CardioRVAR: A New R Package and Shiny Application for the Evaluation of Closed-Loop Cardiovascular Interactions
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector Autoregressive Models
2.2. Causal Coherence and Noise Contribution
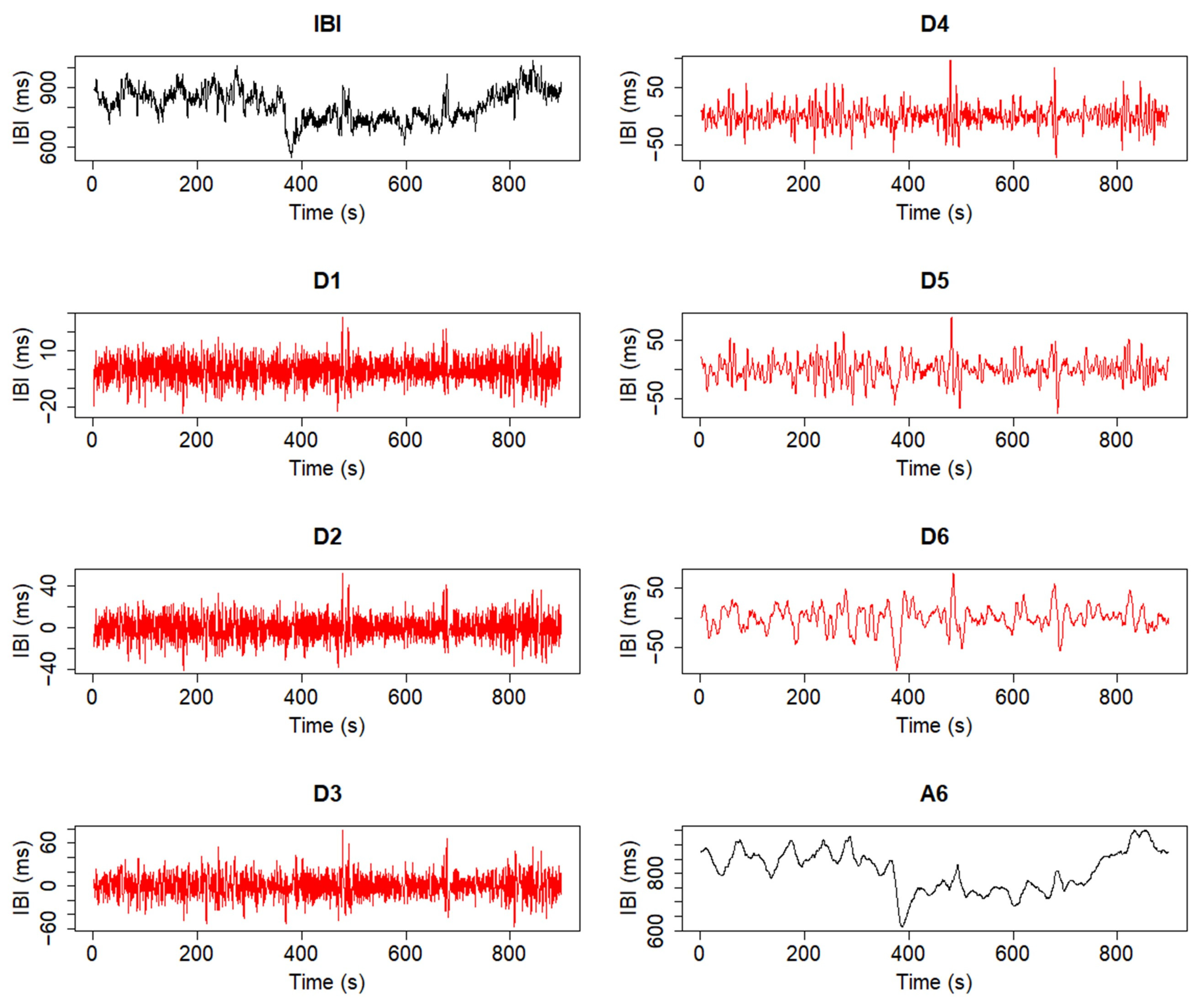
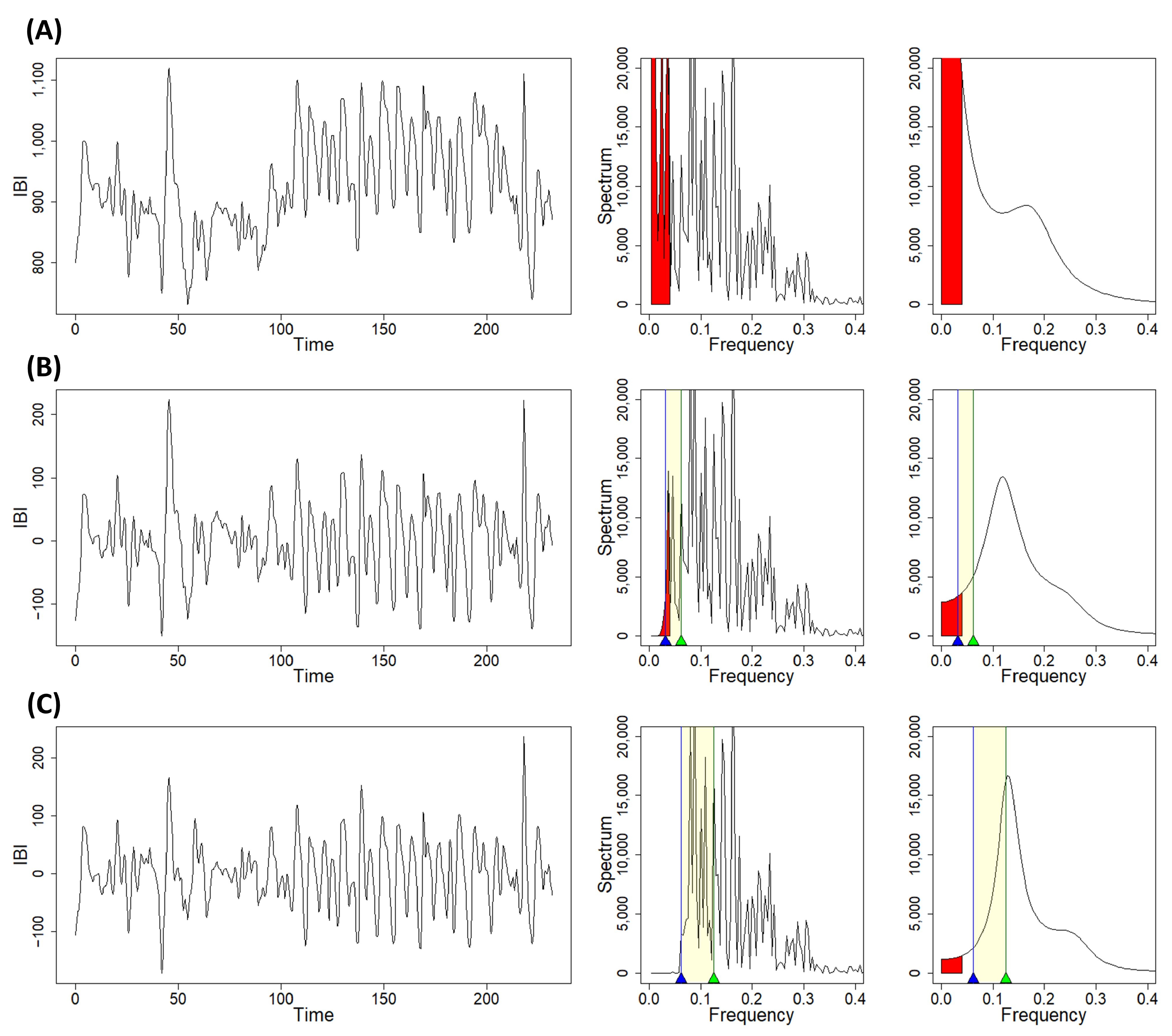
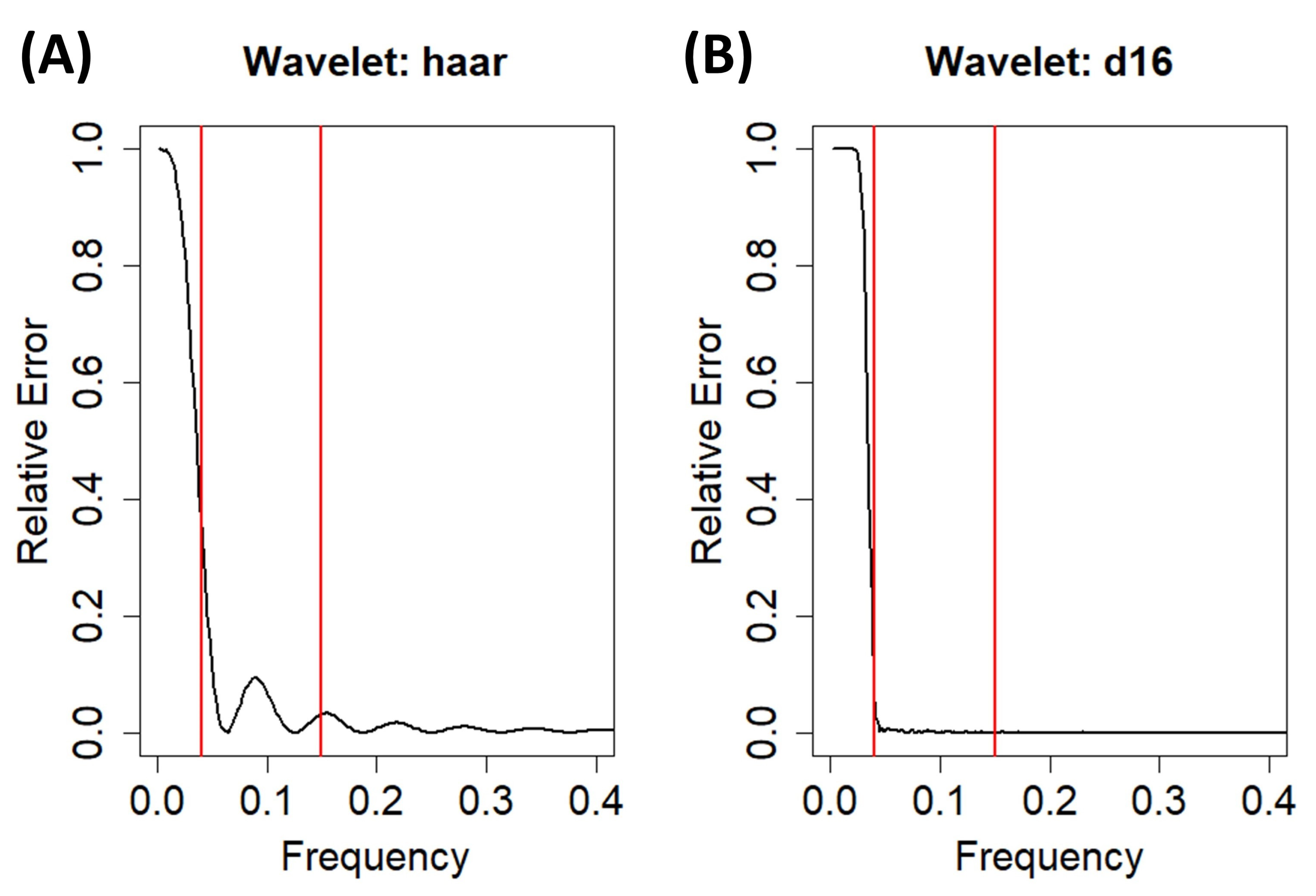
2.3. Trend Removal with the Discrete Wavelet Transform
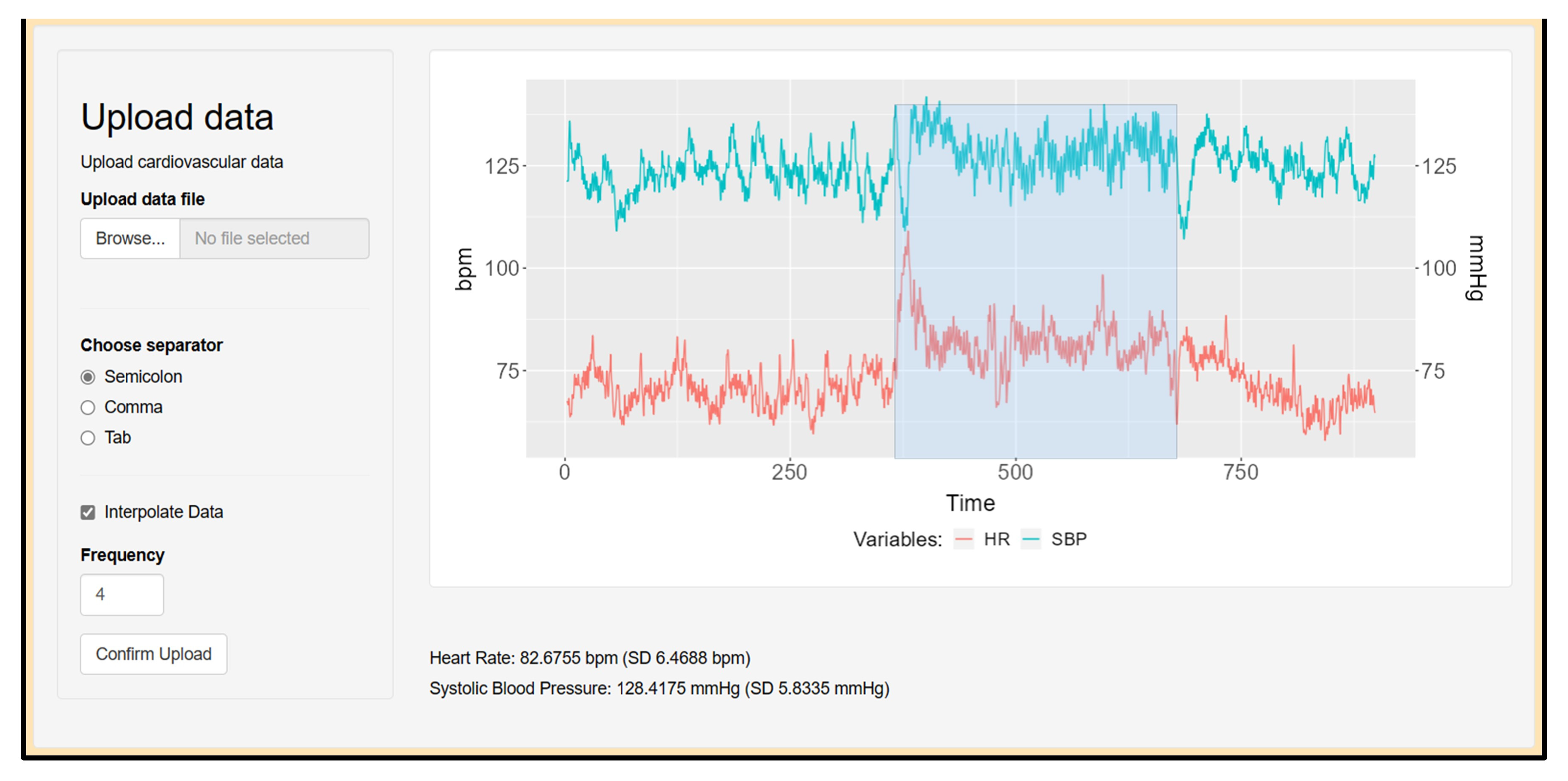
2.4. CardioRVAR Workflow
- Select a data file with CardioRVARapp and upload it into the software structure;
- Resample the uploaded time series after selecting a certain frequency, if needed;
- Manually select from the estimated HR and BP recordings a specific time window of interest;
- Transform the model into the frequency domain;
- Extract instantaneous unidirectional interactions from this frequency-domain representation, given a specific zero-lag-interaction path already chosen by the user, and update the model with such interactions;
- Estimate the most important features of the model and then display and report them;
- Generate and report single-subject indices from the model, allowing the user to choose a method to estimate said indices.
