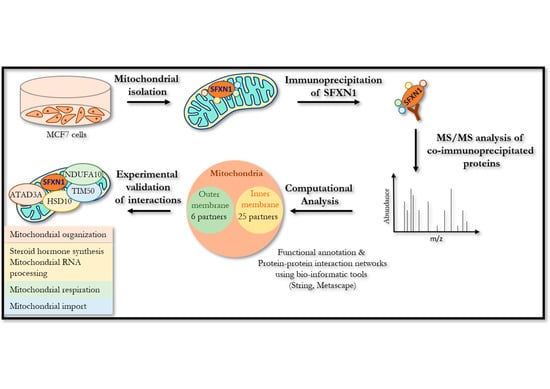
A High-Throughput Search for SFXN1 Physical Partners Led to the Identification of ATAD3, HSD10 and TIM50
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Plasmids and siRNA Transient Transfections
2.3. Protein Extraction and Subcellular Fractionation
2.4. Mitochondria Enrichment from Mammalian Cells and Drosophila Larvae
2.5. Western Blot
2.6. Co-Immunoprecipitation
2.7. Immunofluorescence Staining
2.8. Imaging and Statistical Analysis for Co-Localization
2.9. Sample Preparation for Proteomic Analyses
2.10. Proteomic Data Analysis
2.11. Biological Process Analysis Using PANTHER
2.12. Analysis of SFXN1 Interactome Using STRING
2.13. Metascape Analysis of SFXN1 Physical Partners
2.14. Oxygen Consumption Rate Measurement
2.15. Proximity Ligation Assay
2.16. Biological Resources
3. Results
3.1. SFXN1 Is an Evolutionarily Conserved Mitochondrial Protein Widely Distributed over Human Cell Lines
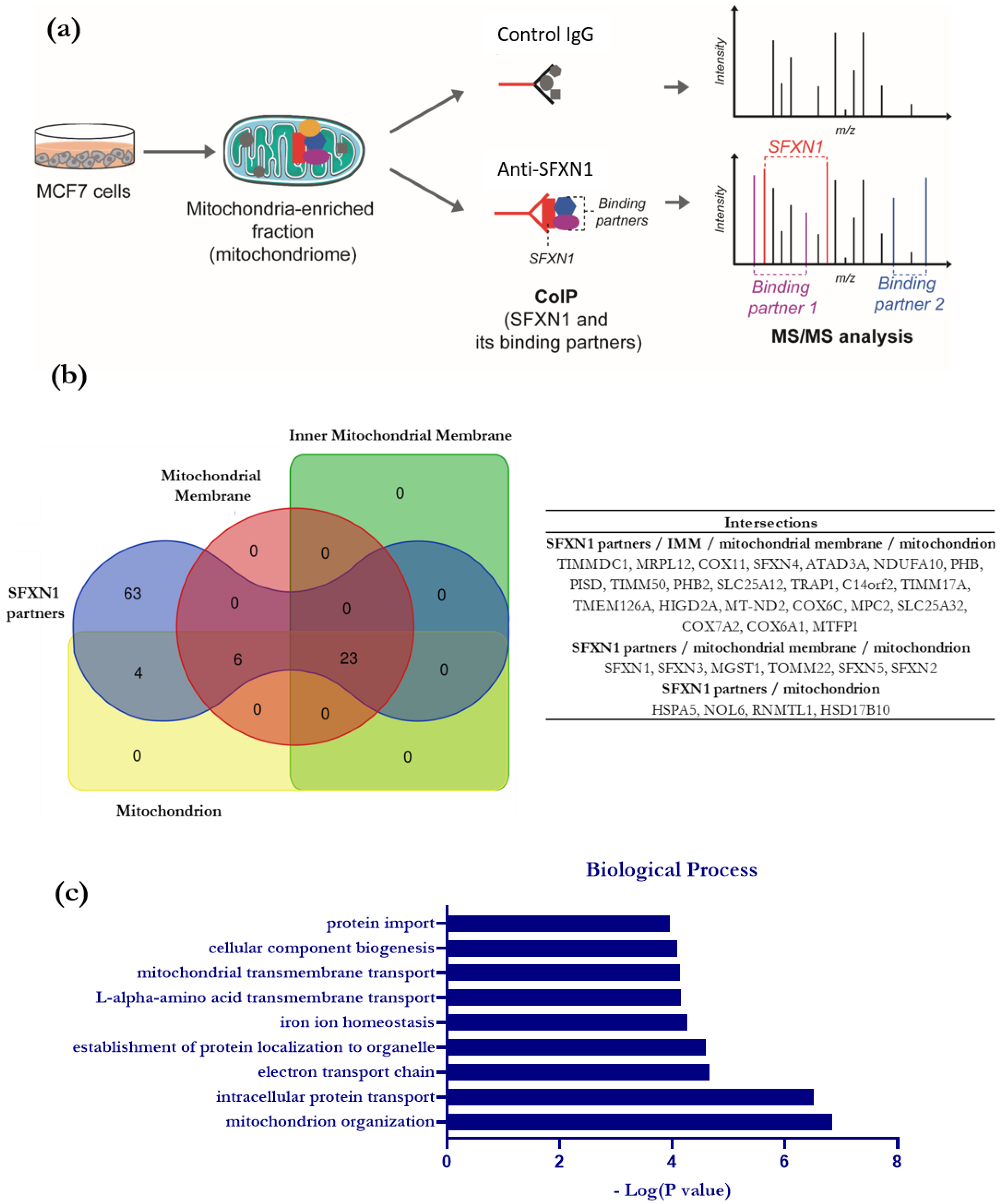
3.2. Identification of SFXN1 Partners by CoIP-MS/MS
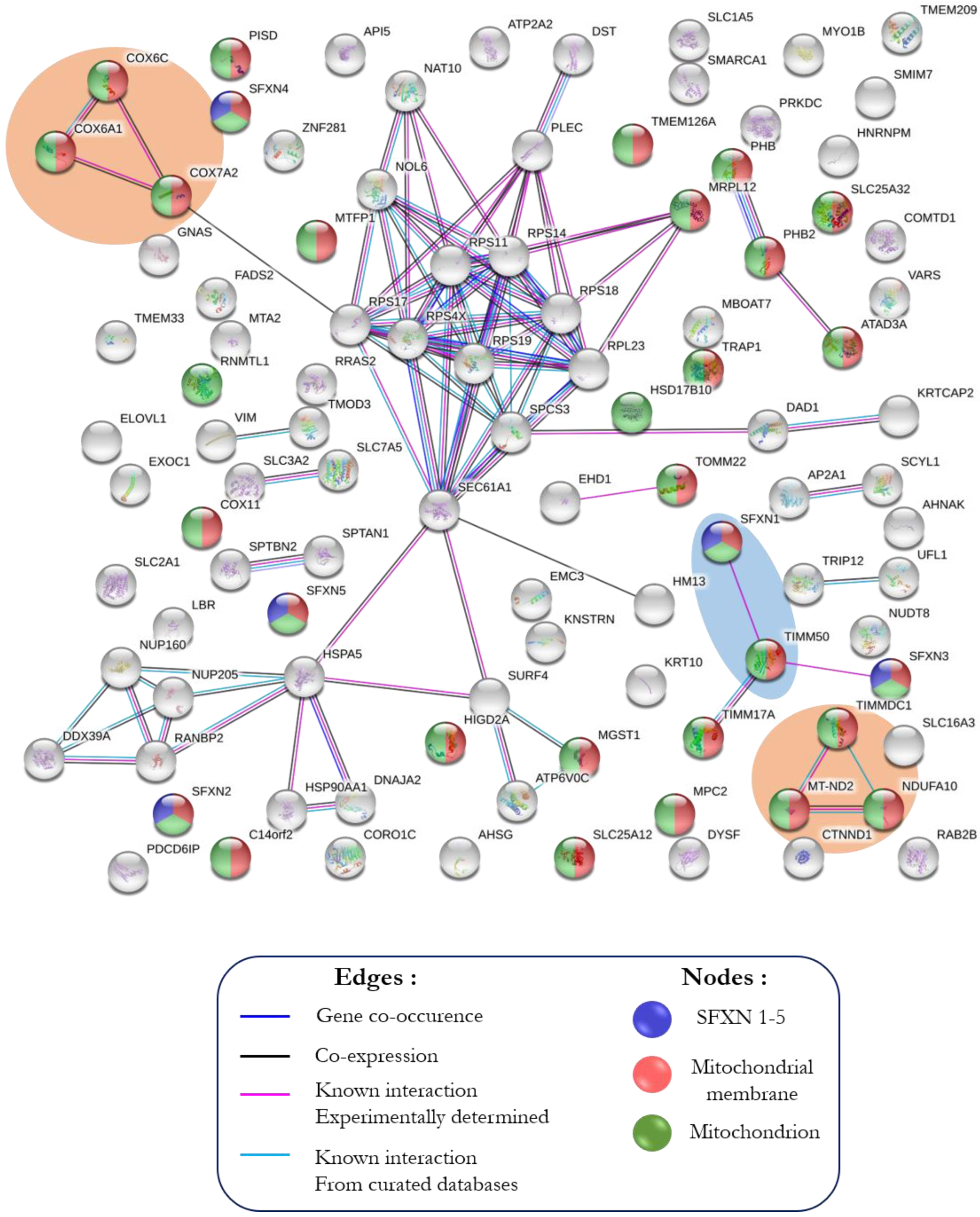
3.3. In Silico Analysis of the Interactome of SFXN1
3.3.1. STRING Analysis
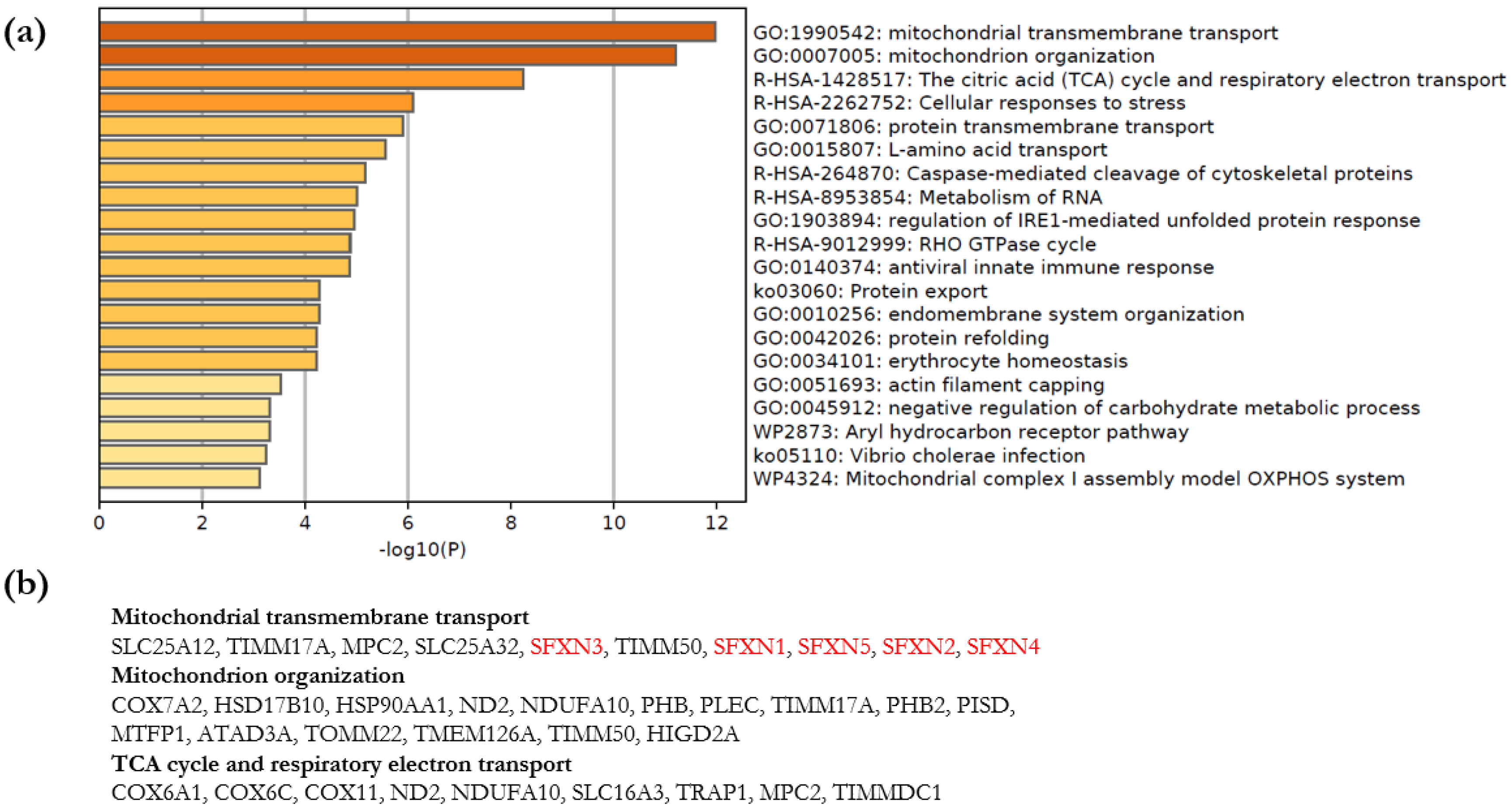
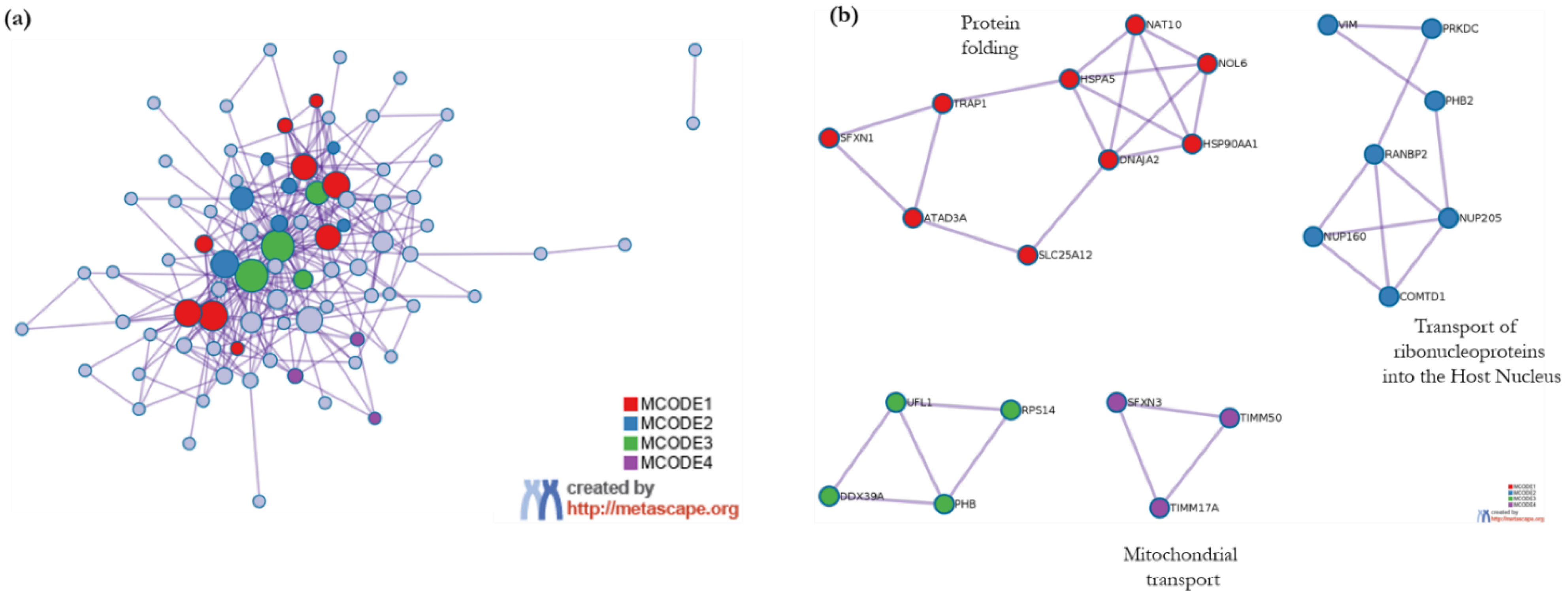
3.3.2. Metascape Analysis
- Enrichment Analysis and Clustering
- Protein–protein interactome network analysis using Metascape
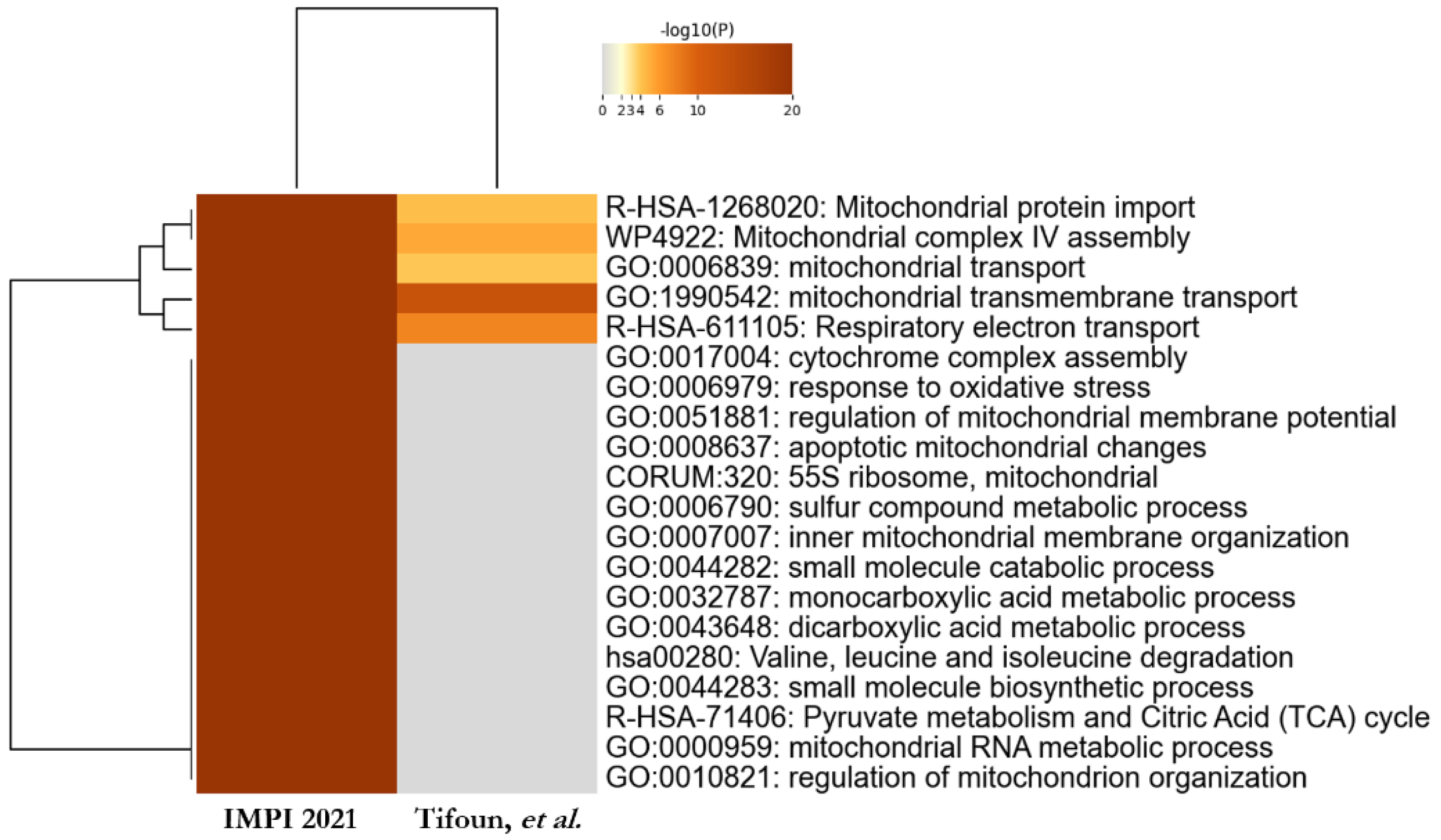
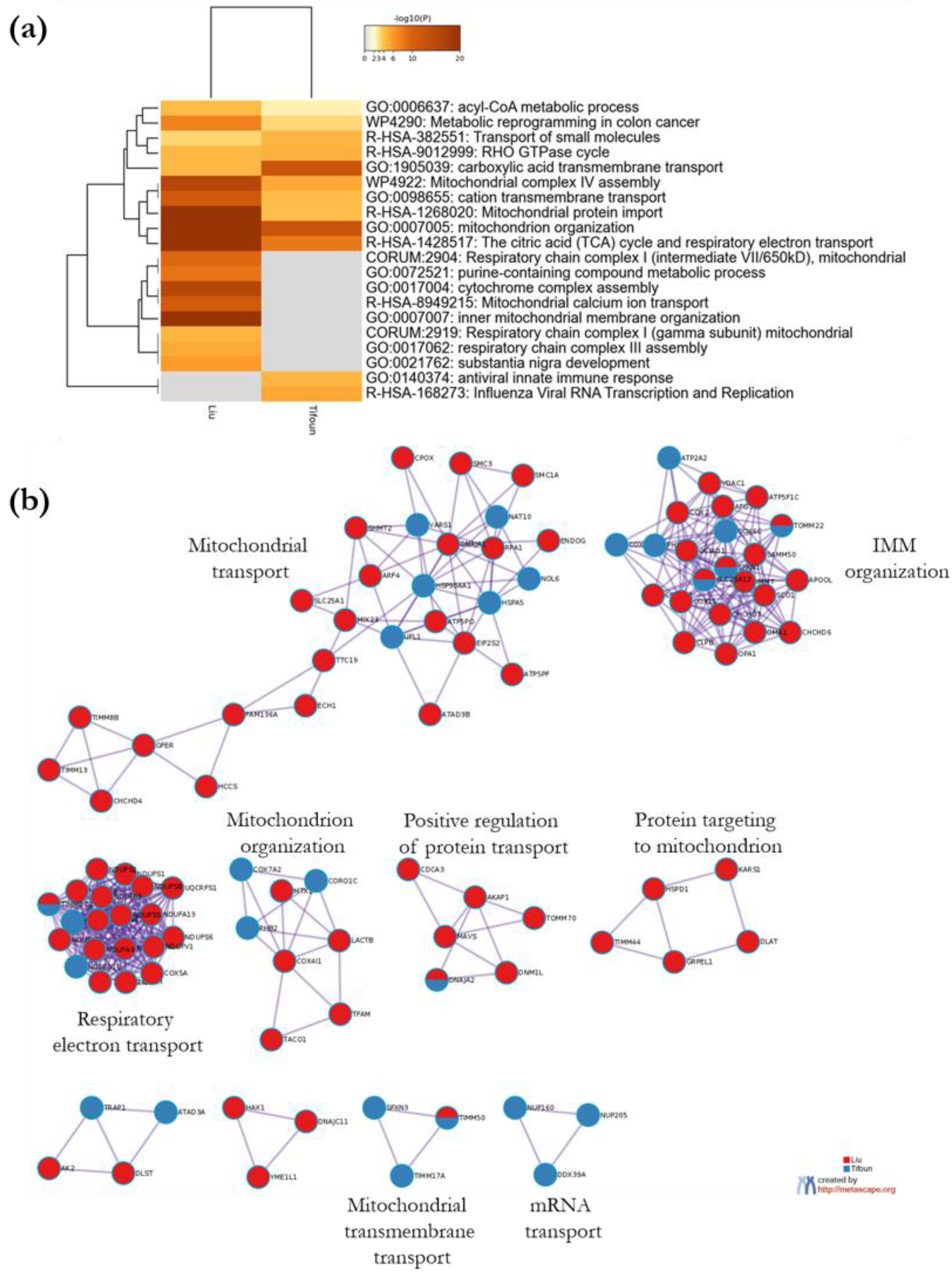
3.4. Comparative Study to Identify Highly Probable SFXN1 Partners
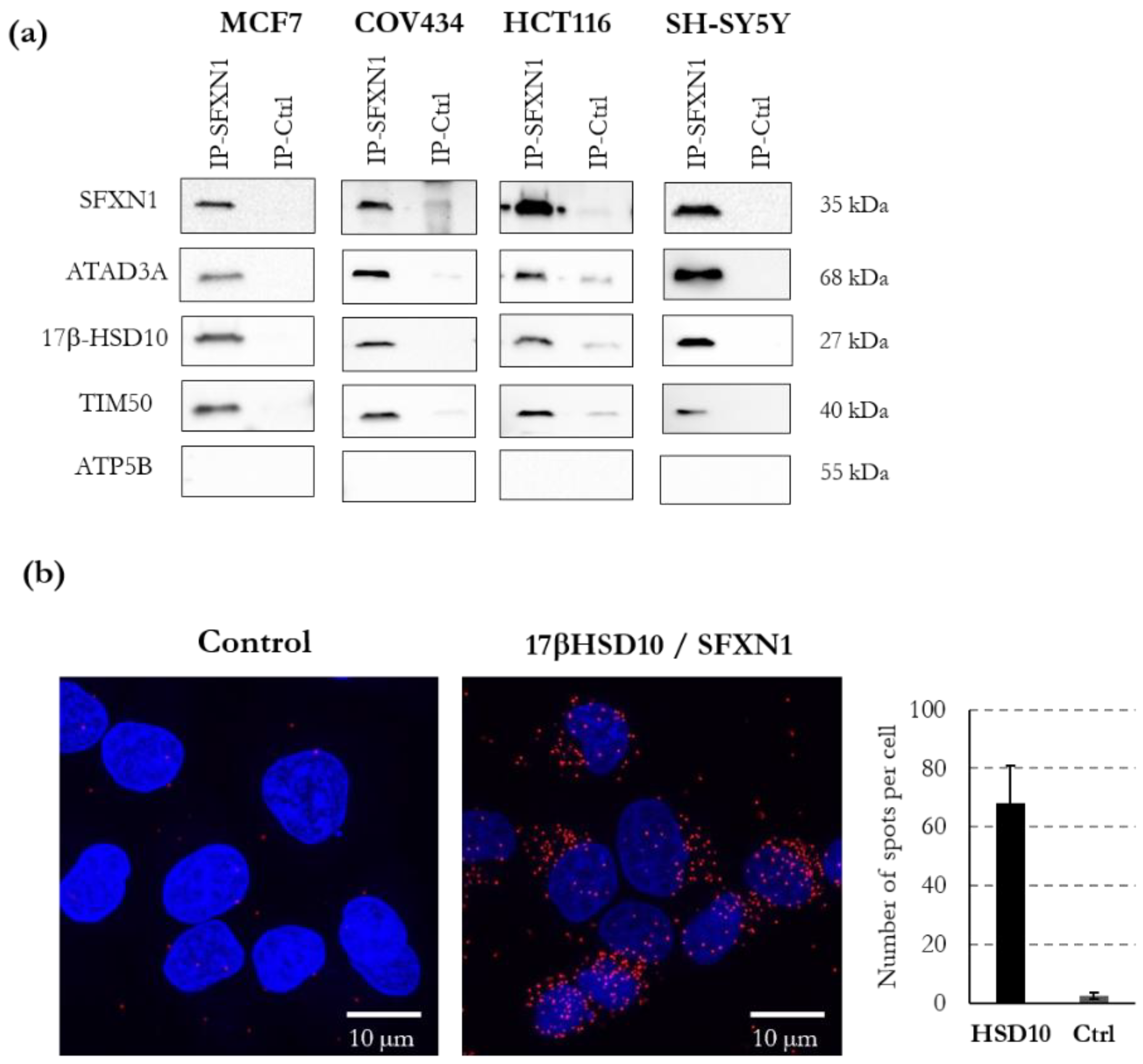
3.5. ATAD3, 17β-HSD10, TIM50 and NDUFA10 Physically Interact with SFXN1
4. Discussion
4.1. Biological Processes Linked to SFXN1 Interactome
4.2. SFXN1 May Interact with the Other Members of the SFXN Family
4.3. SFXN1, ATAD3A and 17β-HSD10
4.4. Physical Interaction between SFXN1 and Mitochondrial Translocases
4.5. Other Partners
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Armstrong, J.F.; Faccenda, E.; Harding, S.D.; Pawson, A.J.; Southan, C.; Sharman, J.L.; Campo, B.; Cavanagh, D.R.; Alexander, S.P.; Davenport, A.P.; et al. The IUPHAR/BPS guide to pharmacology in 2022: Curating pharmacology for COVID-19, malaria and antibacterials. Nucl. Acids Res. 2022, 50, D1282–D1294. [Google Scholar] [CrossRef]
