Simple Summary
This review comprehensively analyzes the molecular mechanism of Suaeda species under salt stress from aspects of physiology, biochemistry, transcriptomics, proteomics, and metabolomics, providing a theoretical basis for understanding the salt tolerance of Suaeda. The unique genetic and physiological characteristics of Suaeda support their high potential for utilization as promising biological resources to improve agriculture under saline conditions.
Abstract
Plant growth and development are inevitably affected by various environmental factors. High salinity is the main factor leading to the reduction of cultivated land area, which seriously affects the growth and yield of plants. The genus Suaeda is a kind of euhalophyte herb, with seedlings that grow rapidly in moderately saline environments and can even survive in conditions of extreme salinity. Its fresh branches can be used as vegetables and the seed oil is rich in unsaturated fatty acids, which has important economic value and usually grows in a saline environment. This paper reviews the progress of research in recent years into the salt tolerance of several Suaeda species (for example, S. salsa, S. japonica, S. glauca, S. corniculata), focusing on ion regulation and compartmentation, osmotic regulation of organic solutes, antioxidant regulation, plant hormones, photosynthetic systems, and omics (transcriptomics, proteomics, and metabolomics). It helps us to understand the salt tolerance mechanism of the genus Suaeda, and provides a theoretical foundation for effectively improving crop resistance to salt stress environments.
1. Introduction
Soil salinization is the main limitation for agricultural economic development, and globally about 10% of soil is affected by high salinity [1]. Salt stress is an important type of abiotic stress, and can cause both osmotic and ionic toxicity in cells, seriously affecting the growth and yield of crops. However, as a halophyte, the genus Suaeda can survive and even grow healthily in high-salinity environments (salt concentrations of 200 mM or greater) [2,3,4]. Therefore, the investigation of the salt tolerance mechanism of the genus Suaeda will provide a molecular basis for better utilization of saline alkali land.
The genus Suaeda, an annual succulent herb of the Amaranthaceae/Chenopodiaceae [5], is a typical halophyte that usually grows in coastal, lakeside, desert, or swamp saline alkali environments [6]. Studies have shown that Suaeda seeds contain oil at approximately 20%, and are rich in unsaturated fatty acids, which have extremely high economic value and health benefits [7]. Suaeda species contain abundant protein, amino acids, minerals, and other essential micronutrients [8]. The tender seedlings are not only nutritious but also taste good [9]. In addition, the genus Suaeda has strong resistance to extreme environments such as cold, drought, and high salinity [10]. It can grow in desert alkaline soil and arid grassland, which means it has been considered a symbolic vegetation in maintaining the ecology of saline–alkali desert areas. As a natural polysalt plant, Suaeda absorbs soluble salt from saline soil to reduce the salt content of the soil [11]. Therefore, the genus Suaeda spp. is a priority plant for the reconstruction of saline and alkaline land.
In order to adapt to the stress of soil salinization and to reduce damage to their growth and reproduction, plants have evolved complex response mechanisms. These include, for instance, upregulating the genes and proteins that participate in salt tolerance, and promoting the production of phytohormones and metabolites that alleviate the toxic effects of salinity. In this review, we summarize the mechanisms of several Suaeda species response to salt stress. This review focuses on recent advances including ion regulation and compartmentation, osmotic adjustment of organic solutes, antioxidant regulation, plant hormones, changes in the pathway of photosynthetic system, transcription factors (transcriptomics), various stress-inducible proteins (proteomics), and the role of metabolites (metabolomics).
2. Ion Regulation and Compartmentation
Salt stress can disturb the ion balance in plants. Halophytes compartmentalize inorganic ions into the vacuolar cytoplasm mainly through transmembrane transport, thus increasing the osmotic pressure in the vacuolar, so that organelles can be protected from the toxic effects of ions, especially salt ions [12]. Suaeda species form an enhanced transmembrane ion gradient through tonoplast Na+/H+ antiporter (NHX), vacuolar membrane ATPase (V-H+-ATPase), vacuolar membrane proton pyrophosphatase (V-H+-PPase), K+ transporter, and chloride channels, maintaining the stability of Na+, K+, and Cl− concentration, to protect Suaeda from salt ions and play an important role in the adapting to a high salt environment (Figure 1).
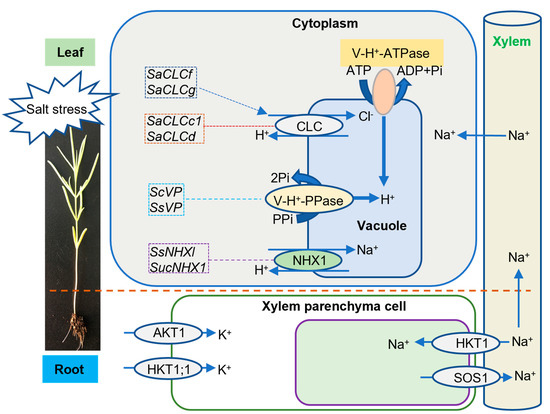
Figure 1.
Schematic diagram of transmembrane transporters transporting Na+, H+, K+, and Cl− in Suaeda.
