1. Introduction
Globally, diabetes mellitus (DM) is one of the most common chronic diseases, and its prevalence has increased significantly over the last three decades [
1]. At present, 460 million individuals have DM; this number is expected to increase to 700 million by 2045. It is a chronic and metabolic condition characterized by elevated blood glucose levels [
2]. The three most common types of diabetes are type 1 diabetes mellitus (T1DM), type 2 diabetes mellitus (T2DM), and gestational diabetes mellitus (GDM). According to WHO, Saudi Arabia has the seventh highest diabetes rate in the world and the second highest rate in the Middle East, with an estimated 7 million diabetes and 3 million individuals with diabetes [
3]. Saudi Arabia is one of the 21 countries and territories of the International Defense Force (IDF) Middle East and North Africa (MENA) Region. Of the 537 million individuals with diabetes worldwide, 73 million are in the MENA Region; this number is expected to increase to 135.7 million by 2045 [
4]. Currently, diabetes is the most prevalent serious health concern in Saudi Arabia. According to the Ministry of Health in Saudi Arabia, the prevalence of diabetes increased from 0.9 million to 2.5 million between 1992 and 2010. However, the prevalence is expected to increase 2.7 times within the next two decades [
5].
Type 2 diabetes mellitus (T2DM) is characterized by hyperglycemia and is caused by a combination of deficient insulin secretion, inadequate insulin resistance, and misleading glucagon secretion [
6]. Since 1960, the prevalence of T2DM has increased in tandem with obesity. T2DM is caused by a combination of genetic and lifestyle factors [
7]. Ethnicity, age, body mass index (BMI), obesity, blood pressure, fasting blood glucose (FBG), and lipid concentrations have all been identified as risk factors for T2DM development [
8]. T2DM has been linked to several gene variants and a number of single nucleotide polymorphisms (SNPs; monogenic and polygenic) forms [
9]. SNPs were discovered through the human genome sequence in 2003, and genome-wide association studies (GWAS) have detected associations between disease and the genetic loci [
10].
Toll-like receptors (TLRs) play an important role in the activation of innate immunity and regulatory adaptive immunological responses. TLRs, in humans, are classified into ten subtypes [
11]. Effector molecules, such as cytokines and chemokines, regulate adaptive immune responses and are expressed when TLRs recognize pathogen-associated molecular patterns [
12]. TLR9 is linked to autoimmune diseases and has been associated with high-risk asthma and low-risk diabetes [
13]. TLR9 deficiency enhances pancreatic islet formation as well as β-cell differentiation and function [
14]. Bizarrely, TLR9 deficiency was found to boost glucose tolerance and insulin sensitivity in individuals with diabetes. Han et al. have confirmed a connection between TLR9 and T2DM osteoporosis via the NF-
KB pathway [
15]. One of the foot ulcer infections in individuals with diabetes is linked to the relationship between infection and diabetes [
16].
The relationship between
TLR9 and diabetes, specifically T2DM, is well established [
12,
13,
17]. The rs352140 SNP was studied in T1DM, and it was confirmed that the rs352140 SNP was connected with residual β-cell function [
11]. There are no direct studies between rs187084 SNP and T2DM, but Liu et al. [
17] carried out studies on the CAD individuals who developed T2DM. Finally, the rs5743836 SNP was studied in individuals with T2DM and diabetic foot [
12]. The main purpose of designing this study specifically in T2DM patients was due to the documentation of the previous studies with these three SNPs in different human diseases in Saudi Arabia [
18,
19]. Altogether, the relation between the
TLR9 gene and T2DM was connected to β-cell function, and additionally, CD73 expression was significantly upregulated and the immunosuppressive function of CD73+ T cells was improved in NOD mice with a TLR9 deficit. As a result, the development of diabetes was slowed while the levels of proinflammatory cytokines were reduced and the expression levels of more anti-inflammatory cytokines were elevated [
20]. In this study, we aimed to determine the relationship between three TLR9 SNPs, namely rs187084, rs352140, and rs5743836, and T2DM. We aimed to determine the molecular role of rs187084, rs352140, and rs5743836 in Saudi Arabian patients, with a familial history of diabetes, diagnosed with T2DM.
