The Evolutionary Dynamics of the Mitochondrial tRNA in the Cichlid Fish Family
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Bioinformatics Analysis
2.3. Primer Design, PCR, and Wet-Lab Validation
3. Results
3.1. Characterization of Cichlids Mitochondrial Genomes
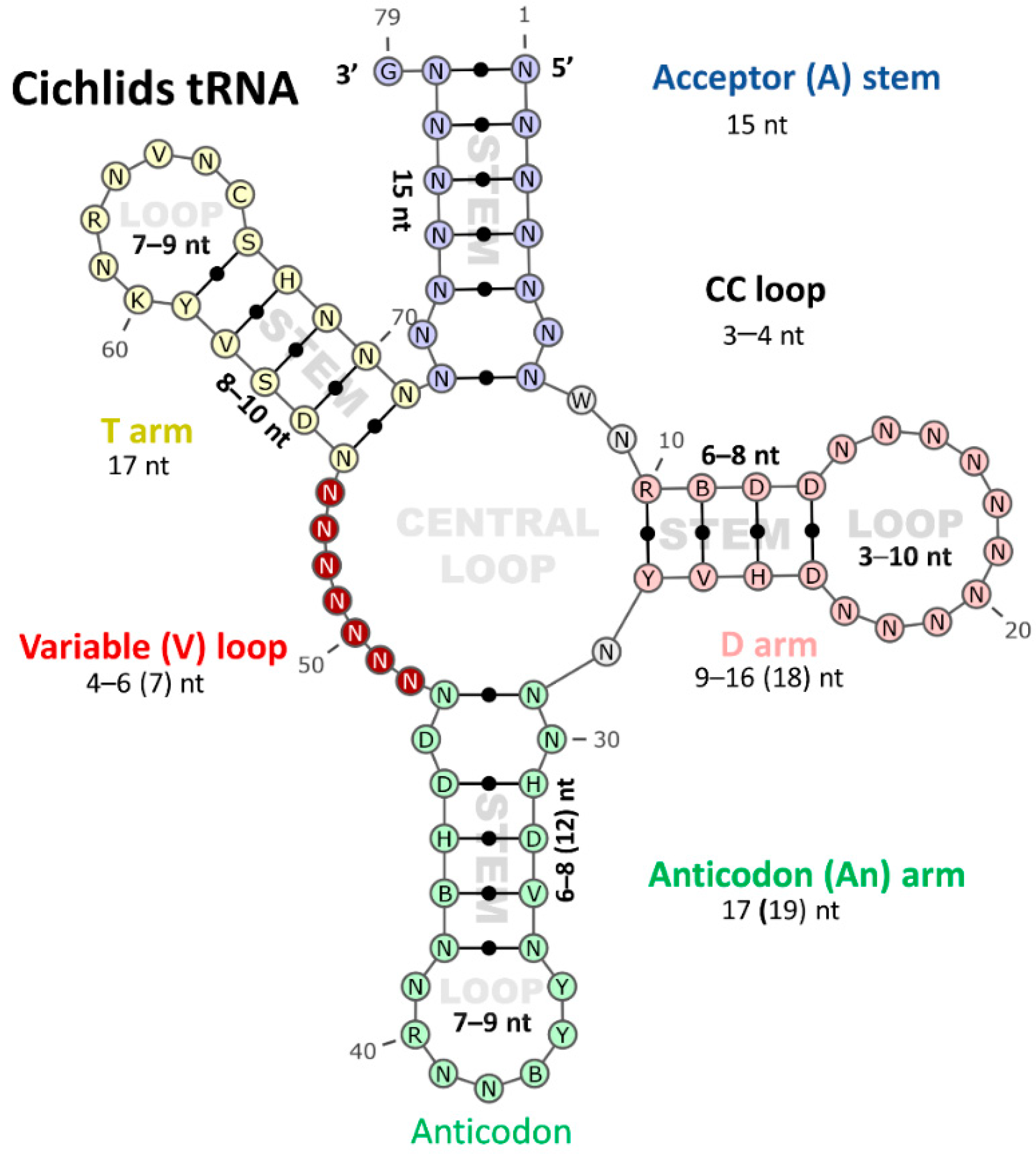
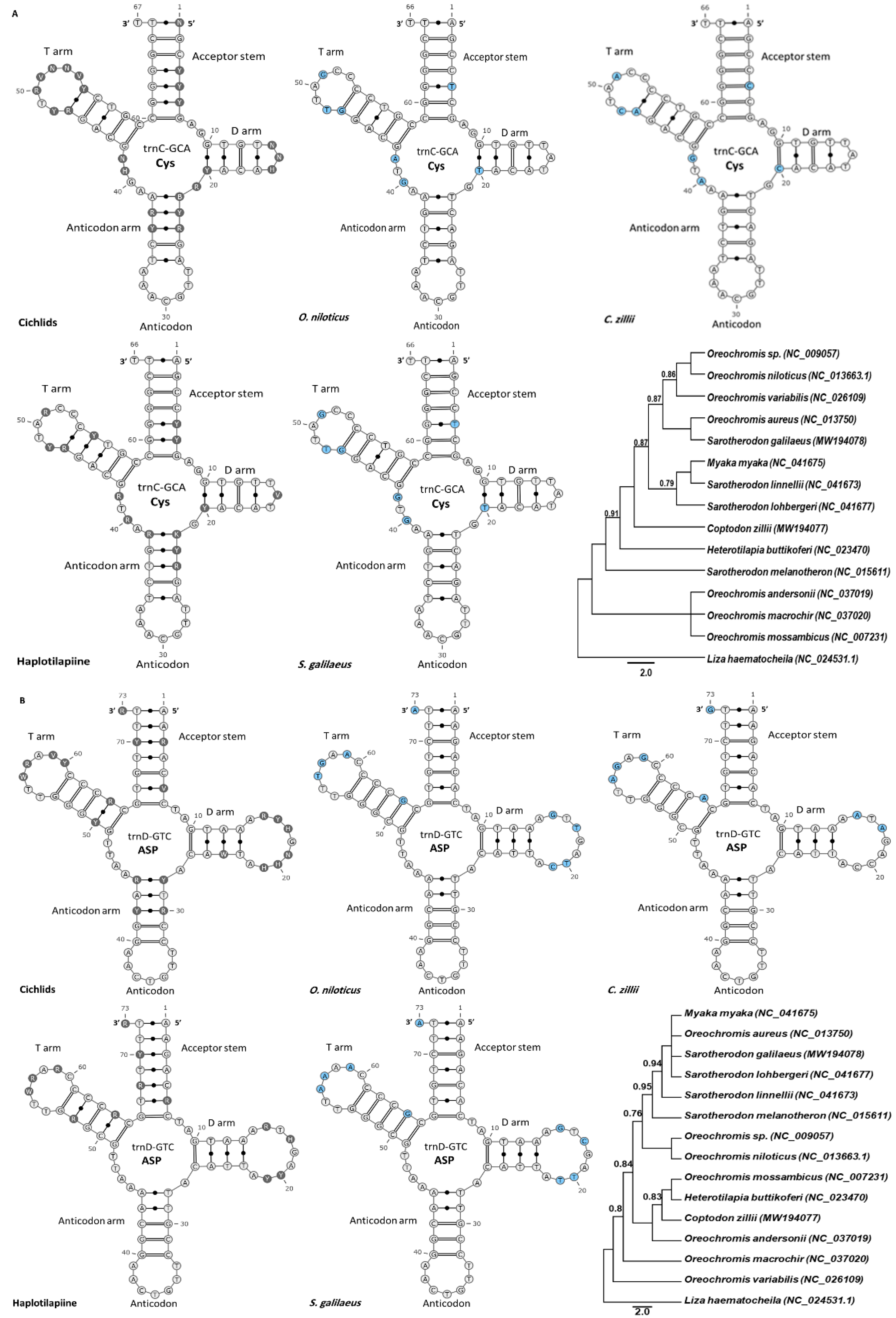
3.2. Diversification of tRNA Secondary Structure
3.3. Mitochondrial tRNAs Polymorphism among Cichlid Species
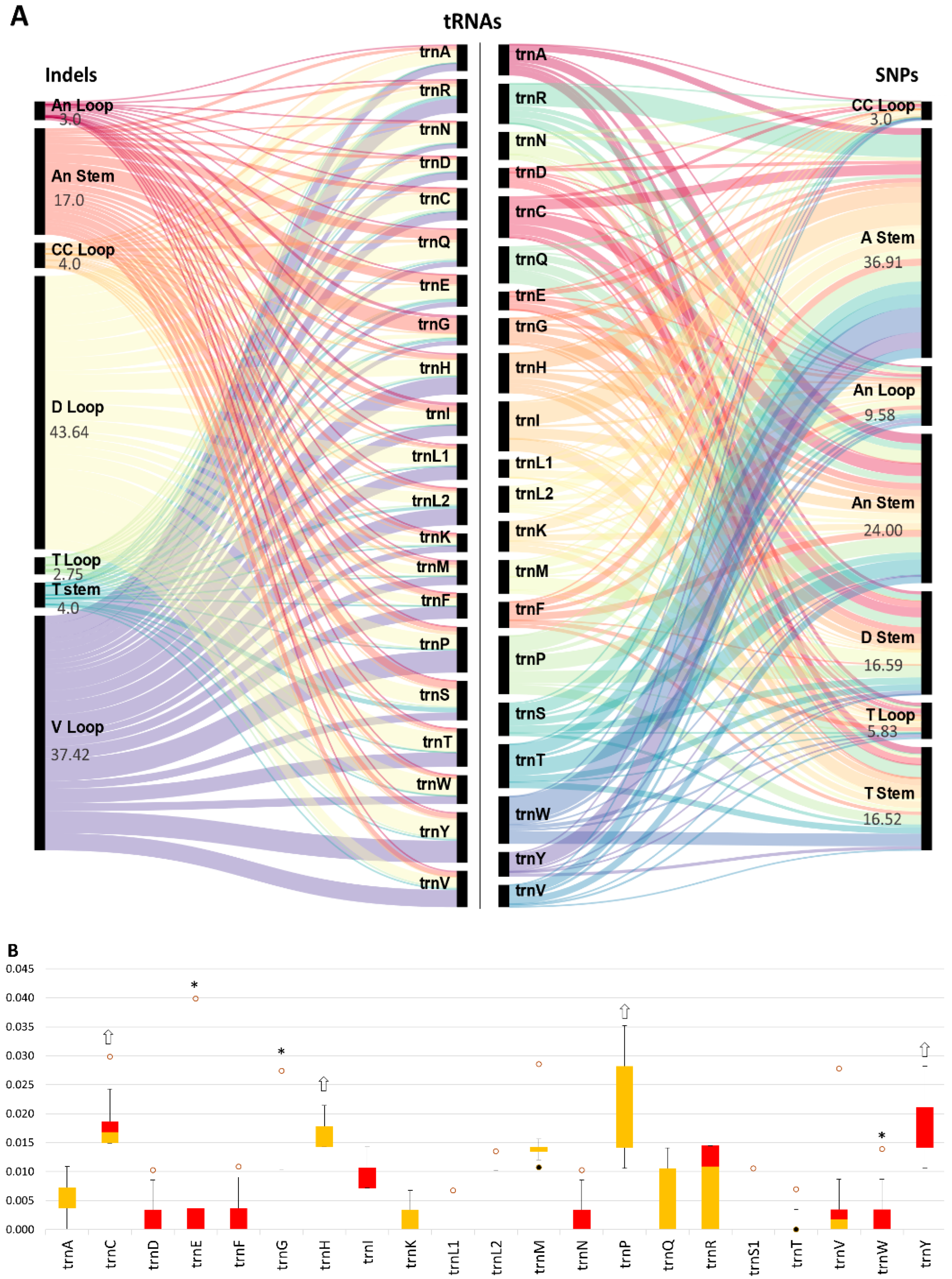
3.3.1. Quantified Polymorphism among mt-tRNAs
3.3.2. Quantified Polymorphism of Each mt-tRNA among Cichlids
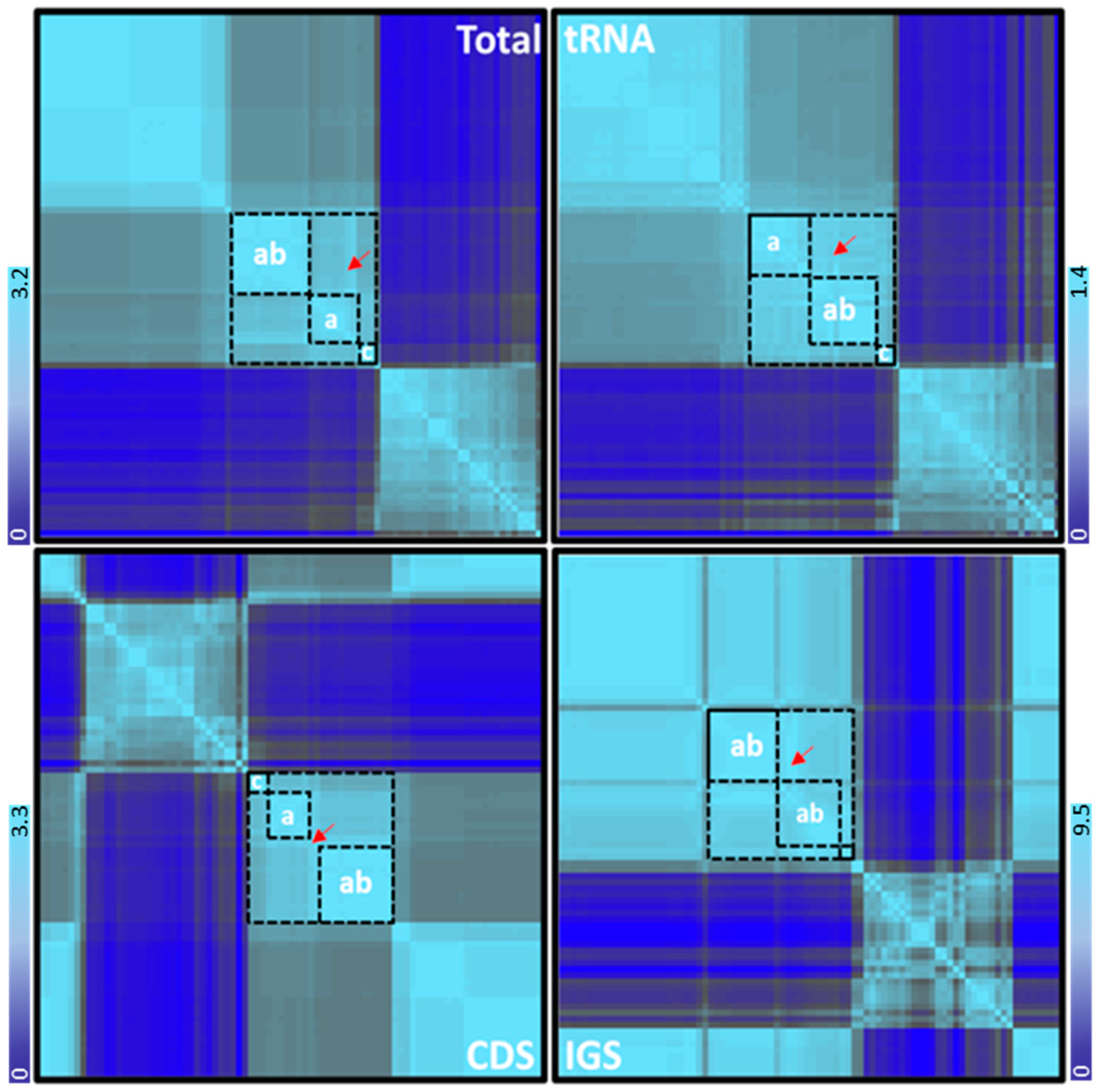
3.3.3. The Transition/Transversion Bias and Phylogenetics
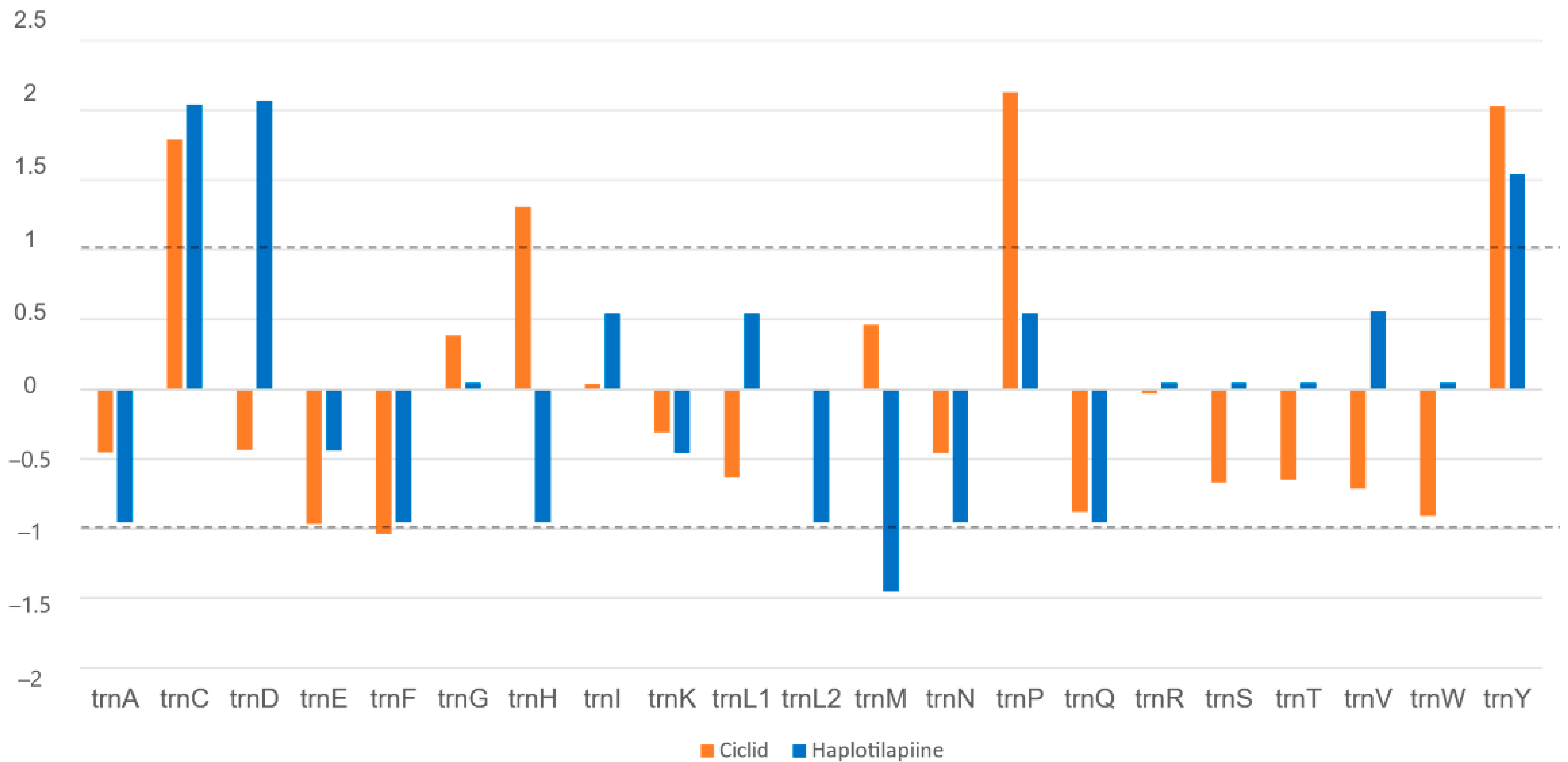
3.4. Cichlids vs. Haplotilapiine
3.5. The tRNA in Egyptian Haplotilapiine
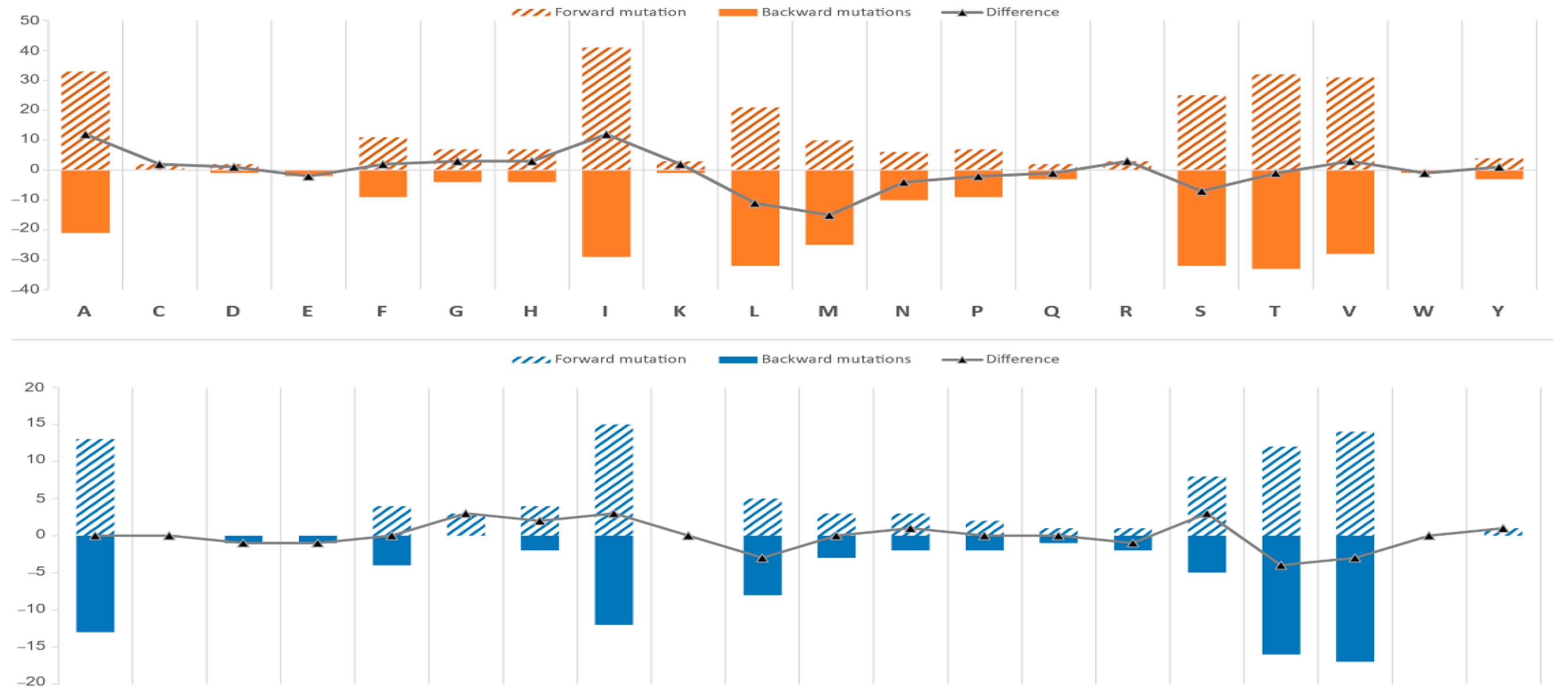
3.6. Amino Acid Change
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klett, V.; Meyer, A. What, If Anything, Is a Tilapia?—Mitochondrial ND2 Phylogeny of Tilapiines and the Evolution of Parental Care Systems in the African Cichlid Fishes. Mol. Biol. Evol. 2002, 19, 865–883. [Google Scholar] [CrossRef] [PubMed]
