Environmental DNA Metabarcoding: A Novel Contrivance for Documenting Terrestrial Biodiversity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Research Methodology
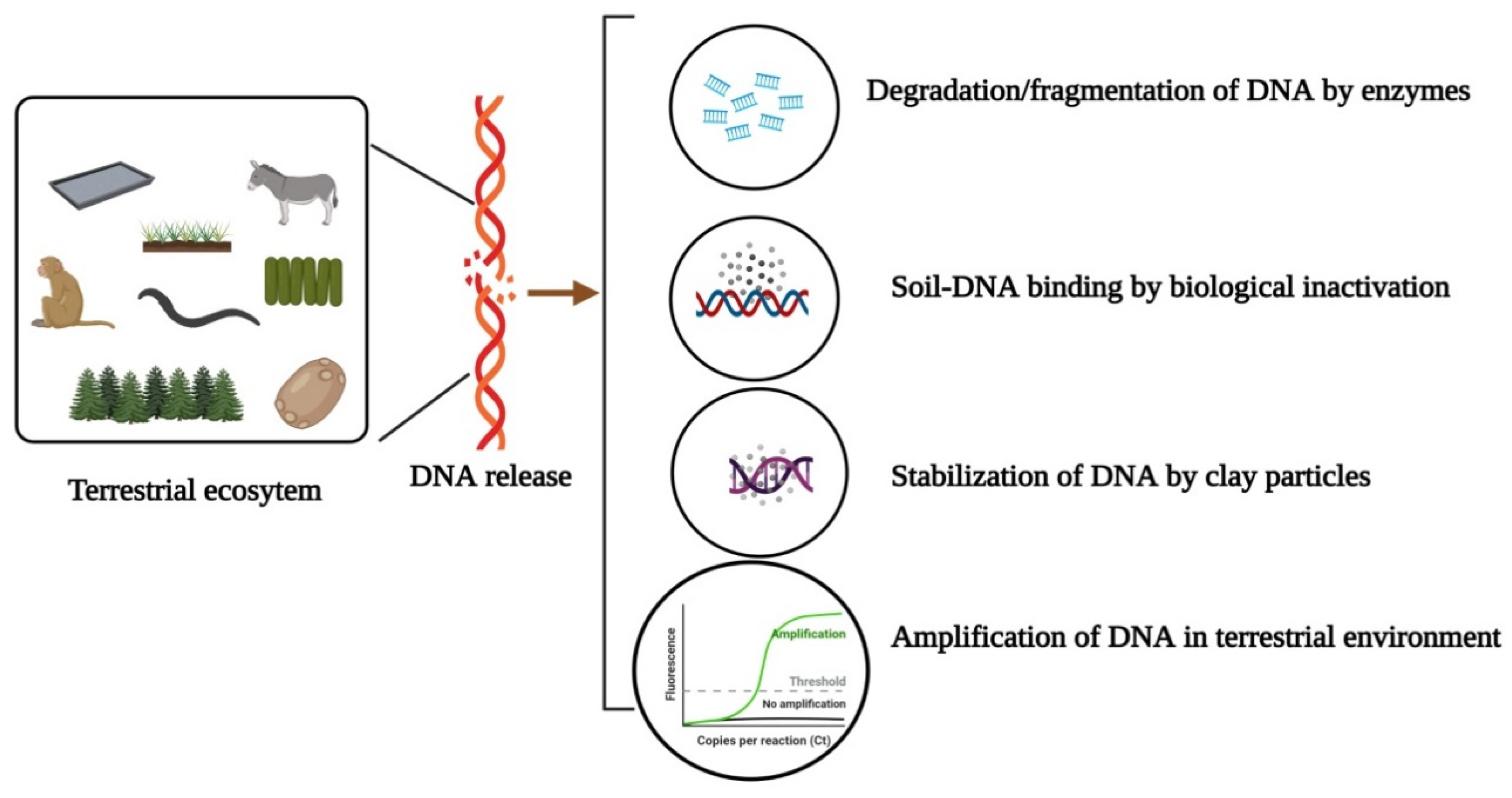
3. Persistence, Detectability and Mobility of Terrestrial eDNA
4. Exertion of Environmental DNA in Terrestrial Ecosystems
4.1. Plant Community Characterization
4.2. Earthworm Community Characterization
4.3. Bacterial Community Characterization
4.4. Multi-Taxa Diversity Surveys
4.5. Endangered Species
4.6. Bulk Specimens
| Reference | Ecosystem | Species/Sample | Utility | Major Findings |
|---|---|---|---|---|
| Ryan et al. (2022) [106] | Terrestrial | Vertebrate species | Suitability of fallen log hollowed sediment as a source of vertebrate eDNA | Environmental DNA (eDNA) monitoring is affected by substrate selection, sampling frequency and size of the animal. |
| Lyman et al. (2022) [99] | Herbaceous vegetation | Endangered rodent (Zapus hudsonius luteus) | Validation of qPCR assay | The feasibility of detecting eDNA from the vegetation is imperative to the life history of New Mexico meadow jumping mice. Environmental DNA can corroborate site occupancy or aid in population density inferences. |
| Campbell et al. (2022) [91] | Terrestrial | Cryptic insects (Cochlicella acuta, Sarcophaga villeneuveana) | Cryptic biological control agent | From a small vegetation sample, the eDNA technique has the potential to detect to infer the presence of cryptic species. |
| Peterson et al. (2022) [105] | Terrestrial | Invasive terrestrial insect (Lycorma delicatula) | Species monitoring | For the detection of lanternfly (Lycorma delicatula) insects, deploying roller surface eDNA methods can provide improved guidance for surveillance and monitoring programs. |
| Lunghi et al. (2022) [57] | Terrestrial | Springtails and insects | Metabarcoding | Environmental DNA from cave soils/sediments acts as a conveyer belt of biodiversity information. |
| Kirtane et al. (2022) [96] | Forest | Adelges tsugae, Leucotaraxis piniperda, L argenticollis, L nigrinus | Forest pest and biological control predators | Environmental DNA as a sensitive biodiversity monitoring tool has greater efficacy for the early detection of Adelges tsugae and its biological control predators. |
| Guerrieri et al. (2021) [31] | Soil | bacteria, fungi and eukaryotes | Effects of soil preservation for biodiversity monitoring | A preserved soil sample can be utilized in metabarcoding research focusing on inaccessible or difficult-to-reach places. |
| Allen et al. (2021) [35] | Agricultural ecosystem | Invasive pest insect (Lycorma delicatula) | Terrestrial eDNA survey | When spotted lanternflies were present in a plot, the likelihood of finding them with eDNA was 84%, more than twice as likely as using visual surveys (36 percent). |
| Valentin et al. (2021) [53] | Terrestrial | Arthropods | Above-ground terrestrial eDNA | An increase in filter pore size had no discernible impact on the amount of intracellular eDNA that was captured, indicating that a variety of feasible pore sizes are available for targeting intracellular eDNA. |
| Ladin et al. (2021) [50] | Forest ecosystem | Mycoplasma sp., Spirosoma sp., Roseomonas sp., Lactococcus sp. Spiroplasma sp., Methylobacterium sp., Massilia sp., Pantoea sp., Sphingomonas sp. | Microbial biodiversity | This innovative technique has the potential to be used to quantify both prokaryotic and eukaryotic lifeforms by evaluating the variety of microbes in forest ecosystems |
| Mena et al. (2021) [113] | Tropical forests | Mammal diversity | Metabarcoding | This work is one of the first to demonstrate the enormous potential of eDNA metabarcoding for evaluating Amazonian mammal ecosystems.. |
| Leempoel et al. (2020) [28] | Terrestrial | Mammal diversity | Diversity assessment | Ecosystem surveys could benefit from eDNA-based monitoring; however, enriching mitochondrial reference datasets is necessary first. |
| Rota et al. (2020) [2] | Soil | Alpine biodiversity | Metabarcoding | This research gave a description of the soil fauna of alpine habitats, produced a description of the community composition for each habitat, and revealed the relationship between the study area’s topographic features, flora, and soil characteristics |
| Thomsen and Sigsgaard (2019) [22] | Wildflowers | Arthropod communities | eDNA metagenomics | Genomic markers like 16S rRNA and COI can be utilized to obtain data related to arthropods from different ecological groups. |