To relieve harmful accumulation of Na+, the Na+/H+ exchanger and V-H+-ATPase were upregulated in Suaeda salsa (L.) Pall. and Suaeda maritima (L.) Dumort., after NaCl treatment [13,14,15,16]. Similarly, the Na+ influx transporter of Suaeda fruticosa Forssk. [17], the plasma membrane H+-ATPase (PM-H+-ATPase) of S. maritima [15], and the V-H+-PPase in S. salsa leaves and roots [18] were significantly upregulated under saline conditions. Furthermore, the expression levels of V-H+-PPase genes VP in S. salsa and S. corniculate (C.A.Mey.) Bunge were upregulated in roots, stems, and leaves after salt stress induction. After transferring the SsVP gene into Arabidopsis thaliana, the activity of V-H+-PPase was significantly enhanced; overexpression of ScVP gene also increased the accumulation of Na+ in leaves and roots, lengthened the roots, and improved the salt tolerance of transgenic plants [19,20]. These results indicated that the enhanced activity of V-H+-PPase can translocate more Na+ to vacuoles, which plays an important role in protecting plants from salt ions and adapting to a high salt environment.
NHX1, high-affinity K+ Transporter (HKT), and Salt Overly Sensitive 1 (SOS1) are the major transporters involved in Na+ accumulation in plants [21]. Vacuolar NHX1 is a membrane protein that plays an important role in the exchange of Na+ for H+ across the vacuolar membrane, and the segregation of Na+ into vacuoles [22]. It was found that the tonoplast NHX1 was upregulated in S. fruticosa, S. salsa, S. maritima, and S. corniculata after salt stress [15,17,23,24]. After the SsNHX1/SucNHX1 gene was transferred into rice, maize, and Arabidopsis thaliana, the salt tolerance of the transgenic plants was significantly improved [21,23,24,25]. Heterologous expression of the gene in poplars also enhanced the salt tolerance of transgenic poplar trees, which may result from the high expression of SsNHX1/SucNHX1 gene, promoting Na+ accumulation in vacuoles, thus alleviating the effect of salt stress on cells [20,26]. These results suggested that Suaeda species might share similar mechanisms underlying ionic balance (Na+ and H+) in response to saline stress.
HKT1 and SOS1 are located on the cellular plasma membrane, and have opposite roles in controlling Na+ influx and efflux, respectively, across the plasma membranes of xylem parenchyma cells in the roots [21]. SsHKT1 encodes an Na+-selective transporter that is preferentially expressed in the root xylem parenchyma and pericycle cells in S. salsa [21]. SsSOS1 was highly expressed by high concentrations of NaCl (300 mM) in S. salsa roots [27]. SsHKT1 coordinated with SsNHX1 and SsSOS1 to maintain Na+ accumulation under salt stress conditions by reducing Na+ retrieval from the xylem sap in S. salsa [21].
Although Na+ has been shown to suppress K+ influx in many plants, external Na+ treatment (25–400 mM) enhanced the growth of Suaeda species, including Suaeda glauca (Bunge) Bunge, S. salsa, S. fruticosa, and S. maritima; K+ concentrations in these plants were differential [28,29,30,31,32,33,34,35]. With increasing external NaCl concentrations, Na+ concentrations increased in the leaves of S. fruticosa [28] and S. maritima [32], K+ concentration decreased in S. fruticosa leaves [28], and K+ content was relatively stable in S. maritima shoots [32,34,35]. S. salsa plant tissues can accumulate large amounts of Na+ and K+ under high salinity conditions [29,30,31]. For S. glauca, there was no competitive inhibition between Na+ and K+ absorptions [33]. Therefore, the role of Na+ and K+ in the Suaeda tissues may be to maintain water absorption by maintaining osmolality, which is essential for Suaeda plants’ survival in high-salinity conditions. Thus, the Na+ and K+ concentrations might be key requirements for growth of Suaeda species in highly saline soils.
Shao et al. [36] found that SsHKT1;1, a K+ transporter within S. salsa, was involved in salt tolerance by participating in cytosolic cation homeostasis, particularly mediating root K+ uptake and transport under salinity. HKT family and shaker AKT1-like channels in plants are considered the main channels that mediate K+ influx into root cells and correlate with salt tolerance [37,38,39]. The SsAKT1 gene, encoding the inward rectifying K+ channel in S. salsa, significantly increased the transcript levels in roots with the increase of external Na+ concentration (25–250 mM) for 6 h [40]. Therefore, it may play an important role in salt tolerance of S. salsa by mediating both high- and low-affinity K+ uptake across different K+ concentration conditions and to the maintenance of K+ nutrition under salinity [40].
Although one HKT1-like transporter of S. fruticosa was found to be downregulated under salinity conditions, NHX, HKT, and PM-H+-ATPase showed no significantly different expression between salt-treated and control samples in S. glauca [41], suggesting that different Suaeda species may have different proteins regulating the ion balance. Therefore, Suaeda species may have certain similar and particular pathways that help them adapt to saline conditions.