- Gyimesi, G.; Hediger, M.A. Sequence Features of Mitochondrial Transporter Protein Families. Biomolecules 2020, 10, 1611. [Google Scholar] [CrossRef] [PubMed]
- Tifoun, N.; De las Heras, J.M.; Guillaume, A.; Bouleau, S.; Mignotte, B.; Le Floch, N. Insights into the Roles of the Sideroflexins/SLC56 Family in Iron Homeostasis and Iron-Sulfur Biogenesis. Biomedicines 2021, 9, 103. [Google Scholar] [CrossRef]
- Hildick-Smith, G.J.; Cooney, J.D.; Garone, C.; Kremer, L.S.; Haack, T.B.; Thon, J.N.; Miyata, N.; Lieber, D.S.; Calvo, S.E.; Akman, H.O.; et al. Macrocytic Anemia and Mitochondriopathy Resulting from a Defect in Sideroflexin 4. Am. J. Hum. Genet. 2013, 93, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Tulinius, M.; Kollberg, G.; Darin, N.; Oldfors, A.; Asin-Cayuela, J. Congenital Mitochondrial Encephalomyopathy with Complex I Deficiency Due to Mutations in Sideroflexin 4 (SFXN4). Neuromuscul. Disord. 2016, 26, S175. [Google Scholar] [CrossRef]
- Sofou, K.; Hedberg-Oldfors, C.; Kollberg, G.; Thomsen, C.; Wiksell, Å.; Oldfors, A.; Tulinius, M. Prenatal onset of mitochondrial disease is associated with sideroflexin 4 deficiency. Mitochondrion 2019, 47, 76–81. [Google Scholar] [CrossRef]
- Minjarez, B.; Calderón-González, K.G.; Rustarazo, M.L.V.; Herrera-Aguirre, M.E.; Labra-Barrios, M.L.; Rincon-Limas, D.E.; Del Pino, M.M.S.; Mena, R.; Luna-Arias, J.P. Identification of Proteins That Are Differentially Expressed in Brains with Alzheimer’s Disease Using ITRAQ Labeling and Tandem Mass Spectrometry. J. Proteom. 2016, 139, 103–121. [Google Scholar] [CrossRef]
- Simunovic, F.; Yi, M.; Wang, Y.; Macey, L.; Brown, L.T.; Krichevsky, A.M.; Andersen, S.L.; Stephens, R.M.; Benes, F.M.; Sonntag, K.C. Gene Expression Profiling of Substantia Nigra Dopamine Neurons: Further Insights into Parkinson’s Disease Pathology. Brain 2009, 132, 1795–1809. [Google Scholar] [CrossRef]
- Amorim, I.S.; Graham, L.C.; Carter, R.N.; Morton, N.M.; Hammachi, F.; Kunath, T.; Pennetta, G.; Carpanini, S.M.; Manson, J.C.; Lamont, D.J.; et al. Sideroflexin 3 Is an α-Synuclein-Dependent Mitochondrial Protein That Regulates Synaptic Morphology. J. Cell Sci. 2017, 130, 325–331. [Google Scholar] [CrossRef]