| Seeber et al. (2019) [25] | Terrestrial | African mammal | Hybridization capture of eDNA | Hybridization capture enrichment of environmental DNA can be an effective technique for monitoring terrestrial mammal species. |
| Ficetola et al. (2019) [26] | Terrestrial | Amphibians and reptiles | Species distribution | Environmental DNA can be a valuable method to investigate terrestrial organisms, assess the relative abundance of species and distinguish amphibians and reptiles |
| Valentin et al. (2018) [24] | Crop surfaces | Halyomorpha halys | Invasive exotic insect infestations | The knowledge of environmental DNA for the surveillance of exotic species in terrestrial ecosystems can provide high sensitivity and detection. |
| Chang et al. (2018) [43] | Terrestrial | Migratory species | Pollinators and migratory species | The concept of environmental DNA can aid in comprehending the pollen from the migratory pollinators and the distances and geographic assortment of migratory species. |
| Banchi et el. (2018) [60] | Air borne and terrestrial | Fungal diversity | Species monitoring | The study revealed that diversity analysis using environmental DNA showed ten times more inclusive taxa detection than microscopic identification. |
| Khaliq et al. (2018) [63] | soil | Phytophthora diversity | Biodiversity analysis | The study revealed that environmental DNA is suitable for documenting the Phytophthora at the species level. |
| Liu et al. (2018) [70] | Terrestrial | Archeal diversity | Biodiversity monitoring | Environmental DNA can be utilized to estimate community composition based on the mcrA gene studies. |
| Galan et al. (2018) [84] | Terrestrial | bats | Diet variability | The study used the metabarcoding approach to detect the variability of the bats’ diet to cognize the bats’ biology and conservation strategies. |
| Gellie et al. (2017) [85] | Soil | Bacterial diversity | Species composition changes | Environmental DNA based on amplicon sequencing offers consistent ecological monitoring and cost-effective detection |
| Parducci et al. (2017) [64] | Soil | Palaeofloras reconstruction | Sediment profiling | Environmental DNA can be employed as a contrivance for identifying indigenous vegetation. |
| Bitok et al. (2017) [65] | Soil | Biosynthetic gene clusters (BGC) | BGC identification | Soil environmental DNA screening provides identification of gene clones embedding BGCs |
| Wakelin et al. (2016) [68] | Soil | Soil environmental genomics | High-density functional gene microarray analysis | The study revealed that environmental DNA defines the alterations in soil functional ecology and nitrogen cycling metalloenzymes. |
| Katz et al. (2016) [95] | Soil | Microbial diversity | Secondary metabolite screening | Environmental DNA, in combination with metagenomics, provides an alternative for natural product discovery. |
| Drummond et al. (2015) [101] | Soil | Alpha, beta and gamma diversity | Biodiversity assessment | Diversity estimations are affected by the number of sequence reads. Soil beta diversity exhibited the strongest response regarding elevation variation of environmental DNA markers. |
| Hunter et al. (2015) [97] | Terrestrial | Python diversity | Biomonitoring | The study revealed that species-specific environmental DNA assays could be used to detect python diversity. |
| Gibson et al. (2014) [110] | Bulk sample | Tropical arthropods | Assessment of macro and microbiomes in a bulk sample | Next-generation sequencing (NGS) can detect species in a bulk sample of terrestrial arthropods |
| Ramirez et al. (2014) [93] | Soil cores | Bacterial and archeal diversity | Biodiversity and biographic patterns | The study revealed that environmental DNA could be utilized to detect unscripted below-ground diversity, much of which has never been explored and explained in public databases. |
| Calvignac-Spencer et al. (2013) [54] | Carrion-fly derived DNA | Mammalian diversity | Biodiversity assessment | The investigation revealed that Carrion flies represent an unexploited resource of mammal DNA. |
| Bienert et al. (2012) [79] | Soil | Earthworms | Species identification | The study illustrates the potential of environmental DNA as a contrivance to assess the soil-dwelling diversity of animal taxa. |
| Anderson et al. (2012) [32] | Soil | Camels | Vertebrate diversity | The deeper portions of soil strata preserve DNA that can be an excellent indicator of the above-ground composition of the vertebrate community. |
5. Challenges and Drawbacks
6. Knowledge Gaps and Future Perspectives
- Protocols should be standardized so they can be implemented globally in diverse locations of a particular habitat type.