In Suaeda altissima (L.) Pall., the expression of chloride channel (CLC) family genes SaCLCc1, SaCLCd, SaCLCf and SaCLCg in leaves increased with the increase of salt concentration, consistent with the accumulation of Cl− in leaf cells. The results indicated that SaCLCc1, SaCLCd, SaCLCf and SaCLCg proteins may be involved in the separation of Cl− in organelles, and may be involved in the mechanism of salt tolerance [42,43]. In addition, the complementation assay and bioinformatic analyses indicated that SaCLCc1 and SaCLCd proteins are Cl−/H+ antiporters, while the SaCLCf and SaCLCg proteins are likely Cl− channels [42,43].
3. Osmotic Adjustment of Organic Solutes
In saline environments, halophytes can maintain intracellular osmotic balance by accumulating organic solutes such as sugars, alcohols, amino acids and their derivatives (proline, betaine, etc.), in addition to inorganic ions. These substances are soluble in water, are not toxicity, and do not intervene with cells’ biochemical reactions or various metabolic processes, even if accumulated at high concentrations. In this way, they not only protect the activity of enzymes in cells, but are also used as osmotic agents to maintain the osmotic balance of plant cells and improve the resistance of plants to salt stress [44].
Betaine is an important secondary metabolite that can be synthesized by cells for protection against osmotic stresses associated with high salinity [45]. In high-salt environments, plants store most of their NaCl in vacuoles through the Na+/H+ antiporter, and achieve equivalent osmotic potential by synthesizing compatible solutes such as glycine betaine in the cytoplasm. In the presence of NaCl, glycine betaine accumulated to maintain osmotic adjustment, playing an important role for Suaeda plants grown under high Na+ concentrations, inluding S. salsa [46], S. maritima [47], S. fruticosa [28], Suaeda aralocaspica (Bunge) Freitag & Schütze [48], Suaeda eltonica lljin [48], and Suaeda heterophylla (Kar. & Kir.) Bunge ex Boiss [48]. Thus, glycine betaine functions as an osmolyte to lower the plant’s water potential in order to protect membranes under high salinity conditions [48].
Under salinity conditions, proline also accumulated in S. maritima [47], Suaeda physophora Pall. [49], and S. salsa [50]. It was found that species with glycine betaine accumulators exhibited low proline content, and vice versa [51]. The enhancement of proline and glycine betaine may stimulate the expression of salt-tolerance proteins such as SKP1A in S. maritima [52]. In S. salsa, glycine betaine may play a more important role than proline in osmotic adjustment under high-salinity conditions [49]. Thus, as compatible solutes, betaine and proline appear to have different osmotic adjustment effects among Suaeda species.
Betaine is synthesized through converting choline into betaine aldehyde by choline monooxygenase (CMO), then catalyzing with betaine aldehyde dehydrogenase (BADH) [53]. Phosphoethanolamine methyltransferase gene PEAMT is related to betaine synthesis in S. salsa [54]. The CMO gene was upregulated in S. salsa [14,55,56], Suaeda aegyptiaca (Hasselq.) Zohary [57], and S. maritima [15] after salt stress, suggesting that Suaeda accumulates betaine to maintain osmotic balance. Under salt conditions, the BADH gene was induced to express in S. salsa [58,59], S. maritima [15], and S. corniculate [60] seedlings, which was involved in the biosynthesis of betaine to maintain the osmotic balance and enhance salt tolerance. Therefore, the CMO, BADH, and PEAMT genes involved in the synthesis of osmolytes are upregulated under salt stress. They may be positive regulators in response to NaCl [60], which can improve the tolerance of Suaeda cells to salt stress by maintaining the cells’ osmotic balance.
It has been reported that the expression levels of proline synthesis key enzyme gene SsP5CS (Δ1-dihydropyrrole-5 carboxylic acid synthase) and inositol synthesis key enzyme gene SsINPS are significantly increased in S. salsa in saline environments [61,62] Under high salinity (500 mM), S. salsa can accumulate organic acids, soluble sugars, lipid metabolites, and unsaturated fatty acids [63], as well as sucrose [56], helping the plant to deal with osmotic stress and increasing its nutritional value. However, in S. corniculata, the soluble sugars were downregulated after salt stress [64,65]. These results suggest that Suaeda plants enhance their resistance to osmotic stress by regulation of osmolytes under salinity conditions [56].
4. Antioxidant Capacity Regulation
Under salt-stress conditions, excessive reactive oxygen species (ROS) including hydrogen peroxide, hydroxyl radicals, and oxygen radicals are accumulated in plant cells, which can damage the cells’ macromolecules and membrane structures. Antioxidants such as superoxide dismutase (SOD), glutathione transferase (GST), ascorbic acid peroxidase (APX), glutathione reductase (GR), peroxidase reductase (PrxR), ascorbic acid glutathione (ASA)-glutathione (GSH) cycle enzyme, or catalase (CAT) can remove all kinds of free radicals and enhance the defense ability of cells against oxidative stress [66]. Suaeda was found to be able to maintain the balance between the formation and elimination of ROS by increasing the activity of antioxidant enzymes, for instance SOD [67,68], CAT [56,69], APX [69] and GPX [56].