- Turner, G.F. Adaptive Radiation of Cichlid Fish. Curr. Biol. 2007, 17, R827–R831. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, M.C.M.; McAndrew, B.J. (Eds.) Tilapias: Biology and Exploitation; Springer: Dordrecht, The Netherlands, 2000; ISBN 9780792363910. [Google Scholar]
- Fitzsimmons, K. Future Trends of Tilapia Aquaculture in the Americas. In Tilapia Aquaculture in the Americas; The World Aquaculture Society: Baton Rouge, LA, USA, 2000; Volume 2, pp. 252–264. [Google Scholar]
- Fineman-Kalio, A.S. Preliminary Observations on the Effect of Salinity on the Reproduction and Growth of Freshwater Nile Tilapia, Oreochromis niloticus (L.), Cultured in Brackishwater Ponds. Aquac. Res. 1988, 19, 313–320. [Google Scholar] [CrossRef]
- Dunz, A.R.; Schliewen, U.K. Molecular Phylogeny and Revised Classification of the Haplotilapiine Cichlid Fishes Formerly Referred to as “Tilapia”. Mol. Phylogenetics Evol. 2013, 68, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Elson, J.; Lightowlers, R. Mitochondrial DNA Clonality in the Dock: Can Surveillance Swing the Case? Trends Genet. 2006, 22, 603–607. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the Mitochondrial Genome of Metazoa as Exemplified by Comparison of Congeneric Species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef]