- Rivell, A.; Petralia, R.S.; Wang, Y.-X.; Mattson, M.P.; Yao, P.J. Sideroflexin 3 Is a Mitochondrial Protein Enriched in Neurons. Neuromol. Med. 2019, 21, 314–321. [Google Scholar] [CrossRef]
- Ledahawsky, L.M.; Terzenidou, M.E.; Edwards, R.; Kline, R.A.; Graham, L.C.; Eaton, S.L.; Hoorn, D.; Chaytow, H.; Huang, Y.; Groen, E.J.N.; et al. The Mitochondrial Protein Sideroflexin 3 (SFXN3) Influences Neurodegeneration Pathways in Vivo. FEBS J. 2022, 289, 3894–3914. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Aredo, B.; Ding, Y.; Zhong, X.; Zhu, Y.; Zhao, C.X.; Kumar, A.; Xing, C.; Gautron, L.; Lyon, S.; et al. Forward Genetic Analysis Using OCT Screening Identifies Sfxn3 Mutations Leading to Progressive Outer Retinal Degeneration in Mice. Proc. Natl. Acad. Sci. USA 2020, 117, 12931–12942. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, G.Y.; Bazykin, G.A.; Brudno, M.; Gelfand, M.S. Comparative genomics of transcriptional regulation in yeasts and its application to identification of a candidate alpha-isopropylmalate transporter. J. Bioinform. Comput. Biol. 2006, 04, 981–998. [Google Scholar] [CrossRef]
- Fleming, M.D. A Mutation in a Mitochondrial Transmembrane Protein Is Responsible for the Pleiotropic Hematological and Skeletal Phenotype of Flexed-Tail (f/f) Mice. Genes Dev. 2001, 15, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Mon, E.E.; Wei, F.-Y.; Ahmad, R.N.R.; Yamamoto, T.; Moroishi, T.; Tomizawa, K. Regulation of Mitochondrial Iron Homeostasis by Sideroflexin 2. J. Physiol. Sci. 2019, 69, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.T.; Tesfay, L.; Winkler, C.R.; Torti, F.M.; Torti, S.V. Sideroflexin 4 Affects Fe-S Cluster Biogenesis, Iron Metabolism, Mitochondrial Respiration and Heme Biosynthetic Enzymes. Sci. Rep. 2019, 9, 19634. [Google Scholar] [CrossRef] [PubMed]
- Kory, N.; Wyant, G.A.; Prakash, G.; uit de Bos, J.; Bottanelli, F.; Pacold, M.E.; Chan, S.H.; Lewis, C.A.; Wang, T.; Keys, H.R.; et al. SFXN1 Is a Mitochondrial Serine Transporter Required for One-Carbon Metabolism. Science 2018, 362, eaat9528. [Google Scholar] [CrossRef] [PubMed]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef]