With the increase of salt concentration, activities of Mn-SOD, Fe-SOD, and CuZn-SOD were detected in S. salsa leaves [67]. The activity of SOD in S. salsa and S. maritima increased significantly under saline conditions [67,68]. In medium containing 400 mM NaCl, the SOD activity of Suaeda japonica Makino leaves increased, resulting from the concentration of the substrate superoxide anion and the production of O2•− under salt stress [70,71].
GST gene expression levels were greatly increased in roots of S. maritima upon salt treatment [72]. However, in S. fruticosa, the level of glutathione increased with high salt (900 mM) treatment, and also with 0 mM NaCl treatment, while it decreased with 300 mM NaCl [73]. After transforming Arabidopsis with S. salsa’s GST gene, the salt tolerance of transgenic Arabidopsis was improved, possibly due to the overexpression of the S. salsa GST gene in Arabidopsis plants alleviating the effect of reactive oxygen free radicals and enhancing the tolerance of cells to salt stress [74].
Dehydrins (DHN), known to be chaperones, could bind to the hydrophilic sites of proteins to scavenge oxygen free radicals, thereby reducing the peroxidation damage caused by stress conditions and enhancing the resistance of plants [75,76]. The expression of the dehydrin gene SsDHN in S. salsa is induced by salt stress [77]. Overexpression of the S. glauca DHN gene in yeast could enhance tolerance to salt stress [78]. These results indicated that S. salsa can scavenge various reactive oxygen free radicals through antioxidant substances, and enhance the tolerance of cells to salt stress, enabling adaptation to the saline–alkali environment.
After the application of NaCl, the activity of CAT increased significantly in S. salsa and S. maritima [68,69]. The expression levels of the Sscat1 and APX genes in S. salsa increased significantly with salt stress [79,80]. Overexpression of the APX gene in Arabidopsis can increase APX activity, lower the H2O2 content, and reduce cell membrane damage caused by salt stress [3,81].
It has also been reported that in S. salsa after treatment with 200 mmol/L NaCl for seven days, the activities of GR in the chloroplast matrix and thylakoid were increased, along with ASA and GSH content, while H2O2 content and membrane lipid peroxidation decreased [69]. These results indicated that the increase of GR activity promoted the production of GSH, enabling the scavenging of reactive oxygen species. Therefore, increased GR content may be an important reason for the decrease of H2O2 content in S. salsa leaves [67].
In conclusion, by increasing the activity of its antioxidant enzymes, Suaeda can maintain the balance between the formation and elimination of ROS. Plant antioxidant systems are generally classified into enzymatic and non-enzymatic systems. The enzymatic defense systems in Suaeda include SOD [67,68], CAT [56,69], APX [69], and GPX [56]. Non-enzymatic defense systems in Suaeda are AsA and GSH [71], etc. Under salinity conditions, the increase of ROS results in the relatively inadequate antioxidant scavenging capacity of antioxidant enzymes, leading to oxidative stress. Suaeda scavenges ROS produced by salt stress mainly through the synergy of SOD activation, the CAT, GPX, and PrxR pathways, and the ASA-GSH cycle enzyme [71]. Therefore, the activities of various enzymes and the content of non-enzyme substances involved in the process of scavenging H2O2 can reflect the salt resistance of Suaeda under salt stress.
5. Secretion of Plant Hormones
Phytohormones integrate various signals in maintaining responses to salt stress and other stresses [82]. Salt stress affects the reproductive growth and yield of plants by regulating the secretion of plant hormones including indole acetic acid (IAA), gibberellin (GA), cytokinins (CTK), ethylene (ETH), abscisic acid (ABA), etc., but promotes the reproductive growth of euhalophytes. CTK and IAA can increase the salt tolerance of seeds in Arabidopsis [62]. Under salt stress, the inhibition of hormone synthase activity led to reduction or cessation of IAA and CTK synthesis, which delayed plant growth, but increased the content of ABA and ETH in rapeseed [83].
Exogenous ABA pretreatment can increase chlorophyll pigment content and accumulation of inorganic osmolytes, thus reducing the damage of salt stress and increasing the general growth rate of S. maritima [52]. Salt stress can also induce the accumulation of ABA in S. salsa seeds. Under salt stress, 1-aminocyclopropane-1-carboxylate (ACC, the direct precursor of ethylene), GA4, and 6-benzyladenine (BA) can promote the germination of seeds, which indicates that these hormones can reduce the impact of salt stress on seeds and improve the ecological adaptability of S. salsa, S. maritima, and Suaeda prostrata Pall. seeds to a saline alkali environment [84,85,86]. High salinity inhibited seed germination by decreasing the levels of GA4 in S. salsa [85]. During the later stages of plant vegetative growth, NaCl treatment can significantly increase the content of endogenous GA3, GA4, ABA, and brassinolide (BR) in the stems of S. salsa plants [87]. In the flowering stage, NaCl treatment significantly increased the content of GA3, GA4, IAA, and zeatin (ZR) in floral organs compared with the control [87]. In response to salt stress, ETH-related pathways are upregulated in S. glauca and S. maritima [15,41]. The auxin, ETH, and jasmonic acid (JA) signaling transduction pathways were all upregulated in S. salsa after saline treatment, and are important to gene regulations of ion transport and antioxidation [87]. In addition, genes related to the biosynthesis of ZR, IAA, GA, BR, and ABA, and to plant hormone signal transduction, including genes encoding CYP735A, CYP85A, GID1, NCED, PIF4, AHP, TCH4, SnRK2, and ABF, were upregulated in S. salsa treated with NaCl. Downregulation of gibberellin 2-oxidase 2 was observed in S. fruticose after 300 mM salt treatment [17].
