from Samut Sakhon Province, Thailand. Zootaxa 2012, 3522, 49–60. [Google Scholar] [CrossRef]
- Kowasupat, C.; Panijpan, B.; Ruenwongsa, P. Betta siamorientalis, a new species of bubble-nest building fighting fish (Teleostei: Osphronemidae) from eastern Thailand. Vertebr. Zool. 2012, 62, 387–397. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Robertson, B.C.; Stanton, J.A.L.; Gemmell, N.J. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. BioTechniques 2009, 46, 185–192. [Google Scholar] [CrossRef]
- Fischer, M.C.; Rellstab, C.; Leuzinger, M.; Roumet, M.; Gugerli, F.; Shimizu, K.K.; Holderegger, R.; Widmer, A. Estimating genomic diversity and population differentiation–an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genom. 2017, 18, 69. [Google Scholar] [CrossRef]
- Fola, A.A.; Kattenberg, E.; Razook, Z.; Lautu-Gumal, D.; Lee, S.; Mehra, S.; Bahlo, M.; Kazura, J.; Robinson, L.J.; Laman, M.; et al. SNP barcodes provide higher resolution than microsatellite markers to measure Plasmodium vivax population genetics. Malar. J. 2020, 19, 375. [Google Scholar] [CrossRef]
- Egito, A.A.; Paiva, S.R.; Albuquerque, M.D.S.M.; Mariante, A.S.; Almeida, L.D.; Castro, S.R.; Grattapaglia, D. Microsatellite based genetic diversity and relationships among ten Creole and commercial cattle breeds raised in Brazil. BMC Genet. 2007, 8, 83. [Google Scholar] [CrossRef]
- Cortes, O.; Cañon, J.; Gama, L.T. Applications of microsatellites and single nucleotide polymorphisms for the genetic characterization of cattle and small ruminants: An overview. Ruminants 2022, 2, 456–470. [Google Scholar] [CrossRef]
- Department of Fisheries. Meeting to Discuss Action Guidelines for Crocodiles in Thailand. Available online: https://www4.fisheries.go.th/dof/news_local/131/138388 (accessed on 16 January 2023).
- Kanwatanakid-Savini, C.; Pliosungnoen, M.; Pattanavibool, A.; Thorbjarnarson, J.B.; Limlikhitaksorn, C.; Platt, S.G. A survey to determine the conservation status of Siamese crocodiles in Kaeng Krachan National Park, Thailand. Herpetol. Conserv. Biol. 2012, 7, 157–168. [Google Scholar]
- Platt, S.G. Community-Based Crocodile Conservation in Lao PDR; Wildlife Conservation Society: Bronx, NY, USA, 2012. [Google Scholar]
- Starr, J.C.D.A. Development of a re-introduction and re-enforcement program for Siamese crocodiles in Cambodia. In Global Re-Introduction Perspectives: Additional Case Studies from Around the Globe; IUCN: Gland, Switzerland, 2010; p. 118. [Google Scholar]
- Hata, A.; Nunome, M.; Suwanasopee, T.; Duengkae, P.; Chaiwatana, S.; Chamchumroon, W.; Suzuki, T.; Koonawootrittriron, S.; Matsuda, Y.; Srikulnath, K. Origin and evolutionary history of domestic chickens inferred from a large population study of Thai red junglefowl and indigenous chickens. Sci. Rep. 2021, 11, 2035. [Google Scholar] [CrossRef]
- Singchat, W.; Chaiyes, A.; Wongloet, W.; Ariyaraphong, N.; Jaisamut, K.; Panthum, T.; Ahmed, S.F.; Chaleekarn, W.; Suksavate, W.; Inpota, M.; et al. Red junglefowl resource management guide: Bioresource reintroduction for sustainable food security in Thailand. Sustainability 2022, 14, 7895. [Google Scholar] [CrossRef]
- Adams, R.H.; Blackmon, H.; Reyes-Velasco, J.; Schield, D.R.; Card, D.C.; Andrew, A.L.; Waynewood, N.; Castoe, T.A. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 2016, 59, 295–310. [Google Scholar] [CrossRef]
- Wattanadilokchatkun, P.; Panthum, T.; Jaisamut, K.; Ahmad, S.F.; Dokkaew, S.; Muangmai, N.; Duengkae, P.; Singchat, W.; Srikulnath, K. Characterization of microsatellite distribution in Siamese fighting fish genome to promote conservation and genetic diversity. Fishes 2022, 7, 251. [Google Scholar] [CrossRef]
- Srikulnath, K.; Thapana, W.; Muangmai, N. Role of chromosome changes in Crocodylus evolution and diversity. Genom. Inform. 2015, 13, 102–111. [Google Scholar] [CrossRef]
- Kawagoshi, T.; Nishida, C.; Ota, H.; Kumazawa, Y.; Endo, H.; Matsuda, Y. Molecular structures of centromeric heterochromatin and karyotypic evolution in the Siamese crocodile (Crocodylus siamensis) (Crocodylidae, Crocodylia). Chromosome Res. 2008, 16, 1119–1132. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Ziegler, T.; Rauhaus, A.; Nguyen, T.Q.; Tran, D.T.A.; Wayakone, S.; Luu, V.Q.; Vences, M.; Le, M.D. Genetic screening of Siamese crocodiles (Crocodylus siamensis) in Laos and Vietnam: Identifying purebred individuals for conservation and release programs. Crocodile Spec. Group Newsl. 2018, 37, 8–14. [Google Scholar]
- Goudet, J.F. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.W.; Thompson, E.A. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Welch, B.L. The generalization of student’s’ problem when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef]
- Van, O.C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Ph.D. Thesis, University of Dublin, Dublin, Ireland, 2001. [Google Scholar]
- Præstgaard, J.T. Permutation and bootstrap Kolmogorov-Smirnov tests for the equality of two distributions. Scand. J. Stat. 1995, 22, 305–322. [Google Scholar]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A. Structure harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Weir, B.; Cockerham, C. Estimating F-Statistics for the analysis of population-structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. Parallel Distrib. Process. Symp. Int. Proc. 2002, 2, 184. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).