- Thornlow, B.P.; Hough, J.; Roger, J.M.; Gong, H.; Lowe, T.M.; Corbett-Detig, R.B. Transfer RNA Genes Experience Exceptionally Elevated Mutation Rates. Proc. Natl. Acad. Sci. USA 2018, 115, 8996–9001. [Google Scholar] [CrossRef]
- Florentz, C.; Sohm, B.; Tryoen-Tóth, P.; Pütz, J.; Sissler, M. Human Mitochondrial TRNAs in Health and Disease. Cell. Mol. Life Sci. (CMLS) 2003, 60, 1356–1375. [Google Scholar] [CrossRef]
- Kogelnik, A.M.; Lott, M.T.; Brown, M.D.; Navathe, S.B.; Wallace, D.C. MITOMAP: An Update on the Status of the Human Mitochondrial Genome Database. Nucleic Acids Res. 1997, 25, 196–199. [Google Scholar] [CrossRef][Green Version]
- Söll, D.; RajBhandary, U.L. (Eds.) TRNA: Structure, Biosynthesis, and Function; ASM Press: Washington, DC, USA, 1994; ISBN 9781683672739. [Google Scholar]
- Helm, M.; Brulé, H.; Friede, D.; Giegé, R.; Pütz, D.; Florentz, C. Search for Characteristic Structural Features of Mammalian Mitochondrial TRNAs. RNA 2000, 6, 1356–1379. [Google Scholar] [CrossRef]
- Stamatis, C.; Giannouli, S.; Suchentrunk, F.; Sert, H.; Stathopoulos, C.; Mamuris, Z. Recruitment of Mitochondrial TRNA Genes as Auxiliary Variability Markers for Both Intra- and Inter-Species Analysis: The Paradigm of Brown Hare (Lepus Europaeus). Gene 2008, 410, 154–164. [Google Scholar] [CrossRef]