- Development of portable instruments (qPCR, Biomeme, DNA sequencers) for rapid filed analysis to avoid the errors that may occur during sample preservation and handling.
- The data generated during environmental DNA should be properly mined to avoid reiteration. Furthermore, the mined data can be analyzed by specialists to countercheck the outcomes.
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bairoliya, S.; Koh Zhi Xiang, J.; Cao, B. Extracellular DNA in environmental samples: Occurrence, extraction, quantification, and impact on microbial biodiversity assessment. Appl. Environ. Microbiol. 2022, 88, e01821–e01845. [Google Scholar] [CrossRef] [PubMed]
- Rota, N.; Canedoli, C.; Ferrè, C.; Ficetola, G.F.; Guerrieri, A.; Padoa-Schioppa, E. Evaluation of soil biodiversity in alpine habitats through eDNA metabarcoding and relationships with environmental features. Forests 2020, 11, 738. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Ip, Y.C.A.; Tay, Y.C.; Gan, S.X.; Ang, H.P.; Tun, K.; Chou, L.M.; Huang, D.; Meier, R. From marine park to future genomic observatory? Enhancing marine biodiversity assessments using a biocode approach. Biodivers. Data J. 2019, 7, e46833. [Google Scholar] [CrossRef] [PubMed]
- Cowart, D.A.; Murphy, K.R.; Cheng, C.-H.C. Environmental DNA from Marine Waters and Substrates: Protocols for Sampling and eDNA Extraction. In Marine Genomics; Springer: Berlin/Heidelberg, Germany, 2022; pp. 225–251. [Google Scholar]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: New York, NY, USA, 2018. [Google Scholar]
- Hinz, S.; Coston-Guarini, J.; Marnane, M.; Guarini, J.-M. Evaluating eDNA for Use within Marine Environmental Impact Assessments. J. Mar. Sci. Eng. 2022, 10, 375. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Nelson, N.M.; Jerde, C.L.; Luikart, G. Are environmental DNA methods ready for aquatic invasive species management? Trends Ecol. Evol. 2020, 35, 668–678. [Google Scholar] [CrossRef]
- Darling, J.A. How to learn to stop worrying and love environmental DNA monitoring. Aquat. Ecosyst. Health Manag. 2019, 22, 440–451. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Barkay, T. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Britschgi, T.B.; Moyer, C.L.; Field, K.G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 1990, 345, 60–63. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef] [Green Version]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.P.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Pompanon, F.; Deagle, B.E.; Symondson, W.O.; Brown, D.S.; Jarman, S.N.; Taberlet, P. Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef]
- Sønstebø, J.; Gielly, L.; Brysting, A.; Elven, R.; Edwards, M.; Haile, J.; Willerslev, E.; Coissac, E.; Rioux, D.; Sannier, J. Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Resour. 2010, 10, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Tringe, S.G.; Von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Yoccoz, N.G.; Bråthen, K.A.; Gielly, L.; Haile, J.; Edwards, M.E.; Goslar, T.; von Stedingk, H.; Brysting, A.; Coissac, E.; Pompanon, F. DNA from soil mirrors plant taxonomic and growth form diversity. Mol. Ecol. 2012, 21, 3647–3655. [Google Scholar] [CrossRef]
- Baldwin, D.S.; Colloff, M.J.; Rees, G.N.; Chariton, A.A.; Watson, G.O.; Court, L.N.; Hartley, D.M.; Morgan, M.J.; King, A.J.; Wilson, J.S. Impacts of inundation and drought on eukaryote biodiversity in semi-arid floodplain soils. Mol. Ecol. 2013, 22, 1746–1758. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for methods utilizing environmental DNA for detection of fish species. Genes 2020, 11, 296. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Sigsgaard, E.E. Environmental DNA metabarcoding of wild flowers reveals diverse communities of terrestrial arthropods. Ecol. Evol. 2019, 9, 1665–1679. [Google Scholar] [CrossRef] [Green Version]
- Dopheide, A.; Xie, D.; Buckley, T.R.; Drummond, A.J.; Newcomb, R.D. Impacts of DNA extraction and PCR on DNA metabarcoding estimates of soil biodiversity. Methods Ecol. Evol. 2019, 10, 120–133. [Google Scholar] [CrossRef]
- Valentin, R.E.; Fonseca, D.M.; Nielsen, A.L.; Leskey, T.C.; Lockwood, J.L. Early detection of invasive exotic insect infestations using eDNA from crop surfaces. Front. Ecol. Environ. 2018, 16, 265–270. [Google Scholar] [CrossRef]
- Seeber, P.A.; McEwen, G.K.; Löber, U.; Förster, D.W.; East, M.L.; Melzheimer, J.; Greenwood, A.D. Terrestrial mammal surveillance using hybridization capture of environmental DNA from African waterholes. Mol. Ecol. Resour. 2019, 19, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Manenti, R.; Taberlet, P. Environmental DNA and metabarcoding for the study of amphibians and reptiles: Species distribution, the microbiome, and much more. Amphibia-Reptilia 2019, 40, 129–148. [Google Scholar] [CrossRef]