- Acoba, M.G.; Alpergin, E.S.S.; Renuse, S.; Fernández-del-Río, L.; Lu, Y.-W.; Khalimonchuk, O.; Clarke, C.F.; Pandey, A.; Wolfgang, M.J.; Claypool, S.M. The Mitochondrial Carrier SFXN1 Is Critical for Complex III Integrity and Cellular Metabolism. Cell Rep. 2021, 34, 108869. [Google Scholar] [CrossRef]
- Bao, B.; An, W.; Lu, Q.; Wang, Y.; Lu, Z.; Tu, J.; Zhang, H.; Duan, Y.; Yuan, W.; Zhu, X.; et al. Sfxn1 Is Essential for Erythrocyte Maturation via Facilitating Hemoglobin Production in Zebrafish. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2021, 1867, 166096. [Google Scholar] [CrossRef]
- Manousakidi, S.; Guillaume, A.; Pirou, C.; Bouleau, S.; Mignotte, B.; Renaud, F.; Le Floch, N. FGF1 Induces Resistance to Chemotherapy in Ovarian Granulosa Tumor Cells through Regulation of P53 Mitochondrial Localization. Oncogenesis 2018, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Manders, E.M.M.; Verbeek, F.J.; Aten, J.A. Measurement of Co-Localization of Objects in Dual-Colour Confocal Images. J. Microsc. 1993, 169, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Aaron, J.S.; Taylor, A.B.; Chew, T.-L. Image Co-Localization—Co-Occurrence versus Correlation. J. Cell Sci. 2018, 131, jcs211847. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER Version 14: More Genomes, a New PANTHER GO-Slim and Improvements in Enrichment Analysis Tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Bandrowski, A.; Brush, M.; Grethe, J.S.; Haendel, M.A.; Kennedy, D.N.; Hill, S.; Hof, P.R.; Martone, M.E.; Pols, M.; Tan, S.; et al. The Resource Identification Initiative: A Cultural Shift in Publishing. F1000Research 2015, 4, 134. [Google Scholar] [CrossRef] [Green Version]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A Subcellular Map of the Human Proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Kang, M.-G.; Park, J.-S.; Lee, G.; Ting, A.Y.; Rhee, H.-W. APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest. Cell Rep. 2016, 15, 1837–1847. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Salokas, K.; Tamene, F.; Jiu, Y.; Weldatsadik, R.G.; Öhman, T.; Varjosalo, M. An AP-MS- and BioID-Compatible MAC-Tag Enables Comprehensive Mapping of Protein Interactions and Subcellular Localizations. Nat. Commun. 