- Wang, X.; Lavrov, D.V. Gene Recruitment—A Common Mechanism in the Evolution of Transfer RNA Gene Families. Gene 2011, 475, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Loher, P.; Kirino, Y.; Rigoutsos, I. Nuclear and Mitochondrial TRNA-Lookalikes in the Human Genome. Front. Genet. 2014, 5, 344. [Google Scholar] [CrossRef] [PubMed]
- Trewavas, E.; British Museum (Natural History). Tilapiine Fishes of the Genera Sarotherodon, Oreochromis, and Danakilia; British Museum (Natural History): London, UK, 1983. [Google Scholar]
- Li, Y.; Gul, Y.; Cui, L.; Cao, X.; Wang, W. Comparative Analysis of Different Protocols for Extraction of DNA from Fish Scales of Cyprinus carpio. Indian J. Biotechnol. 2015, 14, 382–387. [Google Scholar]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Wilson, D.J.; McVean, G. Estimating Diversifying Selection and Functional Constraint in the Presence of Recombination. Genetics 2006, 172, 1411–1425. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. TRNAscan-SE On-Line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive Drawing and Editing of the RNA Secondary Structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A Visualisation Platform to Create Open Outputs. In Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter, Cagliari, Italy, 18 September 2017; ACM: New York, NY, USA, 2017; pp. 1–5. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.Q.; Al-Harbi, A.H. Evaluation of Three Species of Tilapia, Red Tilapia and a Hybrid Tilapia as Culture Species in Saudi Arabia. Aquaculture 1995, 138, 145–157. [Google Scholar] [CrossRef]
- Duan, M.; Chen, L.; Ge, Q.; Lu, N.; Li, J.; Pan, X.; Qiao, Y.; Tu, J.; Lu, Z. Evaluating Heteroplasmic Variations of the Mitochondrial Genome from Whole Genome Sequencing Data. Gene 2019, 699, 145–154. [Google Scholar] [CrossRef]
- Kelchner, S.A. The Evolution of Non-Coding Chloroplast DNA and Its Application in Plant Systematics. Ann. Missouri Bot. Gard. 2000, 87, 482. [Google Scholar] [CrossRef]
- Kelchner, S.A.; Clark, L.G. Molecular Evolution and Phylogenetic Utility of the Chloroplast rpl16 Intron in Chusquea and the Bambusoideae (Poaceae). Mol. Phylogenet. Evol. 1997, 8, 385–397. [Google Scholar] [CrossRef]
- Watanabe, Y.; Suematsu, T.; Ohtsuki, T. Losing the Stem-Loop Structure from Metazoan Mitochondrial TRNAs and Co-Evolution of Interacting Factors. Front. Genet. 2014, 5, 109. [Google Scholar] [CrossRef]