Some plant hormones positively regulate salt tolerance, while others play a negative role. GA3 was found to stimulate growth at all salinities for S. maritima and S. rostrata, while kinetin (KT) proved to be inhibitory to plant growth at higher salinities [86]. These results indicate that the synergistic upregulation of genes involved in plant hormone synthesis and signal transduction contributes to the reproductive growth of S. salsa under salt stress [87]. Therefore, in response to salt stress, plants build a defense system by orchestrating the synthesis and signaling pathways of various hormones via multiple crosstalks.
6. Changes in the Pathway of Photosynthetic System
Halophytes can fit into or resist the influence of saline environments by regulating photosynthesis and metabolism. Suaeda is a kind of halophyte that grows in high salt environments and extreme high-tide zones. Photosynthesis plays an important role in the accumulation of protein biomass within halophytes in saline soil. The halophytes Suaeda (Chenopodiaceae) include species with both C3 and C4 photosynthetic pathways [88].
High salt stress (200–500 mM NaCl) prompted decent protection of the light response system in S. salsa, maintaining the structure of the light system, promoting light-energy transmission, and improving the activities of related enzymes [63,89]. After 200 mM NaCl treatment, the expression of carbon-assimilation-related enzyme genes SsFNR, SsRbcl, SsRbcs, SsRCA, SsPGK, and SsGAPDH increased significantly, indicating that carbon-assimilation-related enzymes may play an important role in promoting the photosynthesis of S. salsa [63].
Under salinity conditions, the concentration of photosynthetic pigments chlorophyll a, chlorophyll b, and total chlorophyll in the leaves of Suaeda schimperi Moq., Suaeda vermiculata Forssk. Ex J.F.Gmel., Suaeda monoica Forssk. Ex J.F.Gmel. were different [90]. The chlorophyll content in S. salsa was improved after NaCl stress [90]. When cultured with 200 mM NaCl for 14 days, the photosynthetic capacity including net photosynthetic rate (Pn), electron transfer rate, NADPH level, activities of ferredoxin-NADP oxidoreductase, ribulose-1, 5-bisphosphate carboxylase (Rubisco), and Rubisco activase were improved significantly in S. salsa, and 500 mM NaCl had no adverse effect on those parameters [31,90]. Low salt stress had little effect on the photosynthesis of S. corniculata seedlings, while high salinity inhibited their photosynthesis [91]. The maximum photochemical quantum yield Fv/Fm of photosystem II (PSII), the photochemical quenching coefficient qP, and the non-cyclic photosynthetic electron transfer rate RE, T of PSII were not affected by low salt stress, but showed a downward trend in S. corniculata, S. salsa, and S. aegyptiaca [58,91,92]. All these trends indicate that the seedlings of Suaeda can adapt to a high salt environment by changing their rates of photosynthesis.
SsPsaH is a member of the H subunit of the PSI reaction center in S. salsa, and its expression level increases with salt stress [93]. Overexpression of SsPsaH in recombinant yeast can enhance the tolerance of transformants to salt stress [93]. The expression of S. salsa glycerol3-phosphate acyltransferase (SsGPAT) was also increased by salt stress; compared with the wild type, high salinity prompted a smaller reduction in chlorophyll content, PSII photochemical efficiency, photosystem I (PSI) redox activity (δI/Io), and unsaturated fatty acid content of phosphatidylglycerol (PG) in Arabidopsis thaliana seedlings. This indicates that the overexpression of the SsGPAT gene in Arabidopsis can enhance the salt tolerance of PSII and PSI under salt stress by upregulation of the unsaturated fatty acid content of PG, thereby alleviating the photoinhibition of PSII and PSI [94]. PS II in S. salsa shows a high resistance to low salinity [92], making plant growth more adaptable under low salinity conditions (8 ppt). Therefore, increased photosynthetic activity may play a critical role in the biomass enhancement of Suaeda under saline conditions [90].
7. Omics Approaches
Omics approaches, i.e., transcriptomics, proteomics, and metabolomics, play an important role in the study of plant salt tolerance [95]. Omics techniques, including characterization of transcription factors, proteins and metabolites involved in salt tolerance, have been applied to understand response mechanisms in plants and utilized for generating salt-tolerant crops.