- Hartvig, I.; Kosawang, C.; Kjær, E.D.; Nielsen, L.R. Detecting rare terrestrial orchids and associated plant communities from soil samples with eDNA methods. Biodivers. Conserv. 2021, 30, 3879–3901. [Google Scholar] [CrossRef]
- Leempoel, K.; Hebert, T.; Hadly, E.A. A comparison of eDNA to camera trapping for assessment of terrestrial mammal diversity. Proc. R. Soc. B 2020, 287, 20192353. [Google Scholar] [CrossRef]
- Hassan, S.; Khurshid, Z.; Bali, B.S.; Ganai, B.A.; Sayyed, R.; Poczai, P.; Zaman, M. A Critical Assessment of the Congruency between Environmental DNA and Palaeoecology for the Biodiversity Monitoring and Palaeoenvironmental Reconstruction. Int. J. Environ. Res. Public Health 2022, 19, 9445. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Guerrieri, A.; Bonin, A.; Münkemüller, T.; Gielly, L.; Thuiller, W.; Francesco Ficetola, G. Effects of soil preservation for biodiversity monitoring using environmental DNA. Mol. Ecol. 2021, 30, 3313–3325. [Google Scholar] [CrossRef]
- Andersen, K.; Bird, K.L.; Rasmussen, M.; Haile, J.; Breuning-Madsen, H.; Kjaer, K.H.; Orlando, L.; Gilbert, M.T.P.; Willerslev, E. Meta-barcoding of ‘dirt’DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 2012, 21, 1966–1979. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Colwell, R.K. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 2001, 4, 379–391. [Google Scholar] [CrossRef]
- Dickie, I.A.; Boyer, S.; Buckley, H.L.; Duncan, R.P.; Gardner, P.P.; Hogg, I.D.; Holdaway, R.J.; Lear, G.; Makiola, A.; Morales, S.E. Towards robust and repeatable sampling methods in eDNA-based studies. Mol. Ecol. Resour. 2018, 18, 940–952. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.C.; Nielsen, A.L.; Peterson, D.L.; Lockwood, J.L. Terrestrial eDNA survey outperforms conventional approach for detecting an invasive pest insect within an agricultural ecosystem. Environ. DNA 2021, 3, 1102–1112. [Google Scholar] [CrossRef]
- Ni, B.; Zhao, W.; Zuo, X.; You, J.; Li, Y.; Li, J.; Du, Y.; Chen, X. Deyeuxia angustifolia Kom. encroachment changes soil physicochemical properties and microbial community in the alpine tundra under climate change. Sci. Total Environ. 2022, 813, 152615. [Google Scholar] [CrossRef]
- Zinger, L.; Shahnavaz, B.; Baptist, F.; Geremia, R.A.; Choler, P. Microbial diversity in alpine tundra soils correlates with snow cover dynamics. ISME J. 2009, 3, 850–859. [Google Scholar] [CrossRef]
- Kleber, M.; Bourg, I.C.; Coward, E.K.; Hansel, C.M.; Myneni, S.C.; Nunan, N. Dynamic interactions at the mineral–organic matter interface. Nat. Rev. Earth Environ. 2021, 2, 402–421. [Google Scholar] [CrossRef]
- Van Der Heyde, M.; Bunce, M.; Wardell-Johnson, G.; Fernandes, K.; White, N.E.; Nevill, P. Testing multiple substrates for terrestrial biodiversity monitoring using environmental DNA metabarcoding. Mol. Ecol. Resour. 2020, 20, 732–745. [Google Scholar] [CrossRef]
- Franklin, T.W.; McKelvey, K.S.; Golding, J.D.; Mason, D.H.; Dysthe, J.C.; Pilgrim, K.L.; Squires, J.R.; Aubry, K.B.; Long, R.A.; Greaves, S.E. Using environmental DNA methods to improve winter surveys for rare carnivores: DNA from snow and improved noninvasive techniques. Biol. Conserv. 2019, 229, 50–58. [Google Scholar] [CrossRef]
- McInnes, J.C.; Alderman, R.; Deagle, B.E.; Lea, M.A.; Raymond, B.; Jarman, S.N. Optimised scat collection protocols for dietary DNA metabarcoding in vertebrates. Methods Ecol. Evol. 2017, 8, 192–202. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Chang, H.; Guo, J.; Fu, X.; Liu, Y.; Wyckhuys, K.A.; Hou, Y.; Wu, K. Molecular-assisted pollen grain analysis reveals spatiotemporal origin of long-distance migrants of a noctuid moth. Int. J. Mol. Sci. 2018, 19, 567. [Google Scholar] [CrossRef] [PubMed]
- Polling, M.; Sin, M.; de Weger, L.A.; Speksnijder, A.G.; Koenders, M.J.; de Boer, H.; Gravendeel, B. DNA metabarcoding using nrITS2 provides highly qualitative and quantitative results for airborne pollen monitoring. Sci. Total Environ. 2022, 806, 150468. [Google Scholar] [CrossRef] [PubMed]
- Zinger, L.; Taberlet, P.; Schimann, H.; Bonin, A.; Boyer, F.; De Barba, M.; Gaucher, P.; Gielly, L.; Giguet-Covex, C.; Iribar, A. Body size determines soil community assembly in a tropical forest. Mol. Ecol. 2019, 28, 528–543. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, S.P.; Yang, X.; Zhou, J.; Shu, W.; Jiang, L. Mechanisms of soil bacterial and fungal community assembly differ among and within islands. Environ. Microbiol. 2020, 22, 1559–1571. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, X.; Miller, J.A.; de Blécourt, M.; Ji, Y.; Yang, C.; Harrison, R.D.; Douglas, W.Y. Using metabarcoding to ask if easily collected soil and leaf-litter samples can be used as a general biodiversity indicator. Ecol. Indic. 2014, 46, 379–389. [Google Scholar] [CrossRef]
- Koziol, L.; Schultz, P.A.; House, G.L.; Bauer, J.T.; Middleton, E.L.; Bever, J.D. The plant microbiome and native plant restoration: The example of native mycorrhizal fungi. BioScience 2018, 68, 996–1006. [Google Scholar] [CrossRef]