2018, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Go, C.D.; Knight, J.D.R.; Rajasekharan, A.; Rathod, B.; Hesketh, G.G.; Abe, K.T.; Youn, J.-Y.; Samavarchi-Tehrani, P.; Zhang, H.; Zhu, L.Y.; et al. A Proximity-Dependent Biotinylation Map of a Human Cell. Nature 2021, 595, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Pardo, B.; Lasorsa, F.M.; del Arco, A.; Kobayashi, K.; Iijima, M.; Runswick, M.J.; Walker, J.E.; Saheki, T.; Satrústegui, J.; et al. Citrin and Aralar1 Are Ca2+-Stimulated Aspartate/Glutamate Transporters in Mitochondria. EMBO J. 2001, 20, 5060–5069. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Console, L.; Losso, M.A.; Indiveri, C. The Human SLC1A5 (ASCT2) Amino Acid Transporter: From Function to Structure and Role in Cell Biology. Front. Cell Dev. Biol. 2018, 6, 96. [Google Scholar] [CrossRef]
- Yoo, H.C.; Park, S.J.; Nam, M.; Kang, J.; Kim, K.; Yeo, J.H.; Kim, J.-K.; Heo, Y.; Lee, H.S.; Lee, M.Y.; et al. A Variant of SLC1A5 Is a Mitochondrial Glutamine Transporter for Metabolic Reprogramming in Cancer Cells. Cell Metab. 2020, 31, 267–283. [Google Scholar] [CrossRef]
- Desai, R.; Frazier, A.E.; Durigon, R.; Patel, H.; Jones, A.W.; Dalla Rosa, I.; Lake, N.J.; Compton, A.G.; Mountford, H.S.; Tucker, E.J.; et al. ATAD3 Gene Cluster Deletions Cause Cerebellar Dysfunction Associated with Altered Mitochondrial DNA and Cholesterol Metabolism. Brain 2017, 140, 1595–1610. [Google Scholar] [CrossRef]
- Zschocke, J. HSD10 Disease: Clinical Consequences of Mutations in the HSD17B10 Gene. J. Inherit. Metab. Dis. 2012, 35, 81–89. [Google Scholar] [CrossRef]
- Jackson, T.D.; Hock, D.H.; Fujihara, K.M.; Palmer, C.S.; Frazier, A.E.; Low, Y.C.; Kang, Y.; Ang, C.S.; Clemons, N.J.; Thorburn, D.R.; et al. The TIM22 complex mediates the import of sideroflexins and is required for efficient mitochondrial one-carbon metabolism. Mol. Biol. Cell 2021, 32, 475–491. [Google Scholar] [CrossRef]
- Baudier, J. ATAD3 Proteins: Brokers of a Mitochondria-Endoplasmic Reticulum Connection in Mammalian Cells: Mitochondria-Endoplasmic Reticulum Connection. Biol. Rev. 2018, 93, 827–844. [Google Scholar] [CrossRef]