- Masta, S.E.; Boore, J.L. Parallel Evolution of Truncated Transfer RNA Genes in Arachnid Mitochondrial Genomes. Mol. Biol. Evol. 2008, 25, 949–959. [Google Scholar] [CrossRef]
- Molina, W.F.; Martinez, P.A.; Bertollo, L.A.C.; Bidau, C.J. Evidence for Meiotic Drive as an Explanation for Karyotype Changes in Fishes. Mar. Genom. 2014, 15, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Schliewen, U.; Rassmann, K.; Markmann, M.; Markert, J.; Kocher, T.; Tautz, D. Genetic and Ecological Divergence of a Monophyletic Cichlid Species Pair under Fully Sympatric Conditions in Lake Ejagham, Cameroon. Mol. Ecol. 2001, 10, 1471–1488. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D. Adaptive Evolution and Explosive Speciation: The Cichlid Fish Model. Nat. Rev. Genet. 2004, 5, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Barluenga, M.; Stölting, K.N.; Salzburger, W.; Muschick, M.; Meyer, A. Sympatric Speciation in Nicaraguan Crater Lake Cichlid Fish. Nature 2006, 439, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Cardena, M.M.S.G.; Mansur, A.J.; Pereira, A.D.C.; Fridman, C. A New Duplication in the Mitochondrially Encoded TRNA Proline Gene in a Patient with Dilated Cardiomyopathy. Mitochondrial. DNA 2013, 24, 46–49. [Google Scholar] [CrossRef]