- Adams, C.I.; Knapp, M.; Gemmell, N.J.; Jeunen, G.-J.; Bunce, M.; Lamare, M.D.; Taylor, H.R. Beyond biodiversity: Can environmental DNA (eDNA) cut it as a population genetics tool? Genes 2019, 10, 192. [Google Scholar] [CrossRef]
- Ladin, Z.S.; Ferrell, B.; Dums, J.T.; Moore, R.M.; Levia, D.F.; Shriver, W.G.; D’Amico, V.; Trammell, T.L.; Setubal, J.C.; Wommack, K.E. Assessing the efficacy of eDNA metabarcoding for measuring microbial biodiversity within forest ecosystems. Sci. Rep. 2021, 11, 1629. [Google Scholar] [CrossRef]
- Legendre, P.; Dale, M.R.; Fortin, M.J.; Gurevitch, J.; Hohn, M.; Myers, D. The consequences of spatial structure for the design and analysis of ecological field surveys. Ecography 2002, 25, 601–615. [Google Scholar] [CrossRef]
- Fernandes, K.; Van Der Heyde, M.; Coghlan, M.; Wardell-Johnson, G.; Bunce, M.; Harris, R.; Nevill, P. Invertebrate DNA metabarcoding reveals changes in communities across mine site restoration chronosequences. Restor. Ecol. 2019, 27, 1177–1186. [Google Scholar] [CrossRef]
- Valentin, R.E.; Kyle, K.E.; Allen, M.C.; Welbourne, D.J.; Lockwood, J.L. The state, transport, and fate of aboveground terrestrial arthropod eDNA. Environ. DNA 2021, 3, 1081–1092. [Google Scholar] [CrossRef]
- Calvignac-Spencer, S.; Merkel, K.; Kutzner, N.; Kühl, H.; Boesch, C.; Kappeler, P.M.; Metzger, S.; Schubert, G.; Leendertz, F.H. Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Mol. Ecol. 2013, 22, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Colston, T.J.; Jackson, C.R. Microbiome evolution along divergent branches of the vertebrate tree of life: What is known and unknown. Mol. Ecol. 2016, 25, 3776–3800. [Google Scholar] [CrossRef]
- Parkhurst, T.; Prober, S.M.; Hobbs, R.J.; Standish, R.J. Global meta-analysis reveals incomplete recovery of soil conditions and invertebrate assemblages after ecological restoration in agricultural landscapes. J. Appl. Ecol. 2022, 59, 358–372. [Google Scholar] [CrossRef]
- Lunghi, E.; Valle, B.; Guerrieri, A.; Bonin, A.; Cianferoni, F.; Manenti, R.; Ficetola, G.F. Environmental DNA of insects and springtails from caves reveals complex processes of eDNA transfer in soils. Sci. Total Environ. 2022, 826, 154022. [Google Scholar] [CrossRef]
- Havermans, C.; Dischereit, A.; Pantiukhin, D.; Friedrich, M.; Murray, A. Environmental DNA in an ocean of change: Status, challenges and prospects. Arq. Cienc. Mar. 2022, 55, 298–337. Available online: https://epic.awi.de/id/eprint/56372/ (accessed on 1 March 2022).
- Ruppert, K.M. Development and Assessment of an Environmental DNA (eDNA) Assay for the Rio Grande Siren and Review of eDNA Metabarcoding Applications; The University of Texas Rio Grande Valley: Edinburg, TX, USA, 2020; p. 27829413. [Google Scholar]
- Banchi, E.; Ametrano, C.G.; Stanković, D.; Verardo, P.; Moretti, O.; Gabrielli, F.; Lazzarin, S.; Borney, M.F.; Tassan, F.; Tretiach, M. DNA metabarcoding uncovers fungal diversity of mixed airborne samples in Italy. PLoS ONE 2018, 13, e0194489. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol. Ecol. Resour. 2014, 14, 109–116. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Khaliq, I.; Hardy, G.E.S.J.; White, D.; Burgess, T.I. eDNA from roots: A robust tool for determining Phytophthora communities in natural ecosystems. FEMS Microbiol. Ecol. 2018, 94, fiy048. [Google Scholar] [CrossRef]
- Parducci, L.; Bennett, K.D.; Ficetola, G.F.; Alsos, I.G.; Suyama, Y.; Wood, J.R.; Pedersen, M.W. Ancient plant DNA in lake sediments. New Phytol. 2017, 214, 924–942. [Google Scholar] [CrossRef] [Green Version]
- Bitok, J.K.; Lemetre, C.; Ternei, M.A.; Brady, S.F. Identification of biosynthetic gene clusters from metagenomic libraries using PPTase complementation in a Streptomyces host. FEMS Microbiol. Lett. 2017, 364, fnx155. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Wakelin, S.A.; Cave, V.; Dignam, B.; D’Ath, C.; Tourna, M.; Condron, L.M.; Zhou, J.; Van Nostrand, J.; O’Callaghan, M. Analysis of soil eDNA functional genes: Potential to increase profitability and sustainability of pastoral agriculture. New Zealand J. Agric. Res. 2016, 59, 333–350. [Google Scholar] [CrossRef]
- Yoccoz, N.G. The future of environmental DNA in ecology. Mol. Ecol. 2012, 21, 2031–2038. [Google Scholar] [CrossRef]
- Liu, D.; Nishida, M.; Takahashi, T.; Asakawa, S. Transcription of mcrA gene decreases upon prolonged non-flooding period in a methanogenic archaeal community of a paddy-upland rotational field soil. Microb. Ecol. 2018, 75, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Bass, D.; Stentiford, G.D.; Littlewood, D.; Hartikainen, H. Diverse applications of environmental DNA methods in parasitology. Trends Parasitol. 2015, 31, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Sansupa, C.; Fareed Mohamed Wahdan, S.; Disayathanoowat, T.; Purahong, W. Identifying Hidden Viable Bacterial Taxa in Tropical Forest Soils Using Amplicon Sequencing of Enrichment Cultures. Biology 2021, 10, 569. [Google Scholar] [CrossRef]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Douglas, W.Y.; De Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef]