4. Discussion
Toll-like receptors are found on cells of the innate immune system and contribute to the diagnosis of T2DM by identifying damage-associated molecular patterns. Upon TLR recognition, innate immune cells perform a number of actions, such as phagocytosis, cytokine synthesis, co-stimulatory molecule expression, and adhesion molecule expression [
23]. TLRs are pivotal in the development of diabetes, which is primarily expressed in β-cells. TLR expression in β-cells provides an internal cell mechanism for innate signals regulating adaptive immune responses, and TLR signaling has been shown to be important in β-cell growth and activation [
24]. The relationship between the
TLR9 gene and β-cells is considered to be the specialized cells in the pancreas, which is produced via insulin and also plays a major role in elevating blood glucose levels. However, this is not directly connected with immune cells. The connection between TLR9 and the innate immune system may be influenced by β-cell function and diabetes [
14,
25]. One of the major reasons for the increase in diabetes in Saudi Arabia is the burden of obesity/BMI and an aging population [
5]. Consequently, cardiovascular diseases (CVD) have become the future health concern in Saudi Arabia [
26].
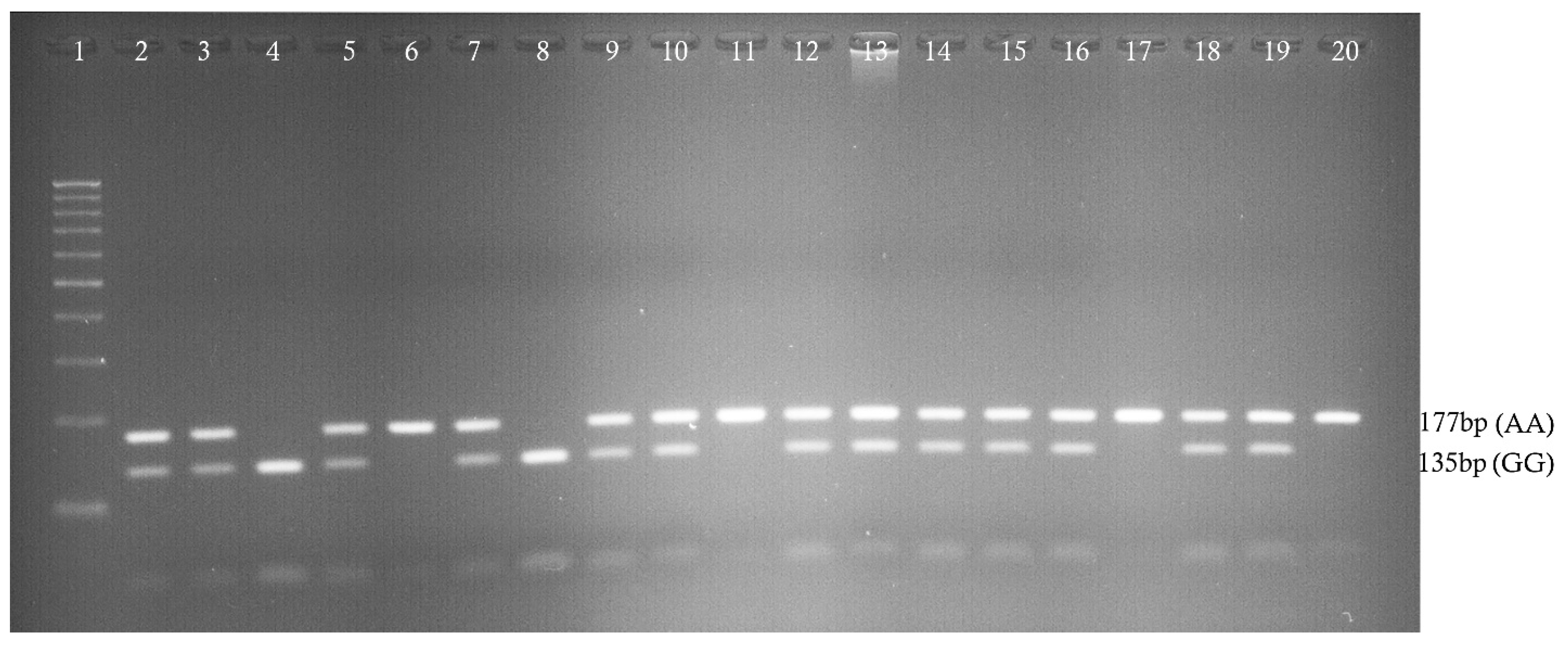
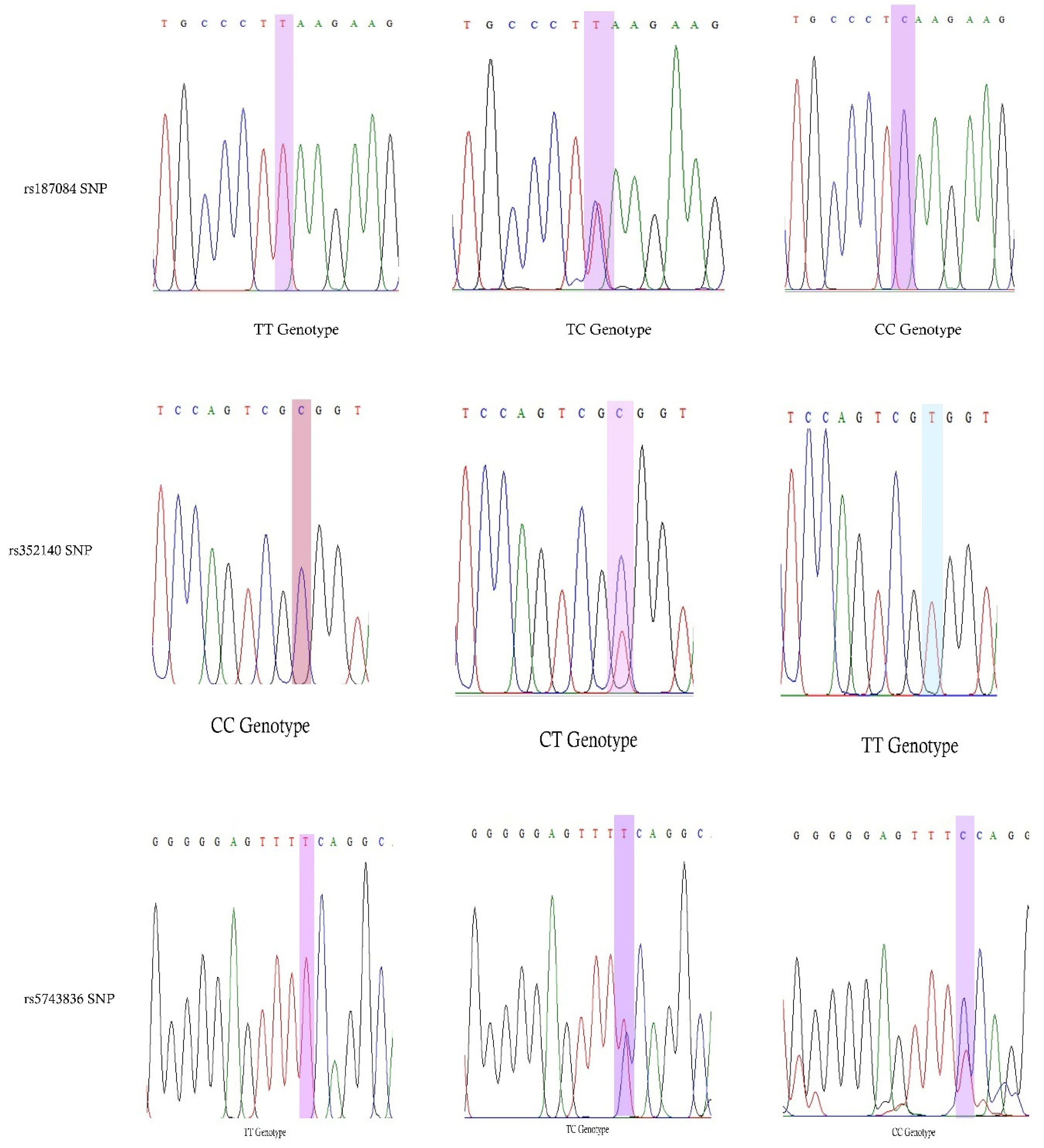
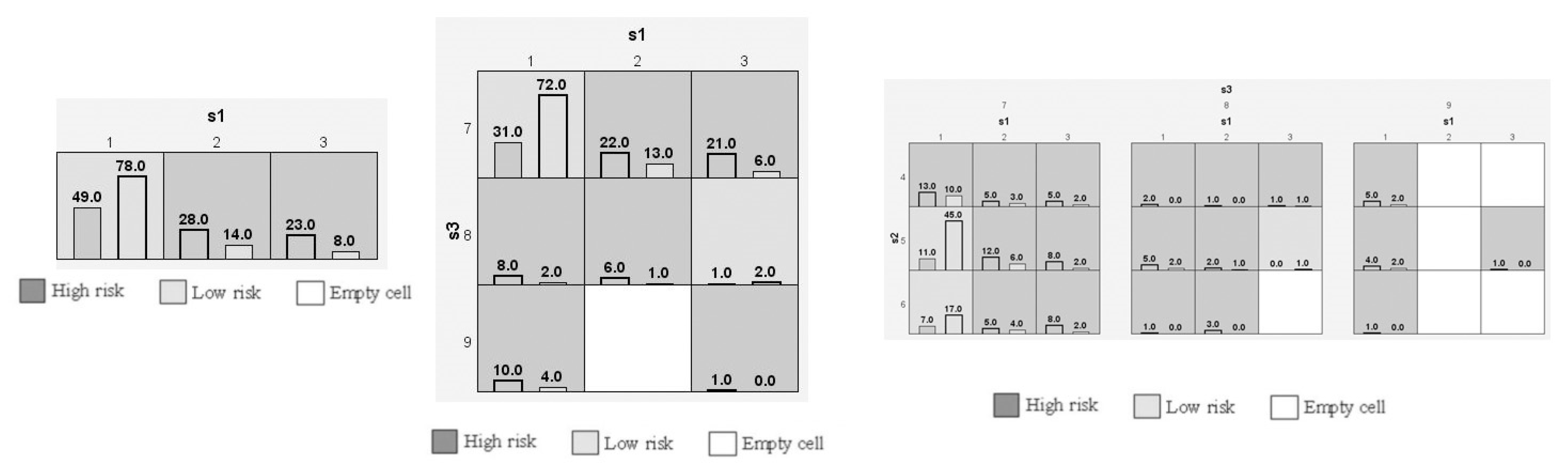
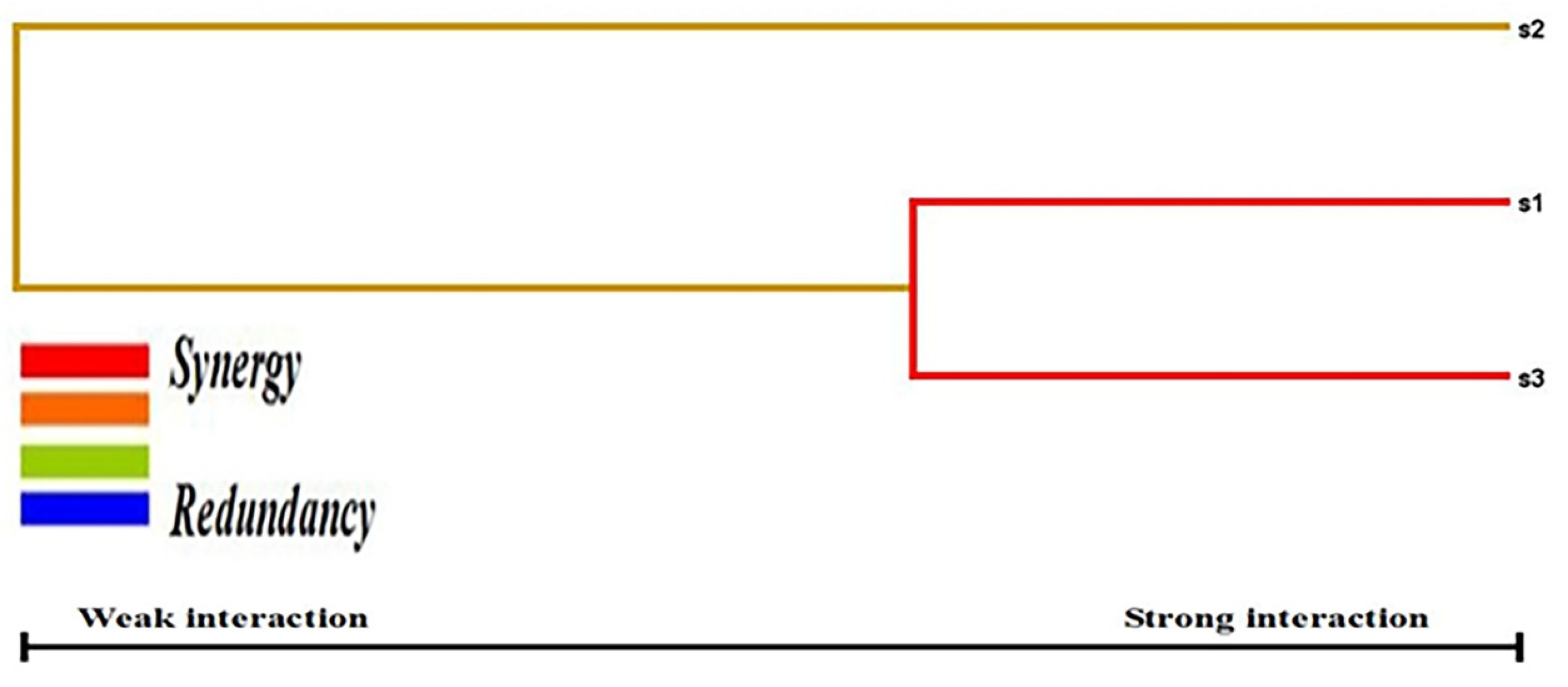
We conducted a case-control study among Saudi Arabian T2DM patients and determined the prevalence of three SNPs (rs187084, rs352140, and rs5743836) in the study population. We confirmed a strong association between T2DM and rs187084 and rs5743836 SNPs. In addition, a negligible association was observed between T2DM and the rs352140 SNP. Additionally, DBP levels (p = 0.02), TC (p = 0.03), and LDLC (p = 0.04) were significantly associated with rs187084. DBP (p = 0.0003), Hb1Ac (p = 0.03), and HDLC (p = 0.04) were significantly associated with rs352140, and waist (p = 0.02) was significantly associated with rs5743836. The C-G-T haplotype increased the risk for developing T2DM (p < 0.003) by approximately four-fold, and the combination of C-G-C/C-A-T haplotypes showed protective role towards T2DM. LD analysis showed a negative association, and both MDR/GMDR analyses showed strong association of gene–gene interaction, dendrogram, and graphical representation (p < 0.05) with T2DM cases. Overall, we confirmed that T2DM development is associated with SNPs present in the TLR9 gene in the Saudi population.