2.5. Data Upload and Preprocessing
| > library(CardioRVAR) |
| > # Data is a list with elements Time, RR, and SBP: |
| > Data <- ResampleData(Data, 4) # Interpolates data |
| > IBI <- DetrendWithCutoff(Data$RR) # Detrends IBI signal |
| > SBP <- DetrendWithCutoff(Data$SBP) # Detrends SBP signal |
| > New_Data <- cbind(SBP = SBP, RR = IBI) |
| > CheckStationarity(New_Data) # Checks stationarity of the data |
| [1] TRUE |
| > # Or alternatively: |
| > Check_stationarity <- CheckStationarity(New_Data, verbose = TRUE) |
| Time series are stationary |
2.6. Analysis of Cardiovascular Closed-Loop Interactions
| > # Data represents a matrix with two interpolated time series, IBI and SBP. |
| > Data[,“IBI”] = DetrendWithCutoff(Data[,“IBI”]) |
| > Data[,“SBP”] = DetrendWithCutoff(Data[,“SBP”]) |
| > # Both signals have been detrended with these commands. |
| > CheckStationarity(Data) |
| [1] TRUE |
| > # A VAR model is estimated from the stationary time series and then validated: |
| > model <- EstimateVAR(Data) |
| > Check_residuals <- DiagnoseResiduals(model, verbose = TRUE) |
| Model residuals are white noise processes |
| > Check_stability <- DiagnoseStability(model, verbose = TRUE) |
| The model is stable |
2.7. Analysis in the Frequency Domain
| > freq_model <- ParamFreqModel(model). |
2.8. Assessment of the Transfer Functions
2.9. Assessment of the Noise Source Contribution and Causal Coherence
2.10. Evaluation of the Tool: Data Sources
3. Results and Discussion
3.1. Descriptive Study of Two Subjects
3.2. Comparison between Normotensive and Hypertensive Subjects
3.3. EUROBAVAR Analysis Results
3.4. Comparison with Other Works
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez-Liñares, L.; Méndez, A.J.; Lado, M.J.; Olivieri, D.N.; Vila, X.A.; Gómez-Conde, I. An Open Source Tool for Heart Rate Variability Spectral Analysis. Comput. Methods Programs Biomed. 2011, 103, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.A.G.; Quintana, A.O.; Vila, X.A.; Touriño, M.J.L.; Rodríguez-Liñares, L.; Presedo, J.M.R.; Penín, A.J.M. Heart Rate Variability Analysis with the R Package RHRV; Springer International Publishing: Cham, Switzerland, 2017; ISBN 978-3-319-65354-9. [Google Scholar]
- Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Heart Rate Variability: Standards of Measurement, Physiological Interpretation and Clinical Use. Circulation 1996, 93, 1043–1065. [Google Scholar] [CrossRef]