- de Groot, G.A.; Geisen, S.; Wubs, E.J.; Meulenbroek, L.; Laros, I.; Snoek, L.B.; Lammertsma, D.R.; Hansen, L.H.; Slim, P.A. The aerobiome uncovered: Multi-marker metabarcoding reveals potential drivers of turn-over in the full microbial community in the air. Environ. Int. 2021, 154, 106551. [Google Scholar] [CrossRef]
- Velásquez-Zapata, V.; Palacio-Rúa, K.; Cano, L.E.; Gaviria-Rivera, A. Assessment of genotyping markers in the molecular characterization of a population of clinical isolates of Fusarium in Colombia. Biomédica 2022, 42, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Kooch, Y.; Jalilvand, H. Earthworms as ecosystem engineers and the most important detritivors in forest soils. Pak. J. Biol. Sci. 2008, 11, 819–825. [Google Scholar] [CrossRef]
- Lemtiri, A.; Colinet, G.; Alabi, T.; Cluzeau, D.; Zirbes, L.; Haubruge, É.; Francis, F. Impacts of earthworms on soil components and dynamics. A review. Biotechnol. Agron. Société Et Environ. 2014, 18. Available online: https://orbi.uliege.be/handle/2268/163467 (accessed on 1 March 2022).
- Huera-Lucero, T.; Labrador-Moreno, J.; Blanco-Salas, J.; Ruiz-Téllez, T. A framework to incorporate biological soil quality indicators into assessing the sustainability of territories in the Ecuadorian Amazon. Sustainability 2020, 12, 3007. [Google Scholar] [CrossRef]
- Bienert, F.; De Danieli, S.; Miquel, C.; Coissac, E.; Poillot, C.; BRUN, J.J.; Taberlet, P. Tracking earthworm communities from soil DNA. Mol. Ecol. 2012, 21, 2017–2030. [Google Scholar] [CrossRef]
- Bartlett, M.D.; Briones, M.J.; Neilson, R.; Schmidt, O.; Spurgeon, D.; Creamer, R.E. A critical review of current methods in earthworm ecology: From individuals to populations. Eur. J. Soil Biol. 2010, 46, 67–73. [Google Scholar] [CrossRef]
- Pansu, J.; Giguet-Covex, C.; Ficetola, G.F.; Gielly, L.; Boyer, F.; Zinger, L.; Arnaud, F.; Poulenard, J.; Taberlet, P.; Choler, P. Reconstructing long-term human impacts on plant communities: An ecological approach based on lake sediment DNA. Mol. Ecol. 2015, 24, 1485–1498. [Google Scholar] [CrossRef]
- Llanos, J. Assessing Earthworm Diversity and Population Dynamics in Agroecosystems; University of Sheffield: Sheffield, UK, 2021. [Google Scholar]
- Jackson, M.; Myrholm, C.; Shaw, C.; Ramsfield, T. Using nested PCR to improve detection of earthworm eDNA in Canada. Soil Biol. Biochem. 2017, 113, 215–218. [Google Scholar] [CrossRef]
- Galan, M.; Pons, J.B.; Tournayre, O.; Pierre, E.; Leuchtmann, M.; Pontier, D.; Charbonnel, N. Metabarcoding for the parallel identification of several hundred predators and their prey: Application to bat species diet analysis. Mol. Ecol. Resour. 2018, 18, 474–489. [Google Scholar] [CrossRef]
- Gellie, N.J.; Mills, J.G.; Breed, M.F.; Lowe, A.J. Revegetation rewilds the soil bacterial microbiome of an old field. Mol. Ecol. 2017, 26, 2895–2904. [Google Scholar] [CrossRef]
- Minamoto, T. Environmental DNA analysis for macro-organisms: Species distribution and more. DNA Res. 2022, 29, dsac018. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.L.; Gibbons, S.M.; Owens, S.M.; Hampton-Marcell, J.; Johnston, E.R.; Jastrow, J.D.; Gilbert, J.A.; Meyer, F.; Antonopoulos, D.A. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 2016, 18, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.M.; Caporaso, J.G.; Pirrung, M.; Field, D.; Knight, R.; Gilbert, J.A. Evidence for a persistent microbial seed bank throughout the global ocean. Proc. Natl. Acad. Sci. USA 2013, 110, 4651–4655. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Nuñez, N.F.; Maggia, L.; Stenger, P.-L.; Lelièvre, M.; Letellier, K.; Gigante, S.; Manez, A.; Mournet, P.; Ripoll, J.; Carriconde, F. Potential of high-throughput eDNA sequencing of soil fungi and bacteria for monitoring ecological restoration in ultramafic substrates: The case study of the New Caledonian biodiversity hotspot. Ecol. Eng. 2021, 173, 106416. [Google Scholar] [CrossRef]
- Campbell, C.D.; Gleeson, D.M.; Furlan, E.M.; Muirhead, K.A.; Caron, V. Detection of a cryptic terrestrial insect using novel eDNA collection techniques. Environ. DNA 2022, 820–829. [Google Scholar] [CrossRef]
- Štursová, M.; Šnajdr, J.; Cajthaml, T.; Bárta, J.; Šantrůčková, H.; Baldrian, P. When the forest dies: The response of forest soil fungi to a bark beetle-induced tree dieback. ISME J. 2014, 8, 1920–1931. [Google Scholar] [CrossRef]
- Ramírez-Pulido, J.; González-Ruiz, N.; Gardner, A.L.; Arroyo-Cabrales, J. List of Recent Land Mammals of Mexico. 2014. Available online: https://repository.si.edu/handle/10088/33974 (accessed on 1 March 2022).