- Arguello, T.; Peralta, S.; Antonicka, H.; Gaidosh, G.; Diaz, F.; Tu, Y.-T.; Garcia, S.; Shiekhattar, R.; Barrientos, A.; Moraes, C.T. ATAD3A Has a Scaffolding Role Regulating Mitochondria Inner Membrane Structure and Protein Assembly. Cell Rep. 2021, 37, 110139. [Google Scholar] [CrossRef] [PubMed]
- Harel, T.; Yoon, W.H.; Garone, C.; Gu, S.; Coban-Akdemir, Z.; Eldomery, M.K.; Posey, J.E.; Jhangiani, S.N.; Rosenfeld, J.A.; Cho, M.T.; et al. Recurrent De Novo and Biallelic Variation of ATAD3A, Encoding a Mitochondrial Membrane Protein, Results in Distinct Neurological Syndromes. Am. J. Hum. Genet. 2016, 99, 831–845. [Google Scholar] [CrossRef]
- van den Ecker, D.; Hoffmann, M.; Müting, G.; Maglioni, S.; Herebian, D.; Mayatepek, E.; Ventura, N.; Distelmaier, F. Caenorhabditis Elegans ATAD-3 Modulates Mitochondrial Iron and Heme Homeostasis. Biochem. Biophys. Res. Commun. 2015, 467, 389–394. [Google Scholar] [CrossRef]
- He, X.-Y.; Isaacs, C.; Yang, S.-Y. Roles of Mitochondrial 17β-Hydroxysteroid Dehydrogenase Type 10 in Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 62, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Aitken, L.; Baillie, G.; Pannifer, A.; Morrison, A.; Jones, P.S.; Smith, T.K.; McElroy, S.P.; Gunn-Moore, F.J. In Vitro Assay Development and HTS of Small-Molecule Human ABAD/17β-HSD10 Inhibitors as Therapeutics in Alzheimer’s Disease. SLAS Discov. Adv. Sci. Drug Discov. 2017, 22, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-Y.; He, X.-Y.; Isaacs, C.; Dobkin, C.; Miller, D.; Philipp, M. Roles of 17β-Hydroxysteroid Dehydrogenase Type 10 in Neurodegenerative Disorders. J. Steroid Biochem. Mol. Biol. 2014, 143, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An Updated Mitochondrial Proteome Now with Sub-Organelle Localization and Pathway Annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef] [Green Version]
- Callegari, S.; Richter, F.; Chojnacka, K.; Jans, D.C.; Lorenzi, I.; Pacheu-Grau, D.; Jakobs, S.; Lenz, C.; Urlaub, H.; Dudek, J.; et al. TIM29 Is a Subunit of the Human Carrier Translocase Required for Protein Transport. FEBS Lett. 2016, 590, 4147–4158. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Stroud, D.A.; Baker, M.J.; De Souza, D.P.; Frazier, A.E.; Liem, M.; Tull, D.; Mathivanan, S.; McConville, M.J.; Thorburn, D.R.; et al. Sengers Syndrome-Associated Mitochondrial Acylglycerol Kinase Is a Subunit of the Human TIM22 Protein Import Complex. Mol. Cell 2017, 67, 457–470.e5. [Google Scholar] [CrossRef] [PubMed]