- Swenne, C.A. Baroreflex Sensitivity: Mechanisms and Measurement. Neth. Heart J. 2013, 21, 58–60. [Google Scholar] [CrossRef]
- Barbieri, R.; Parati, G.; Saul, J.P. Closed- versus Open-Loop Assessment of Heart Rate Baroreflex. IEEE Eng. Med. Biol. Mag. 2001, 20, 33–42. [Google Scholar] [CrossRef]
- Faes, L.; Porta, A.; Antolini, R.; Nollo, G. Role of Causality in the Evaluation of Coherence and Transfer Function between Heart Period and Systolic Pressure in Humans. In Proceedings of the Computers in Cardiology, Chicago, IL, USA, 19–22 September 2004; IEEE: Piscataway, NJ, USA, 2004; pp. 277–280. [Google Scholar]
- Takalo, R.; Saul, J.P.; Korhonen, I. Comparison of Closed-Loop and Open-Loop Models in the Assessment of Cardiopulmonary and Baroreflex Gains. Methods Inf. Med. 2004, 43, 296–301. [Google Scholar] [CrossRef]
- Hytti, H.; Takalo, R.; Ihalainen, H. Tutorial on Multivariate Autoregressive Modelling. J. Clin. Monit. Comput. 2006, 20, 101–108. [Google Scholar] [CrossRef]
- Faes, L.; Erla, S.; Porta, A.; Nollo, G. A Framework for Assessing Frequency Domain Causality in Physiological Time Series with Instantaneous Effects. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2013, 371, 20110618. [Google Scholar] [CrossRef]
- Faes, L.; Nollo, G. Measuring Frequency Domain Granger Causality for Multiple Blocks of Interacting Time Series. Biol. Cybern. 2013, 107, 217–232. [Google Scholar] [CrossRef]
- Faes, L.; Erla, S.; Nollo, G. Block Partial Directed Coherence: A New Tool for the Structural Analysis of Brain Networks. Int. J. Bioelectromagn. 2012, 14, 162–166. [Google Scholar]
- Faes, L.; Erla, S.; Nollo, G. Measuring Connectivity in Linear Multivariate Processes: Definitions, Interpretation, and Practical Analysis. Comput. Math. Methods Med. 2012, 2012, 140513. [Google Scholar] [CrossRef]
- Seth, A.K. A MATLAB Toolbox for Granger Causal Connectivity Analysis. J. Neurosci. Methods 2010, 186, 262–273. [Google Scholar] [CrossRef]
- Seth, A.K. Granger Causal Connectivity Analysis: A MATLAB Toolbox; University of Sussex: Brighton, UK, 2011. [Google Scholar]
- Barnett, L.; Seth, A.K. The MVGC Multivariate Granger Causality Toolbox: A New Approach to Granger-Causal Inference. J. Neurosci. Methods 2014, 223, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Chao-Ecija, A.; Dawid-Milner, M.S. BaroWavelet: An R-Based Tool for Dynamic Baroreflex Evaluation through Wavelet Analysis Techniques. Comput. Methods Programs Biomed. 2023, 242, 107758. [Google Scholar] [CrossRef] [PubMed]