- Reeder, J.; Knight, R. The’rare biosphere’: A reality check. Nat. Methods 2009, 6, 636–637. [Google Scholar] [CrossRef]
- Katz, M.; Hover, B.M.; Brady, S.F. Culture-independent discovery of natural products from soil metagenomes. J. Ind. Microbiol. Biotechnol. 2016, 43, 129–141. [Google Scholar] [CrossRef]
- Kirtane, A.; Dietschler, N.J.; Bittner, T.D.; Lefebvre, M.B.; Celis, S.; O’Connor, K.; Havill, N.; Whitmore, M.C. Sensitive environmental DNA (eDNA) methods to detect hemlock woolly adelgid and its biological control predators Leucotaraxis silver flies and a Laricobius beetle. Environ. DNA 2022, 1–14. [Google Scholar] [CrossRef]
- Hunter, M.E.; Oyler-McCance, S.J.; Dorazio, R.M.; Fike, J.A.; Smith, B.J.; Hunter, C.T.; Reed, R.N.; Hart, K.M. Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive Burmese pythons. PLoS ONE 2015, 10, e0121655. [Google Scholar]
- Lynggaard, C.; Bertelsen, M.F.; Jensen, C.V.; Johnson, M.S.; Frøslev, T.G.; Olsen, M.T.; Bohmann, K. Airborne environmental DNA for terrestrial vertebrate community monitoring. Curr. Biol. 2022, 32, 701–707.e705. [Google Scholar] [CrossRef] [PubMed]
- Lyman, J.A.; Sanchez, D.E.; Hershauer, S.N.; Sobek, C.J.; Chambers, C.L.; Zahratka, J.; Walker, F.M. Mammalian eDNA on herbaceous vegetation? Validating a qPCR assay for detection of an endangered rodent. Environ. DNA 2022, 1–11. [Google Scholar] [CrossRef]
- Akre, T.S.; Parker, L.D.; Ruther, E.; Maldonado, J.E.; Lemmon, L.; McInerney, N.R. Concurrent visual encounter sampling validates eDNA selectivity and sensitivity for the endangered wood turtle (Glyptemys insculpta). PLoS ONE 2019, 14, e0215586. [Google Scholar]
- Drummond, A.J.; Newcomb, R.D.; Buckley, T.R.; Xie, D.; Dopheide, A.; Potter, B.C.; Heled, J.; Ross, H.A.; Tooman, L.; Grosser, S. Evaluating a multigene environmental DNA approach for biodiversity assessment. GigaScience 2015, 4, 46. [Google Scholar] [CrossRef]
- Massey, A.L.; Bronzoni, R.V.d.M.; da Silva, D.J.F.; Allen, J.M.; de Lázari, P.R.; dos Santos-Filho, M.; Canale, G.R.; Bernardo, C.S.S.; Peres, C.A.; Levi, T. Invertebrates for vertebrate biodiversity monitoring: Comparisons using three insect taxa as iDNA samplers. Mol. Ecol. Resour. 2022, 22, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Schnell, I.B.; Thomsen, P.F.; Wilkinson, N.; Rasmussen, M.; Jensen, L.R.; Willerslev, E.; Bertelsen, M.F.; Gilbert, M.T.P. Screening mammal biodiversity using DNA from leeches. Curr. Biol. 2012, 22, R262–R263. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.R.; Handley, L.L.; Carpenter, A.I.; Ghazali, M.; Di Muri, C.; Macgregor, C.J.; Logan, T.W.; Law, A.; Breithaupt, T.; Read, D.S. Environmental DNA (eDNA) metabarcoding of pond water as a tool to survey conservation and management priority mammals. Biol. Conserv. 2019, 238, 108225. [Google Scholar] [CrossRef]
- Peterson, D.L.; Allen, M.C.; Vastano, A.; Lockwood, J.L. Evaluation of sample collection and storage protocols for surface eDNA surveys of an invasive terrestrial insect. Environ. DNA 2022, 1–11. [Google Scholar] [CrossRef]
- Ryan, E.; Bateman, P.; Fernandes, K.; van der Heyde, M.; Nevill, P. eDNA metabarcoding of log hollow sediments and soils highlights the importance of substrate type, frequency of sampling and animal size, for vertebrate species detection. Environ. DNA 2022, 940–953. [Google Scholar] [CrossRef]
- van der Loos, L.M.; Nijland, R. Biases in bulk: DNA metabarcoding of marine communities and the methodology involved. Mol. Ecol. 2021, 30, 3270–3288. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.W.; Ji, Y.; Emerson, B.C.; Wang, X.; Ye, C.; Yang, C.; Ding, Z. Biodiversity soup: Metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol. Evol. 2012, 3, 613–623. [Google Scholar] [CrossRef]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecology Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.; Shokralla, S.; Porter, T.M.; King, I.; van Konynenburg, S.; Janzen, D.H.; Hallwachs, W.; Hajibabaei, M. Simultaneous assessment of the macrobiome and microbiome in a bulk sample of tropical arthropods through DNA metasystematics. Proc. Natl. Acad. Sci. USA 2014, 111, 8007–8012. [Google Scholar] [CrossRef]
- Šigut, M.; Kostovčík, M.; Šigutová, H.; Hulcr, J.; Drozd, P.; Hrček, J. Performance of DNA metabarcoding, standard barcoding, and morphological approach in the identification of host–parasitoid interactions. PLoS ONE 2017, 12, e0187803. [Google Scholar] [CrossRef]
- Rodgers, T.W.; Xu, C.C.; Giacalone, J.; Kapheim, K.M.; Saltonstall, K.; Vargas, M.; Yu, D.W.; Somervuo, P.; McMillan, W.O.; Jansen, P.A. Carrion fly-derived DNA metabarcoding is an effective tool for mammal surveys: Evidence from a known tropical mammal community. Mol. Ecol. Resour. 2017, 17, e133–e145. [Google Scholar] [CrossRef]