7.1. Transcriptomics
The responses of plants to salt stress trigger multi-factor synergistic effects. The related transcription factor can regulate the expression of a series of genes, thereby enhancing the resistance of plants to the saline environment. The regulation of transcription factors in genes can be realized through specific binding with cis elements to initiate the transcription expression of the gene [63,96]. According to the different DNA binding domains, plant transcription factors can be divided into different families. Among these, the transcription factor families related to salt-stress response mainly include NAC, AP2/EREBP, HB, MYB, BZIP/HD-Zip, and WRKY. Under saline conditions, transcription factors can be regarded as the virtual switches that directly upregulate or downregulate the expression of salt-stress-related genes.
Transcriptome analysis of S. salsa showed that HB, MYB, and bZIP transcription factors were regulated by salt stress, and the transcriptional regulation of HB-7 and MYB78 was found to alleviate the damage of salt stress in S. salsa [97]. In S. salsa leaves, MYC2 was significantly upregulated after saline treatment compared with the control [87]. RNA sequencing analysis revealed that WRKY and bHLH transcription factors involved in salt tolerance were upregulated in S. glauca and S. rigida [41,98,99]. MYB genes were also elevated in S. maritima and S. glauca in response to salt stress [15,100]. Study of the transcription factors showed that MYB07, MYB37, and BZIP59 played important roles for regulation of salt tolerance in S. fruticosa [101].
Salt stress can induce the expression of transcription factor genes SlNAC1, SlNAC2, SlNAC7, and SlNAC8 in S. salsa. Compared with wild-type Arabidopsis, salt stress can promote the germination and survival rates of SlNAC1 and SlNAC8 transgenic Arabidopsis, respectively, but was found to inhibit root growth in the transformants [102,103,104]. Overexpression of SlNAC2 and SlNAC7 in Arabidopsis can enhance tolerance to salt stress [103,105]. Moreover, the overexpression of SlNAC8 in transgenic plants also enhanced the expression of stress response genes RD20, GSTF6, COR47, RD29A, RD29B, and NYC1 [104]. In brief, SlNAC1, SlNAC2, SlNAC7, and SlNAC8 transcription factors may make contributions to changes in the physiological and biochemical characteristics of plants by regulating the expression of stress-responsive genes, thus enhancing the resistance of plants to salt stress.
The expression levels of AP2 were upregulated in germinating seeds of S. glauca when exposed to different NaCl concentrations [100]. In S. salsa leaves, ERF1/2 were significantly upregulated by saline treatment compared with the control [87]. In S. salsa, DREB protein belongs to the CBF/DREB transcription factor, and salt stress can significantly promote SsDREB gene expression. Overexpression of the SsDREB gene in tobacco can enhance the salt tolerance of transgenic plants [106]. Two DREB genes of S. salsa can respond to high salt stress through independent ABA pathways, while SsCBF1 may be involved in the regulation of high salt stress through ABA signaling [107,108].
In short, transcription factors play a key role in the responses of plants to salt stress. By overexpressing transcription factor genes in transgenic plants, the expression of transcription-factor-specific binding genes can be regulated, in order to obtain stronger salt tolerance. Therefore, salt-stress-responsive transcription factors can be used as an important tool for the genetic engineering of plants’ salt tolerance.
7.2. Proteomics
Proteomics has become a very important technique in the post-genomic era [109], serving as a powerful tool for describing complete protein changes at organ, tissue, cell, and organelle levels under various stress conditions [110]. Therefore, proteomic investigation can reveal the potential associations between protein expression and plant stress acclimation. Proteomic methods have been widely used for investigating specific genes and proteins contributing to salt tolerance and survival in saline conditions [57,67].
The salinity-responsive proteins belong to various functions including ROS scavenging, ionic and osmotic regulation, signal transduction, and photosynthesis [111]. In S. maritima, photosynthesis, heat shock proteins, peroxidase, expansins, signaling processes, and modulation of transcription/translation were modulated by salinity [112]. In S. salsa, three upregulated and six downregulated proteins were identified, involved in photosynthesis, energy metabolism, stress, and defense [109]. The metabolism proteins of S. salsa were involved in the pentose phosphate pathway, polyamine biosynthesis, amino acid biosynthesis, and isoprenoid biosynthetic processes [109]. In S. corniculate, 10 proteins were observed as being differentially expressed under NaCl treatment [64]. According to KEGG pathway analysis, these proteins were involved in carbohydrate metabolism, energy metabolism, photosynthesis, nucleotide metabolism, protein synthesis, stress, and defense, or were unknown [64].
Comparative proteomic analysis was used to determine the proteomic profiles of S. salsa exposed to salinity [109]. The ATP synthase CF1 alpha subunit and ATPase subunit were downregulated by salinity exposure, implying disturbance in energy metabolism [109]. Salt treatment upregulated plasma membrane aquaporins [113], suggesting that S. salsa maintains homeostasis and ion distribution by increasing leaf succulence and compartmenting the ions. Mitogen-activated protein kinase (MPK6), ethylene-insensitive protein 2 (EIN2), and ethylene-insensitive protein 3 (EIN3) were significantly upregulated in S. salsa leaves after saline treatment compared with the control [87]; the result was consistent with the observation that ethylene signaling is indispensable for tolerance to saline stress in plants [114].