- Farnè, M.; Montanari, A. A Bootstrap Method to Test Granger-Causality in the Frequency Domain. Comput. Econ. 2022, 59, 935–966. [Google Scholar] [CrossRef]
- Porta, A.; Furlan, R.; Rimoldi, O.; Pagani, M.; Malliani, A.; van de Borne, P. Quantifying the Strength of the Linear Causal Coupling in Closed Loop Interacting Cardiovascular Variability Signals. Biol. Cybern. 2002, 86, 241–251. [Google Scholar] [CrossRef]
- Faes, L.; Nollo, G. Multivariate Frequency Domain Analysis of Causal Interactions in Physiological Time Series. In Biomedical Engineering, Trends in Electronics, Communications and Software; InTech: Rijeka, Croatia, 2011; pp. 403–428. [Google Scholar]
- Percival, D.B.; Walden, A.T. The Discrete Wavelet Transform. In Wavelet Methods for Time Series Analysis; Cambridge University Press: Cambridge, UK, 2000; pp. 56–158. [Google Scholar]
- Percival, D.B.; Walden, A.T. The Maximal Overlap Discrete Wavelet Transform. In Wavelet Methods for Time Series Analysis; Cambridge University Press: Cambridge, UK, 2000; pp. 159–205. [Google Scholar]
- Percival, D.B.; Walden, A.T. The Discrete Wavelet Packet Transform. In Wavelet Methods for Time Series Analysis; Cambridge University Press: Cambridge, UK, 2000; pp. 206–254. [Google Scholar]
- Tzabazis, A.; Eisenried, A.; Yeomans, D.C.; Hyatt, M.I. V Wavelet Analysis of Heart Rate Variability: Impact of Wavelet Selection. Biomed. Signal. Process. Control 2018, 40, 220–225. [Google Scholar] [CrossRef]
- Quandt, V.I.; Pacola, E.R.; Pichorim, S.F.; Sovierzoski, M.A. Border Extension in the Wavelet Analysis of Lung Sounds. In World Congress on Medical Physics and Biomedical Engineering 26–31 May 2012, Beijing, China; Springer: Berlin/Heidelberg, Germany, 2013; pp. 597–600. [Google Scholar]
- Li, L.; Li, K.; Liu, C.; Liu, C. Comparison of Detrending Methods in Spectral Analysis of Heart Rate Variability. Res. J. Appl. Sci. Eng. Technol. 2011, 3, 1014–1021. [Google Scholar]
- Andrecut, M. Fast Time Series Detrending with Applications to Heart Rate Variability Analysis. Int. J. Mod. Phys. C 2019, 30, 1950069. [Google Scholar] [CrossRef]
- Yoo, C.S.; Yi, S.H. Effects of Detrending for Analysis of Heart Rate Variability and Applications to the Estimation of Depth of Anesthesia. J. Korean Phys. Soc. 2004, 44, 561. [Google Scholar] [CrossRef]
- Gebauer, J.E.; Adler, J. Using Shiny Apps for Statistical Analyses and Laboratory Workflows. J. Lab. Med. 2023, 47, 149–153. [Google Scholar] [CrossRef]