- Moraes, C.T.; Ciacci, F.; Bonilla, E.; Ionasescu, V.; Schon, E.A.; DiMauro, S. A Mitochondrial TRNA Anticodon Swap Associated with a Muscle Disease. Nat. Genet. 1993, 4, 284–288. [Google Scholar] [CrossRef]
- Nadimi, M.; Daubois, L.; Hijri, M. Mitochondrial Comparative Genomics and Phylogenetic Signal Assessment of MtDNA among Arbuscular Mycorrhizal Fungi. Mol. Phylogenet. Evol. 2016, 98, 74–83. [Google Scholar] [CrossRef]
- Smith, J.M.; Haigh, J. The Hitch-Hiking Effect of a Favourable Gene. Genet. Res. 1974, 23, 23–35. [Google Scholar] [CrossRef]
- Lü, J.; Xia, L.; Liu, X.; Ma, Y.; Li, J.; Ye, Y.; Guo, B. The Mitochondrial Genome of Grapsus albolineatus (Decapoda: Brachyura: Grapsidae) and Phylogenetic Associations in Brachyura. Sci. Rep. 2022, 12, 2104. [Google Scholar] [CrossRef]
- Sekiya, T.; Oda, K.-I. The Altered Patterns of Transfer RNA in SV40-Infected and Transformed Cells. Virology 1972, 47, 168–180. [Google Scholar] [CrossRef]
- Schwarzer, J.; Misof, B.; Tautz, D.; Schliewen, U.K. The Root of the East African Cichlid Radiations. BMC Evol. Biol. 2009, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yoder, A.D. Estimation of the Transition/Transversion Rate Bias and Species Sampling. J. Mol. Evol. 1999, 48, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Gregory, T.R. The