Protein pattern analysis revealed that 22 kDa and 55 kDa proteins occurred in salt-treated S. maritima leaves [52]. The enhancement of nontoxic metabolites may stimulate the expression of salt-tolerance proteins in S. maritima [52]. Acetolactate synthase 1 and histone H4 were up- and down-accumulated in S. maritima at the lower (200 mM) and higher (500 mM) NaCl dosages, respectively [112]. In S. maritima, cytochrome b6f complex, cytosolic, expansin-B1, chloroplastic GAPDH, and the chloroplastic ATP synthase subunit α were downregulated with salt treatment, resulting in a reduction of photosynthetic activity [112]. Thus, proteomics is helpful to understand the characterization of interactions and the response to salt stress in Suaeda plants.
7.3. Metabolomics
Metabolites, compatible solutes, and bioactive compounds are biomolecules produced by plants under natural or stressed conditions. Under stress conditions, plant systems need to regulate metabolite levels to maintain basal metabolism and achieve homeostasis [115]. Metabolomics is a potent approach to the identification and quantification of all low-molecular weight metabolites required by plants in response to abiotic stress. It can be used to study metabolic pathways or metabolites associated with salt-stress tolerance [116]. In halophytes, metabolites involved in salt tolerance include glycine betaine, proline, pinitol, mannitol, sorbitol, O-methyl muco-inositol, inositol, and polyamines [116].
In plant metabolomics, metabolites are divided into primary and secondary categories. During plant stress, primary metabolites directly contribute to the accumulation of compatible solutes, such as amino acids, sugars, and sugar alcohols, to cope with osmotic stress [117]. Under saline conditions, 61 primary metabolites were detected in the leaves of S. monoica and S. fruticosa species [117]. These metabolites included amino acids, sugars, sugar acids, fatty acids, different compounds, and flavonoid (kaempferol) groups [117]. Serine concentration was higher in S. monoica compared with S. fruticosa under saline conditions [117]. In S. corniculate, 21 metabolites were identified under conditions of salinity, including amino acids, carbohydrates, organic osmolyte, and intermediates in the tricarboxylic acid (TCA) cycle, among others (e.g., ethanol, dimethylamine O-phosphocholine, and choline) [65].
Under saline conditions, amino acids including valine, alanine, glutamate, tyrosine, leucine, isoleucine, and phenylalanine were decreased in the aboveground parts of S. salsa and S. corniculata seedlings [58,64]. Furthermore, the total protein content in the aboveground parts of S. salsa seedlings decreased with increasing concentrations of salinity (0 mM–170 mM–500 mM) [58]. However, in the S. salsa root tissues, the metabolic responses were different from the aboveground parts of the seedlings. The proline and citrate in the root tissues were uniquely increased, and the branched-chain amino acids, lactate, choline, phosphocholine, glutamine, and fructose were uniquely decreased. The differences of metabolic responses between the roots and aboveground parts of S. salsa seedlings suggest different regulating mechanisms in various tissues under salt treatment [58]. Moreover, studies have shown that high salinity can lead to accumulation of more amino acids (500 mM > 200 mM > 0 mM) in S. salsa [63], and increases in leucine, isoleucine, valine, and glutamine in S. corniculata seedlings [65].
As osmolytes, betaine and proline can be synthesized for protection against salinity stress [45]. In S. salsa [58] and S. corniculata [64], betaine was significantly elevated in the aboveground parts of seedlings under salt stress, and CMO was induced in S. salsa [55] and S. aegyptiaca [57] after salt stress, suggesting that Suaeda accumulates betaine to maintain osmotic balance. The Suaeda species exhibited significant variation in amino acid biosynthesis under similar salinity conditions. For example, S. monoica and S. schimperi accumulated significantly higher foliar proline than S. vermiculata, suggesting that proline is an important compatible osmolyte in S. monoica and S. schimperi species [90].
The metabolic processes of Suaeda include primary and secondary metabolism, and the accumulation of metabolites is related to the osmotic tolerance, energy supply, and nutritional value of Suaeda species. Changes in metabolite content may play an important role in maintaining cell osmotic potential, protecting cell membrane structure, and promoting resistance by destruction of ROS. The related studies showed that there were differences in the accumulation of metabolites among Suaeda species under different salt-stress conditions. Therefore, the metabolic mechanisms of compounds in Suaeda species are complex and deserve to be further investigated.
8. Conclusions and Perspectives
Salinization is an important issue for global agricultural productivity and food security, which seriously affects crop growth and yield. Improving crops’ salt tolerance is the most direct and effective way to solve this problem. As halophytes, the genus Suaeda can grow healthily in high salt environments. Over twenty species of Suaeda have been described for their ability to survive under high salinity environments [64]. For example, S. salsa grew equally well with 400 mM NaCl as with 10 mM NaCl, and its growth optimal concentration was found to be 200 mM NaCl [31]. Furthermore, Suaeda seedlings have important economic value, the fresh branches can be used as vegetables, and its seed oil is edible and rich in unsaturated fatty acids [3]. Therefore, Suaeda is a prime salt-tolerant model plant with great economic value. Elucidating the salt-tolerance mechanism of Suaeda is helpful for developing salt-tolerant plant varieties and making effective use of saline land resources.