- Vukotic, M.; Nolte, H.; König, T.; Saita, S.; Ananjew, M.; Krüger, M.; Tatsuta, T.; Langer, T. Acylglycerol Kinase Mutated in Sengers Syndrome Is a Subunit of the TIM22 Protein Translocase in Mitochondria. Mol. Cell 2017, 67, 471–483.e7. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Song, J.; Pfanner, N. Versatility of Preprotein Transfer from the Cytosol to Mitochondria. Trends Cell Biol. 2019, 29, 534–548. [Google Scholar] [CrossRef]
- Shiota, T.; Mabuchi, H.; Tanaka-Yamano, S.; Yamano, K.; Endo, T. In Vivo Protein-Interaction Mapping of a Mitochondrial Translocator Protein Tom22 at Work. Proc. Natl. Acad. Sci. USA 2011, 108, 15179–15183. [Google Scholar] [CrossRef]
- Kuang, Q.; Purhonen, P.; Ålander, J.; Svensson, R.; Hoogland, V.; Winerdal, J.; Spahiu, L.; Ottosson-Wadlund, A.; Jegerschöld, C.; Morgenstern, R.; et al. Dead-End Complex, Lipid Interactions and Catalytic Mechanism of Microsomal Glutathione Transferase 1, an Electron Crystallography and Mutagenesis Investigation. Sci. Rep. 2017, 7, 7897. [Google Scholar] [CrossRef]
- Bräutigam, L.; Zhang, J.; Dreij, K.; Spahiu, L.; Holmgren, A.; Abe, H.; Tew, K.D.; Townsend, D.M.; Kelner, M.J.; Morgenstern, R.; et al. MGST1, a GSH Transferase/Peroxidase Essential for Development and Hematopoietic Stem Cell Differentiation. Redox Biol. 2018, 17, 171–179. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Fowler, S.L.; Akins, M.; Zhou, H.; Figeys, D.; Bennett, S.A.L. The Liver Connexin32 Interactome Is a Novel Plasma Membrane-Mitochondrial Signaling Nexus. J. Proteome Res. 2013, 12, 2597–2610. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Lin, S.; Yang, S.; Que, H.; Liu, S. Identification of Novel SCIRR69-Interacting Proteins During ER Stress Using SILAC-Immunoprecipitation Quantitative Proteomics Approach. Neuromol. Med. 2017, 19, 81–93. [Google Scholar] [CrossRef]
- Fang, H.; Xie, A.; Du, M.; Li, X.; Yang, K.; Fu, Y.; Yuan, X.; Fan, R.; Yu, W.; Zhou, Z.; et al. SERAC1 Is a Component of the Mitochondrial Serine Transporter Complex Required for the Maintenance of Mitochondrial DNA. Sci. Transl. Med. 2022, 14, eabl6992. [Google Scholar] [CrossRef]