Promise of DNA Barcoding for Taxonomy. Syst. Biol. 2005, 54, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Muir, G.; Filatov, D. A Selective Sweep in the Chloroplast DNA of Dioecious Silene (Section Elisanthe). Genetics 2007, 177, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Hershberg, R. Mutation—The Engine of Evolution: Studying Mutation and Its Role in the Evolution of Bacteria. Cold Spring Harb. Perspect. Biol. 2015, 7, a018077. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiteha, Y.G.; Magdy, M. The Evolutionary Dynamics of the Mitochondrial tRNA in the Cichlid Fish Family. Biology 2022, 11, 1522. https://doi.org/10.3390/biology11101522
Fiteha YG, Magdy M. The Evolutionary Dynamics of the Mitochondrial tRNA in the Cichlid Fish Family. Biology. 2022; 11(10):1522. https://doi.org/10.3390/biology11101522
Chicago/Turabian StyleFiteha, Yosur G., and Mahmoud Magdy. 2022. "The Evolutionary Dynamics of the Mitochondrial tRNA in the Cichlid Fish Family" Biology 11, no. 10: 1522. https://doi.org/10.3390/biology11101522
APA StyleFiteha, Y. G., & Magdy, M. (2022). The Evolutionary Dynamics of the Mitochondrial tRNA in the Cichlid Fish Family. Biology, 11(10), 1522. https://doi.org/10.3390/biology11101522