TLRs are widely expressed in the human body cells, and 12 SNPs are present in the
TLR9 gene. rs187084 is present at the promoter region [
27]. In this study, 23% of CC genotypes, 25% of AA genotypes, and 11% of CC genotypes were present in three different SNPs in the form of homozygous variants in T2DM patients, whereas, in controls, 8%, 23%, 4% were present. This indicates homozygous variants are high in the Saudi Arabian population. However, rs352140 prevalence was similar between both groups. Only the co-dominant model showed a statistical association (
p = 0.02), with 1.9 times increased risk.
A previous study in an Egyptian population has confirmed a significant role of rs5743836 in T2DM (
p = 0.01) and diabetes foot (DF) (
p = 0.02). In T2DM patients, 73.3% and 6.7% of the heterozygous genotypes were observed. In DF patients, TT and TC genotypes were present (50% each), and the CC genotype was not observed. The genotype frequencies observed in our study were different and could be due to a number of factors. First, the Egyptian study contained a small sample size. In addition, differences could be due to the ethnic diversity between the populations [
12]. Another study was carried out in Chinese patients diagnosed with T2DM, CAD, and a combination of T2DM + CAD. The study determined negative associations in T2DM, CAD, and a combination of T2DM + CAD among −1486T>C and −1237T>C SNPs. However, this study was documented over a decade ago [
17]. The rs187084 SNP was studied in other human diseases with different ethnicities, and no significant associations were determined between the SNP and disease development [
28,
29]. However, a positive association was confirmed at the +1174 locus [
29]. In the Iranian population, the rs352140 SNP was not associated with T2DM or DN [
13]; however, this could be a result of small sample size. The rs352140 SNP was studied in Chinese children with T1DM, and a strong association was observed between the SNP and T1DM development [
11].
Different meta-analysis studies were reported with rs187084, rs352140, and rs5743836 SNPs in different human diseases, but there are no meta-analysis studies documented between rs187084, rs352140, and rs5743836 SNPs in the TLR9 gene and T2DM.
Family history is considered as one of the non-modifiable risk factors along with age, gender, and height. Family of T2DM plays a significant role in the Saudi Arabian population. The first degree relative has 30–70% risk of developing T2DM disease. Family history of T2DM has been shown to increase disease risk by two-fold in the future generations [
30]. Tuomilehto et al. [
31] confirmed effectiveness of an intervention program, following that the intervention program lowers T2DM from 58% to 43% in a span of three years. In Saudi Arabia, the combination of family history of T2DM and parental consanguinity will lead to increase the risk of both impaired fasting glucose and diabetes in the offspring [
32]. Nonetheless, frequently checking glucose levels, altering eating habits, and increasing physical exercise can alter the quality of insulin produced in order to fulfill the body’s blood glucose targets and maintain appropriate blood glucose levels.
HTN (both essential and idiopathic) is defined as a chronic condition marked by expanded SBP and DBP levels. In our study, we have found an association between DBP and the
TLR9 gene. The DBP was found to be higher in the T2DM patients (
p = 0.002). These results were confirmed with multiple linear regression; a positive association was observed between DBP and T2DM (
p = 0.02). The association between the rs352140 SNP and DBP was statistically significant (
p = 0.0003). The elevated levels of DBP were found in the rs352140 AA genotype with 81.77 ± 5.97, which indicates there is a relation between HTN and the
TLR9 gene. The relationship between the
TLR9 gene and HTN has been previously documented [
33,
34]. Overall, based on the current study results, we conclude that the
TLR9 gene plays a role in T2DM and HTN development in the Saudi Arabian population.
The major limitation of this study was small population size (100 T2DM patients and 100 healthy controls). The other limitations of this study could be not matching with gender and anthropometric characteristics in T2DM cases and control subjects. However, the number of participants included in this study was based on a sample size calculation. Only three SNPs were screened in this study. Another limitation of this study was not selecting T2DM patients with any of the infectious diseases. The final limitation of this study was missing of functional studies in TLR9. The strength of this study was enrolling T2DM patients with a family history of diabetes. By selecting the patients with complete family history, we were able to determine the molecular role of the TLR9 gene. Additional statistics was another strength.