- Ding, M.; Bressler, S.; Yang, W.; Liang, H. Short-Window Spectral Analysis of Cortical Event-Related Potentials by Adaptive Multivariate Autoregressive Modeling: Data Preprocessing, Model Validation, and Variability Assessment. Biol. Cybern. 2000, 83, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-K.; Hu, Y.-F.; Lin, S.-F. Methodological Considerations in Calculating Heart Rate Variability Based on Wearable Device Heart Rate Samples. Comput. Biol. Med. 2018, 102, 396–401. [Google Scholar] [CrossRef]
- Shibata, R. Selection of the Order of an Autoregressive Model by Akaike’s Information Criterion. Biometrika 1976, 63, 117. [Google Scholar] [CrossRef]
- Boardman, A.; Schlindwein, F.S.; Rocha, A.P.; Leite, A. A Study on the Optimum Order of Autoregressive Models for Heart Rate Variability. Physiol. Meas. 2002, 23, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Bassani, T.; Bari, V.; Marchi, A.; Wu, M.A.; Baselli, G.; Citerio, G.; Beda, A.; de Abreu, M.G.; Güldner, A.; Guzzetti, S.; et al. Coherence Analysis Overestimates the Role of Baroreflex in Governing the Interactions between Heart Period and Systolic Arterial Pressure Variabilities during General Anesthesia. Auton. Neurosci. 2013, 178, 83–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Laude, D.; Elghozi, J.-L.; Girard, A.; Bellard, E.; Bouhaddi, M.; Castiglioni, P.; Cerutti, C.; Cividjian, A.; Rienzo, M.D.; Fortrat, J.-O.; et al. Comparison of Various Techniques Used to Estimate Spontaneous Baroreflex Sensitivity (the EuroBaVar Study). Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2004, 286, R226–R231. [Google Scholar] [CrossRef]
- McLoone, V.; Ringwood, J.V. A System Identification Approach to Baroreflex Sensitivity Estimation. In Proceedings of the IET Irish Signals and Systems Conference (ISSC 2012), Maynooth, Ireland, 28–29 June 2012; Institution of Engineering and Technology: Stevenage, UK, 2012; pp. 301–306. [Google Scholar]
- Bari, V.; Vaini, E.; De Maria, B.; Cairo, B.; Pistuddi, V.; Ranucci, M.; Porta, A. Comparison of Different Strategies to Assess Cardiac Baroreflex Sensitivity Based on Transfer Function Technique in Patients Undergoing General Anesthesia. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 2780–2783. [Google Scholar] [CrossRef]
- Dani, M.; Dirksen, A.; Taraborrelli, P.; Torocastro, M.; Panagopoulos, D.; Sutton, R.; Lim, P.B. Autonomic Dysfunction in “Long COVID”: Rationale, Physiology and Management Strategies. Clin. Med. 2021, 21, e63–e67. [Google Scholar] [CrossRef]