- Jackson, T.D.; Crameri, J.J.; Muellner-Wong, L.; Frazier, A.E.; Palmer, C.S.; Formosa, L.E.; Hock, D.H.; Fujihara, K.M.; Stait, T.; Sharpe, A.J.; et al. Sideroflexin 4 Is a Complex I Assembly Factor That Interacts with the MCIA Complex and Is Required for the Assembly of the ND2 Module. Proc. Natl. Acad. Sci. USA 2022, 119, e2115566119. [Google Scholar] [CrossRef] [PubMed]
- Formosa, L.E.; Reljic, B.; Sharpe, A.J.; Hock, D.H.; Muellner-Wong, L.; Stroud, D.A.; Ryan, M.T. Optic Atrophy–Associated TMEM126A Is an Assembly Factor for the ND4-Module of Mitochondrial Complex I. Proc. Natl. Acad. Sci. USA 2021, 118, e2019665118. [Google Scholar] [CrossRef] [PubMed]
- Guarani, V.; Paulo, J.; Zhai, B.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. TIMMDC1/C3orf1 Functions as a Membrane-Embedded Mitochondrial Complex I Assembly Factor through Association with the MCIA Complex. Mol. Cell. Biol. 2014, 34, 847–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tifoun, N.; Bekhouche, M.; De las Heras, J.M.; Guillaume, A.; Bouleau, S.; Guénal, I.; Mignotte, B.; Le Floch, N. A High-Throughput Search for SFXN1 Physical Partners Led to the Identification of ATAD3, HSD10 and TIM50. Biology 2022, 11, 1298. https://doi.org/10.3390/biology11091298
Tifoun N, Bekhouche M, De las Heras JM, Guillaume A, Bouleau S, Guénal I, Mignotte B, Le Floch N. A High-Throughput Search for SFXN1 Physical Partners Led to the Identification of ATAD3, HSD10 and TIM50. Biology. 2022; 11(9):1298. https://doi.org/10.3390/biology11091298
Chicago/Turabian StyleTifoun, Nesrine, Mourad Bekhouche, José M. De las Heras, Arnaud Guillaume, Sylvina Bouleau, Isabelle Guénal, Bernard Mignotte, and Nathalie Le Floch. 2022. "A High-Throughput Search for SFXN1 Physical Partners Led to the Identification of ATAD3, HSD10 and TIM50" Biology 11, no. 9: 1298. https://doi.org/10.3390/biology11091298
APA StyleTifoun, N., Bekhouche, M., De las Heras, J. M., Guillaume, A., Bouleau, S., Guénal, I., Mignotte, B., & Le Floch, N. (2022). A High-Throughput Search for SFXN1 Physical Partners Led to the Identification of ATAD3, HSD10 and TIM50. Biology, 11(9), 1298. https://doi.org/10.3390/biology11091298