- Mena, J.L.; Yagui, H.; Tejeda, V.; Bonifaz, E.; Bellemain, E.; Valentini, A.; Tobler, M.W.; Sánchez-Vendizú, P.; Lyet, A. Environmental DNA metabarcoding as a useful tool for evaluating terrestrial mammal diversity in tropical forests. Ecol. Appl. 2021, 31, e02335. [Google Scholar] [CrossRef]
- Pilotte, N.; Papaiakovou, M.; Grant, J.R.; Bierwert, L.A.; Llewellyn, S.; McCarthy, J.S.; Williams, S.A. Improved PCR-based detection of soil transmitted helminth infections using a next-generation sequencing approach to assay design. PLoS Negl. Trop. Dis. 2016, 10, e0004578. [Google Scholar] [CrossRef]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; van Rooyen, A.R.; Weeks, A.R.; Tingley, R. Dealing with false-positive and false-negative errors about species occurrence at multiple levels. Methods Ecol. Evol. 2017, 8, 1081–1091. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Taberlet, P.; Coissac, E. How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 2016, 604–607. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Gann, G.D.; McDonald, T.; Walder, B.; Aronson, J.; Nelson, C.R.; Jonson, J.; Hallett, J.G.; Eisenberg, C.; Guariguata, M.R.; Liu, J. International principles and standards for the practice of ecological restoration. Restor. Ecology. 2019, 27 (Suppl. S1), S1–S46. [Google Scholar] [CrossRef] [Green Version]
- Cowart, D.A.; Murphy, K.R.; Cheng, C.-H.C. Metagenomic sequencing of environmental DNA reveals marine faunal assemblages from the West Antarctic Peninsula. Mar. Genom. 2018, 37, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Elbrecht, V.; Leese, F. Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass—sequence relationships with an innovative metabarcoding protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef]
- McLaren, M.R.; Willis, A.D.; Callahan, B.J. Consistent and correctable bias in metagenomic sequencing experiments. eLife 2019, 8, e46923. [Google Scholar] [CrossRef]
- Cristescu, M.E.; Hebert, P.D. Uses and misuses of environmental DNA in biodiversity science and conservation. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 209–230. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Howland, K.; Normandeau, E.; Grey, E.K.; Archambault, P.; Deiner, K.; Lodge, D.M.; Hernandez, C.; Leduc, N.; Bernatchez, L. eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol. Evol. 2018, 8, 7763–7777. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.; Gough, K.C. The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Mauvisseau, Q.; Burian, A.; Gibson, C.; Brys, R.; Ramsey, A.; Sweet, M. Influence of accuracy, repeatability and detection probability in the reliability of species-specific eDNA based approaches. Sci. Rep. 2019, 9, 580. [Google Scholar] [CrossRef]
- Johnson, M.D.; Cox, R.D.; Grisham, B.A.; Lucia, D.; Barnes, M.A. Airborne eDNA reflects human activity and seasonal changes on a landscape scale. Front. Environ. Sci. 2021, 8, 563431. [Google Scholar] [CrossRef]
- Cordier, T.; Forster, D.; Dufresne, Y.; Martins, C.I.; Stoeck, T.; Pawlowski, J. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour. 2018, 18, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Schenekar, T.; Schletterer, M.; Lecaudey, L.A.; Weiss, S.J. Reference databases, primer choice, and assay sensitivity for environmental metabarcoding: Lessons learnt from a re-evaluation of an eDNA fish assessment in the Volga headwaters. River Res. Appl. 2020, 36, 1004–1013. [Google Scholar] [CrossRef] [Green Version]
- Berry, O.; Jarman, S.; Bissett, A.; Hope, M.; Paeper, C.; Bessey, C.; Schwartz, M.K.; Hale, J.; Bunce, M. Making environmental DNA (eDNA) biodiversity records globally accessible. Environ. DNA 2021, 3, 699–705. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, S.; Sabreena; Poczai, P.; Ganai, B.A.; Almalki, W.H.; Gafur, A.; Sayyed, R.Z. Environmental DNA Metabarcoding: A Novel Contrivance for Documenting Terrestrial Biodiversity. Biology 2022, 11, 1297. https://doi.org/10.3390/biology11091297
Hassan S, Sabreena, Poczai P, Ganai BA, Almalki WH, Gafur A, Sayyed RZ. Environmental DNA Metabarcoding: A Novel Contrivance for Documenting Terrestrial Biodiversity. Biology. 2022; 11(9):1297. https://doi.org/10.3390/biology11091297
Chicago/Turabian StyleHassan, Shahnawaz, Sabreena, Peter Poczai, Bashir Ah Ganai, Waleed Hassan Almalki, Abdul Gafur, and R. Z. Sayyed. 2022. "Environmental DNA Metabarcoding: A Novel Contrivance for Documenting Terrestrial Biodiversity" Biology 11, no. 9: 1297. https://doi.org/10.3390/biology11091297
APA StyleHassan, S., Sabreena, Poczai, P., Ganai, B. A., Almalki, W. H., Gafur, A., & Sayyed, R. Z. (2022). Environmental DNA Metabarcoding: A Novel Contrivance for Documenting Terrestrial Biodiversity. Biology, 11(9), 1297. https://doi.org/10.3390/biology11091297