- Westerhof, B.E.; Gisolf, J.; Stok, W.J.; Wesseling, K.H.; Karemaker, J.M. Time-Domain Cross-Correlation Baroreflex Sensitivity: Performance on the EUROBAVAR Data Set. J. Hypertens. 2004, 22, 1371–1380. [Google Scholar] [CrossRef]
- Choi, Y.; Ko, S.; Sun, Y. Effect of Postural Changes on Baroreflex Sensitivity: A Study on the Eurobavar Data Set. In Proceedings of the 2006 Canadian Conference on Electrical and Computer Engineering, Ottawa, ON, Canada, 7–10 May 2006; IEEE: Piscataway, NJ, USA, July, 2006; pp. 110–114. [Google Scholar]
- Parati, G.; Castiglioni, P.; Faini, A.; Di Rienzo, M.; Mancia, G.; Barbieri, R.; Saul, J.P. Closed-Loop Cardiovascular Interactions and the Baroreflex Cardiac Arm: Modulations Over the 24 h and the Effect of Hypertension. Front. Physiol. 2019, 10, 477. [Google Scholar] [CrossRef]
- Chao-Écija, Á.; Carrasco-Gómez, D.; Dawid-Milner, M.S. Effectiveness of an R-Based Software to Detect Closed-Loop Cardio- Vascular Interactions and Baroreflex Impairment in Human subjects from the EUROBAVAR Data Set. J. Physiol. Biochem. 2022, 78, S34. [Google Scholar] [CrossRef]

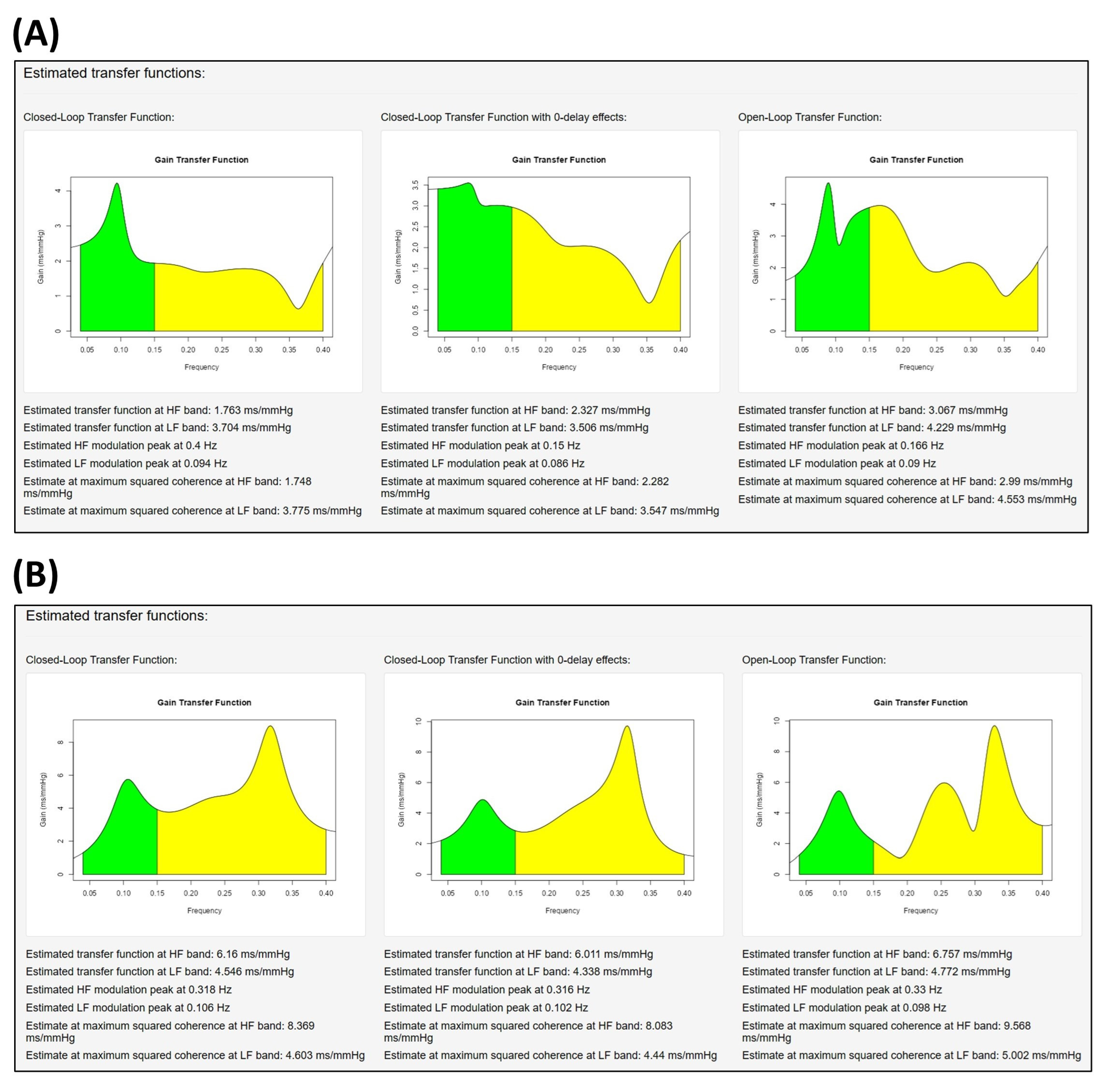
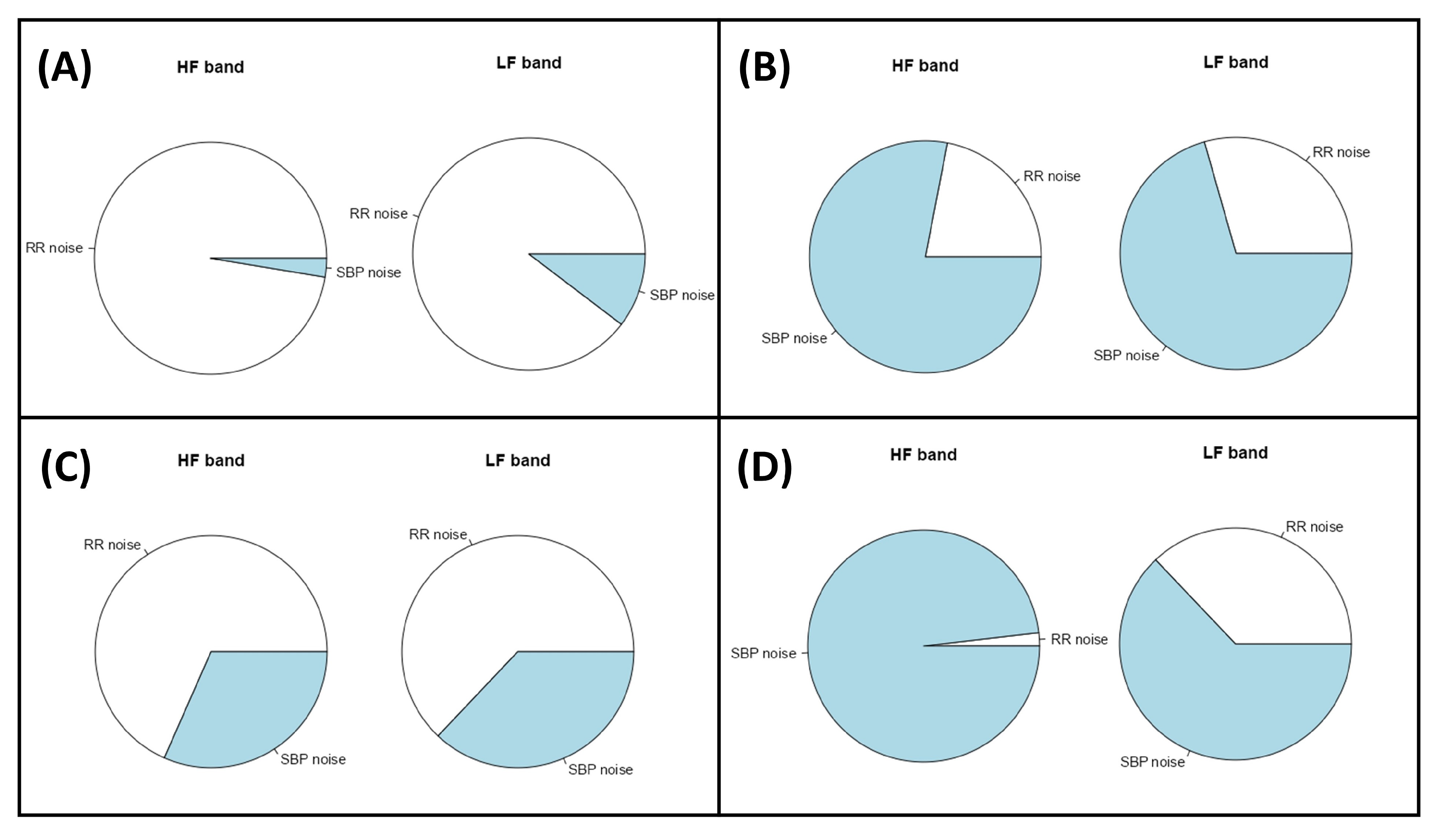
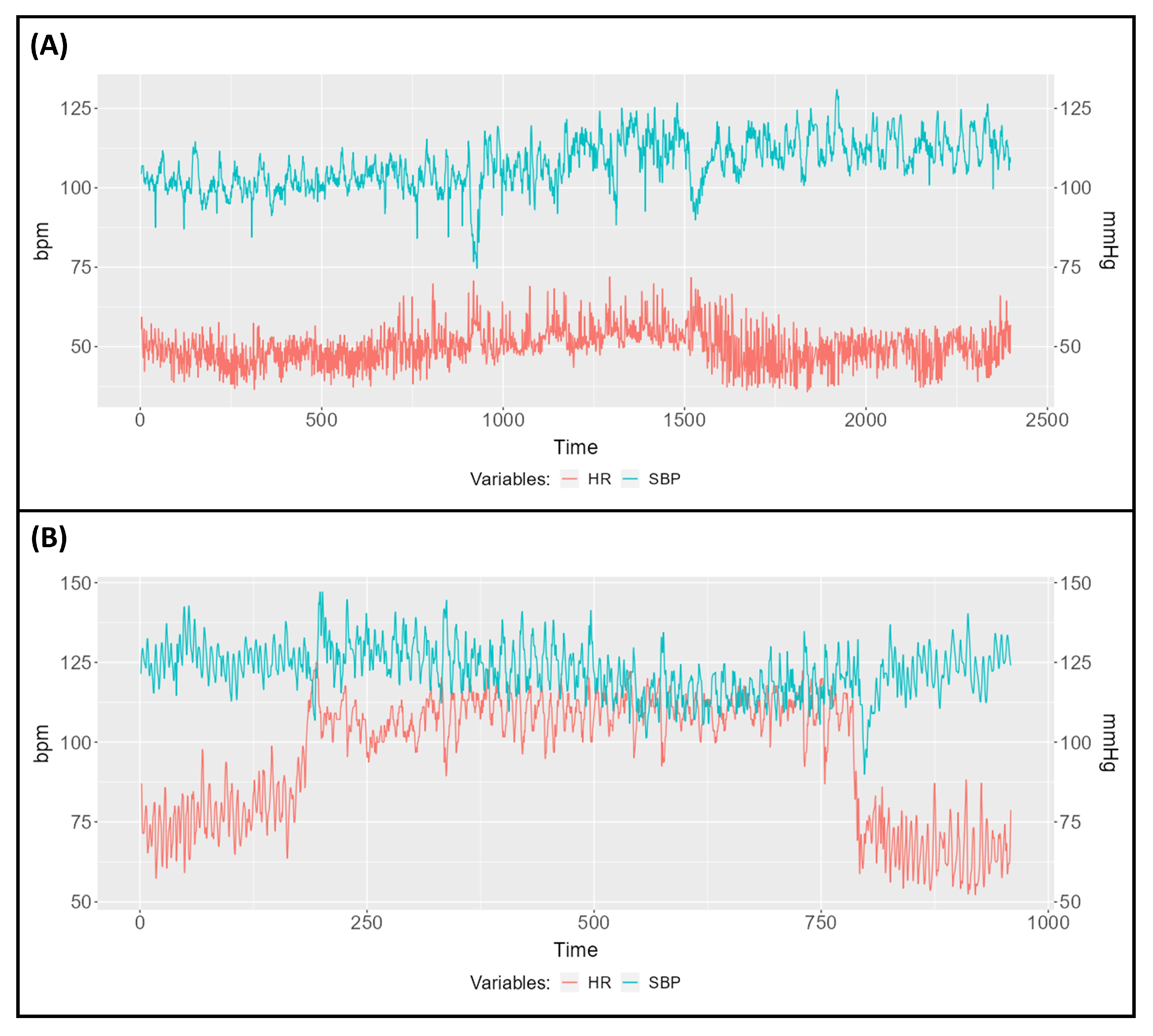

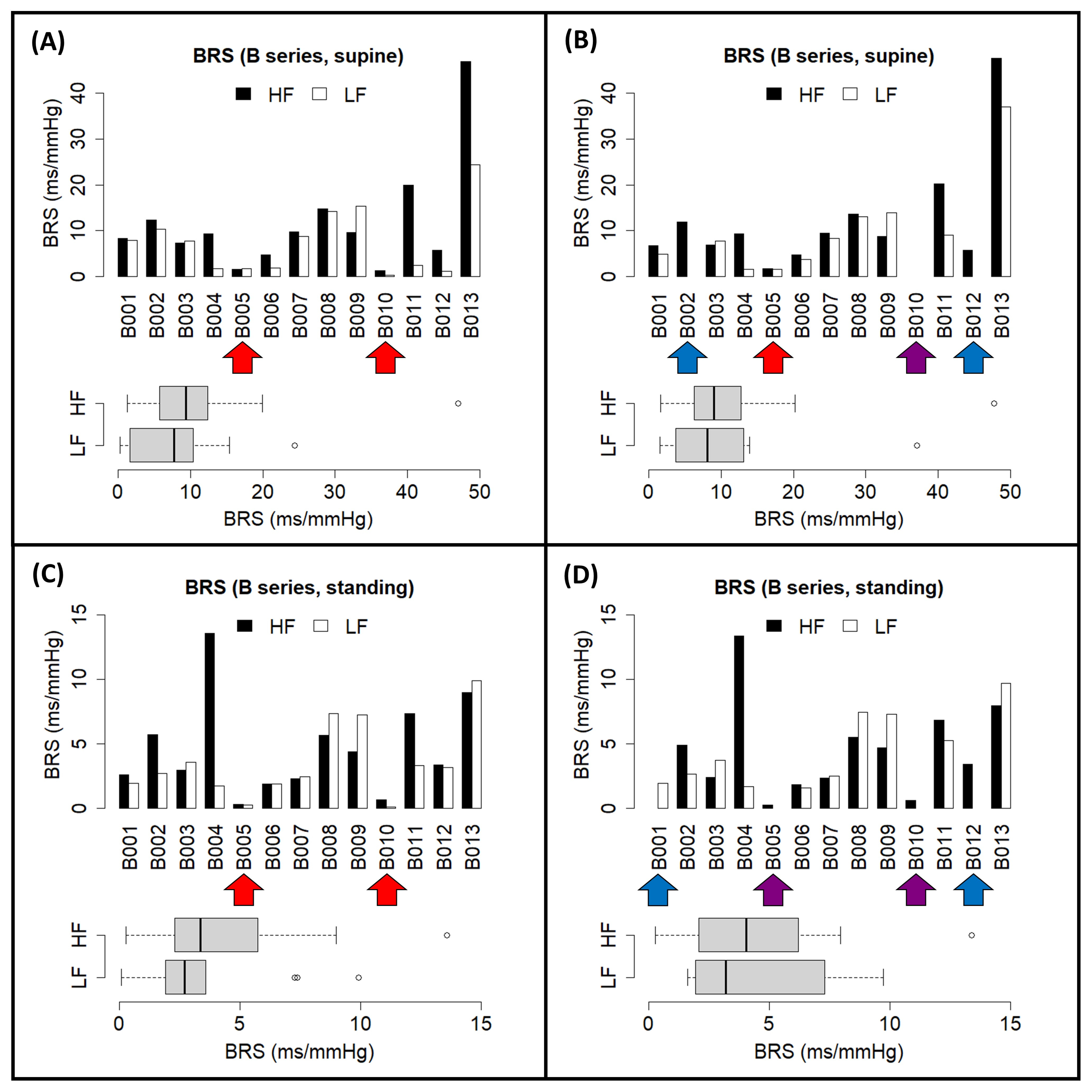
| Subject A | Subject B | |||||
|---|---|---|---|---|---|---|
| Variable | Pre-Tilt Interval | Tilt Interval | Post-Tilt Interval | Pre-Tilt Interval | Tilt Interval | Post-Tilt Interval |
| HR (bpm) | 48.27 | 54.03 | 49.42 | 78.42 | 109.74 | 69.75 |
| SBP (mmHg) | 102.12 | 109.15 | 111.65 | 125.84 | 121.62 | 120.95 |
| CT-HF αc (ms/mmHg) | 26.54 | 11.57 | 35.90 | 13.30 | N/A † | 13.60 |
| WA-HF αc (ms/mmHg) | 24.13 | 9.05 | 31.56 | 9.54 | 1.76 | 12.81 |
| MC-HF αc (ms/mmHg) | 27.34 | 12.50 | 36.33 | 13.10 | 1.78 | 16.33 |
| CT-LF αc (ms/mmHg) | 6.95 | 1.74 | N/A † | 11.78 | 3.98 | 17.88 |
| WA-LF αc (ms/mmHg) | 7.33 | 4.59 | 17.41 | 11.28 | 3.81 | 17.78 |
| MC-LF αc (ms/mmHg) | 7.02 | 1.73 | 1.63 | 12.25 | 4.11 | 16.77 |
| Position | Band | Estimate Type | Normotensive (n = 5) | Hypertensive (n = 7) | p Value |
|---|---|---|---|---|---|
| Supine rest | HF | Weighted-averaged | 9.02 ± 3.88 | 2.03 ± 0.45 | p < 0.01 |
| Estimate at maximum coherence | 10.99 ± 4.14 | 3.10 ± 0.75 | p < 0.05 | ||
| LF | Weighted-averaged | 5.94 ± 1.38 | 2.25 ± 0.39 | p = 0.054 | |
| Estimate at maximum coherence | 6.19 ± 1.32 | 1.69 ± 0.37 | p < 0.05 | ||
| Tilt | HF | Weighted-averaged | 4.34 ± 1.39 | 1.27 ± 0.29 | p = 0.091 |
| Estimate at maximum coherence | 5.01 ± 1.95 | 1.46 ± 0.21 | p = 0.143 | ||
| LF | Weighted-averaged | 4.90 ± 0.64 | 2.06 ± 0.22 | p < 0.01 | |
| Estimate at maximum coherence | 4.69 ± 0.96 | 1.66 ± 0.23 | p < 0.05 |
| Closed-Loop | Open-Loop (Type II) | Open-Loop (Type I) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Band | Method | Supine (ms/mmHg) | Standing (ms/mmHg) | p Value | Supine (ms/mmHg) | Standing (ms/mmHg) | p Value | Supine (ms/mmHg) | Standing (ms/mmHg) | p Value |
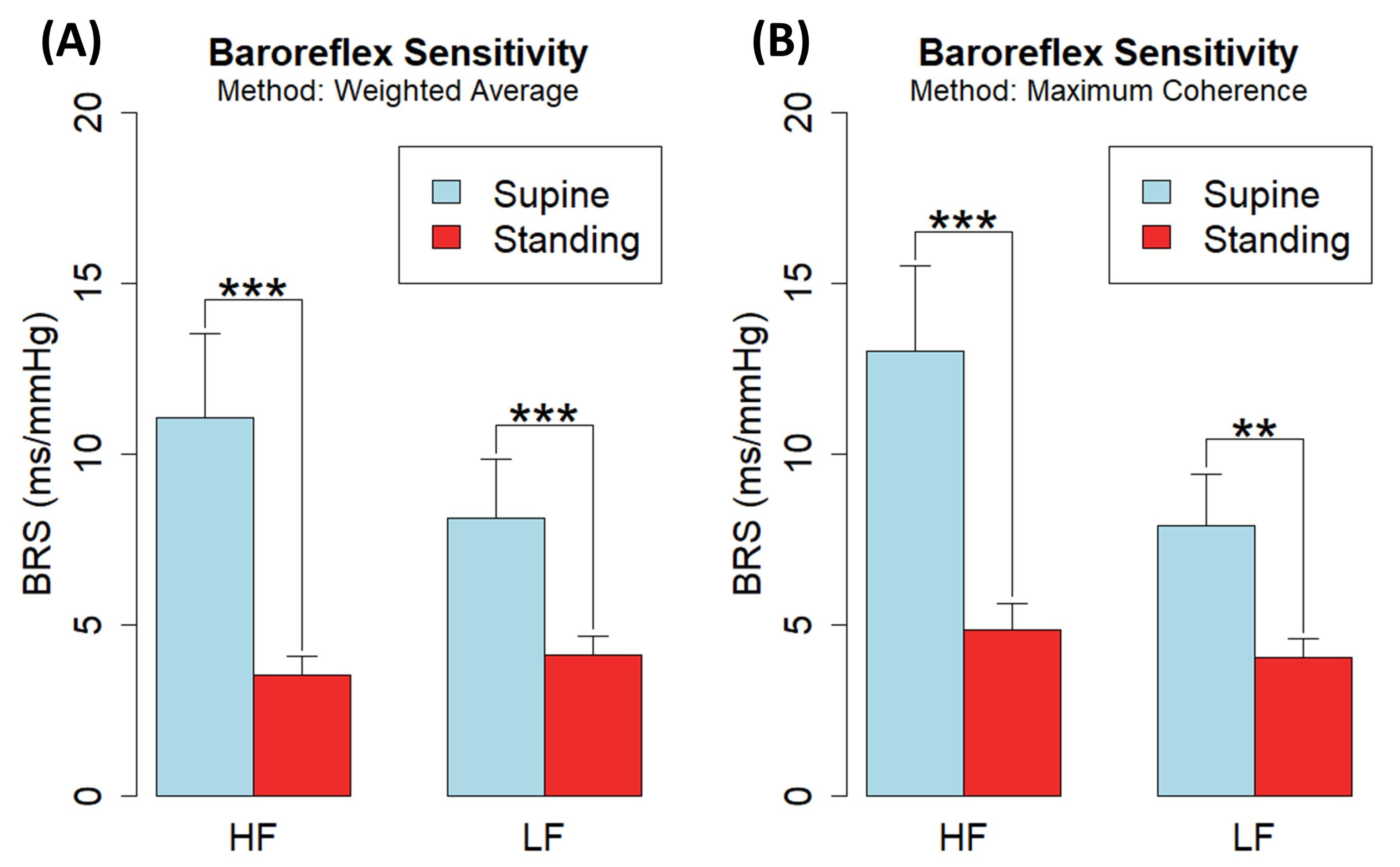
| HF | Weighted average | 11.06 ± 2.46 | 3.54 ± 0.54 | p < 0.001 | 13.82 ± 2.92 | 5.03 ± 0.81 | p < 0.001 | 20.79 ± 3.74 | 7.82 ± 1.27 | p < 0.001 |
| Maximum coherence | 13.03 ± 2.47 | 4.84 ± 0.79 | p < 0.001 | 16.07 ± 2.68 | 6.40 ± 1.07 | p < 0.001 | 17.40 ± 2.92 | 7.51 ± 1.38 | p < 0.001 | |
| LF | Weighted average | 8.12 ± 1.72 | 4.12 ± 0.55 | p < 0.001 | 9.23 ± 2.25 | 5.12 ± 0.77 | p < 0.001 | 12.72 ± 2.75 | 7.12 ± 0.91 | p < 0.001 |
| Maximum coherence | 7.92 ± 1.50 | 4.06 ± 0.54 | p < 0.01 | 10.48 ± 2.04 | 5.42 ± 0.68 | p < 0.01 | 12.43 ± 2.25 | 6.41 ± 0.77 | p < 0.01 | |
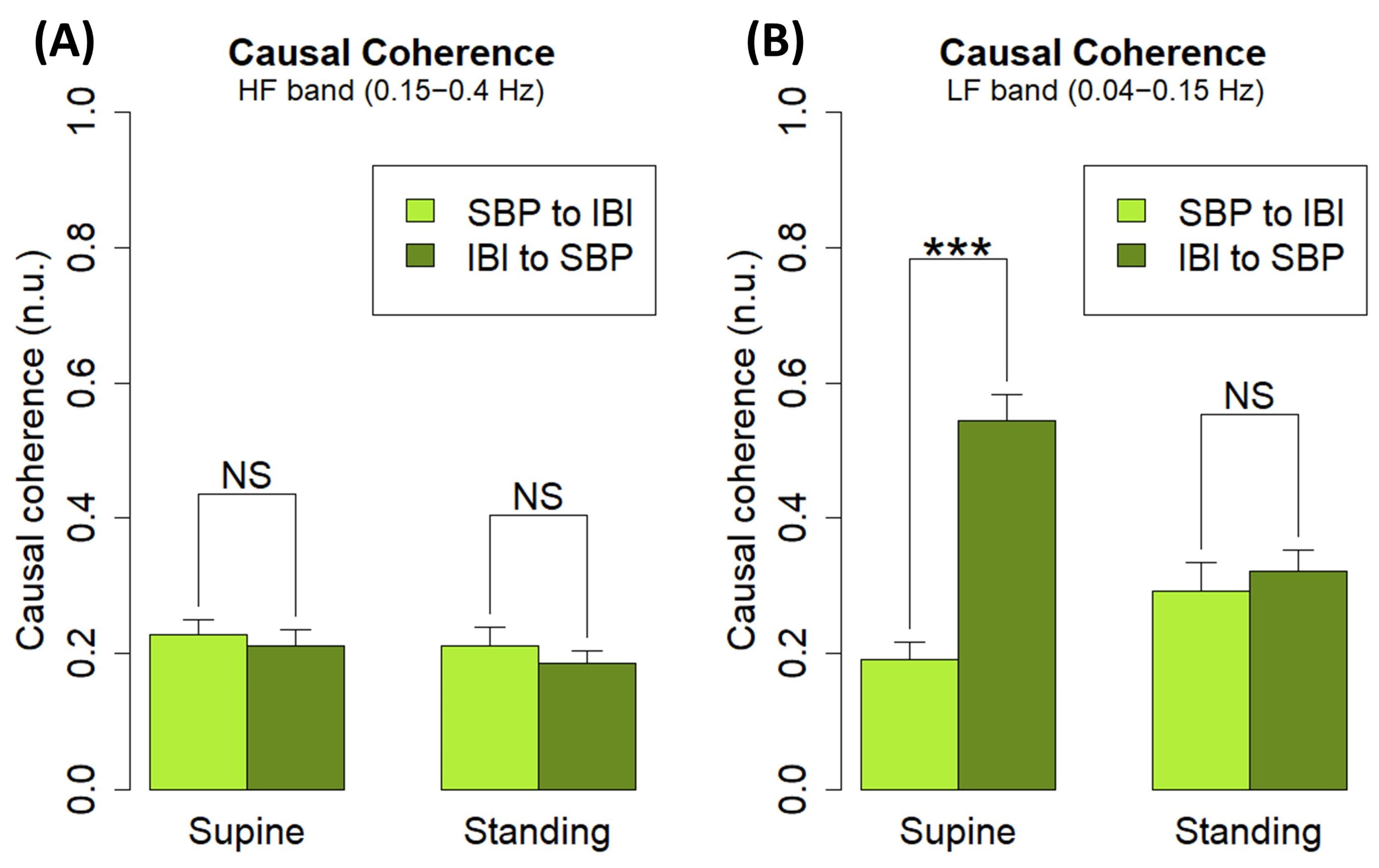
| Position | Band | (n.u.) | (n.u.) | p Value |
|---|---|---|---|---|
| Supine | HF | 0.23 ± 0.02 | 0.21 ± 0.02 | p = 0.644 |
| LF | 0.19 ± 0.03 | 0.54 ± 0.04 | p < 0.001 | |
| Standing | HF | 0.21 ± 0.03 | 0.19 ± 0.02 | p = 0.510 |
| LF | 0.29 ± 0.04 | 0.32 ± 0.03 | p = 0.606 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chao-Écija, A.; López-González, M.V.; Dawid-Milner, M.S. CardioRVAR: A New R Package and Shiny Application for the Evaluation of Closed-Loop Cardiovascular Interactions. Biology 2023, 12, 1438. https://doi.org/10.3390/biology12111438
Chao-Écija A, López-González MV, Dawid-Milner MS. CardioRVAR: A New R Package and Shiny Application for the Evaluation of Closed-Loop Cardiovascular Interactions. Biology. 2023; 12(11):1438. https://doi.org/10.3390/biology12111438
Chicago/Turabian StyleChao-Écija, Alvaro, Manuel Víctor López-González, and Marc Stefan Dawid-Milner. 2023. "CardioRVAR: A New R Package and Shiny Application for the Evaluation of Closed-Loop Cardiovascular Interactions" Biology 12, no. 11: 1438. https://doi.org/10.3390/biology12111438
APA StyleChao-Écija, A., López-González, M. V., & Dawid-Milner, M. S. (2023). CardioRVAR: A New R Package and Shiny Application for the Evaluation of Closed-Loop Cardiovascular Interactions. Biology, 12(11), 1438. https://doi.org/10.3390/biology12111438