The salt-tolerance mechanism of the genus Suaeda is very complicated, involving cells, tissues, and organs, and the whole plant, being the result of the synergy of plant physiology, biochemistry, molecular, transcript, protein, and metabolic level (Figure 2). This review has analyzed the genes (Table 1), enzymes, proteins, and metabolites that participate in salinity adaption, and investigated the roles of these factors in Suaeda ion transport (Figure 1), osmotic regulation, free radical scavenging, hormone regulation, and photosystem regulation, thereby summarizing the molecular mechanisms of salt tolerance in Suaeda (Figure 2). At present, a series of genes related to salt tolerance have been cloned and preliminary functional verification has been carried out. Further research should be undertaken to consider methods of fully improving adaptable properties in terms of salt-stress response, and coordinatively regulating the multiple salt-tolerance genes in the genus Suaeda. The systematic combination of various “omics”, including genomics, transcriptomics, proteomics, and metabolomics, is necessary to understand the molecular networks underlying Suaeda species’ responses to salt stress. The integrated application of multiple “omics” technologies and precise genome editing by the CRISPR/Cas9 system can be further conducted in future studies, which will lay a foundation for elucidating the molecular regulation mechanism of the genus Suaeda, to enable further salt-tolerant plant breeding. Overall, this review might enhance an integrated comprehensive understanding of salt tolerance. These results may provide elite genetic resources for the modification of salinity-resistant crop species, and improve the efficiency of saline–alkali land utilization.
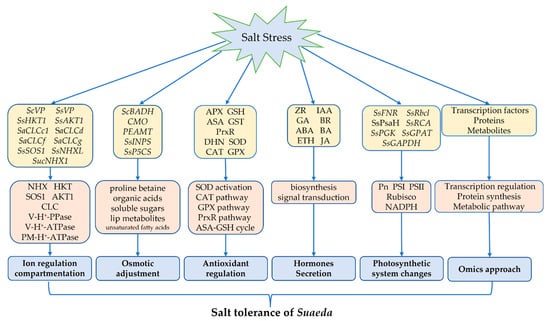
Figure 2.
Researched advances of molecular mechanism in the genus Suaeda.

Table 1.
The genes in Suaeda species and their probable functions.
Author Contributions
The authors W.Y., Q.L. and Y.W. (Yong Wang) discussed and created the review’s outline, W.W., Y.W. (Yiheng Wang), N.Z., L.W. and B.W. collected the related references. W.Y. wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (31201245), and Financial Seed Industry Innovation Project of Tianjin Academy of Agricultural Sciences (2022ZYCX015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Song, J.; Shi, W.; Liu, R.; Xu, Y.; Sui, N.; Zhou, J.; Feng, G. The role of the seed coat in adaptation of dimorphic seeds of the euhalophyte Suaeda salsa to salinity. Plant Species Biol. 2017, 32, 107–114. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, B.S. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 2018, 45, 350–361. [Google Scholar] [CrossRef]
- Wang, X.; Shao, X.; Zhang, W.; Sun, T.; Ding, Y.; Lin, Z.; Li, Y. Genus Suaeda: Advances in phytology, chemistry, pharmacology and clinical application. Pharmacol. Res. 2022, 106203, 1895–2021. [Google Scholar]
- He, Q.; Altieri, A.H.; Cui, B.S. Herbivory drives zonation of stress-tolerant marsh plant. Ecology 2015, 96, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, Y.; Duan, H.; Liu, R.; Song, J. Traits of fatty acid accumulation in dimorphic seeds of the euhalophyte Suaeda salsa in saline conditions. Plant Biosyst. 2019, 153, 514–520. [Google Scholar] [CrossRef]
- Zhao, H.L. Study on edible value of Suaeda salsa l. pall. J. Anhui Agricul. Sci. 2010, 38, 14350–14351. [Google Scholar]
- Zhang, J.Y.; Li, M.H.; Xu, L.M.; Wang, Z.J. Effect of Suaeda seed oil on blood-fat and immunologic function of mouse. Occup. Health 2008, 24, 1529–1530. [Google Scholar]
- Guo, J.; Du, M.; Tian, H.; Wang, B. Exposure to high salinity during seed development markedly enhances seedling emergence and fitness of the progeny of the extreme halophyte Suaeda salsa. Front. Plant Sci. 2020, 11, 1291. [Google Scholar] [CrossRef]
- Huang, W.; Li, Z.; Qiao, H.; Li, C.; Liu, X. Interactive effect of sodium chloride and drought on growth and osmotica of Suaeda salsa. Chin. J. Eco-Agric. 2008, 16, 173–178. [Google Scholar] [CrossRef]
- Guo, F.; Tang, Z. Enhanced H+ transport activity of tonoplast vesicles isolated from roots of salt-tolerant mutant of wheat under NaCl stress. Chin. Sci. Bull. 1999, 44, 1198–1201. [Google Scholar] [CrossRef]
- Qiu, N.; Chen, M.; Guo, J.; Bao, H.; Ma, X.; Wang, B. Coordinate up–regulation of V-H+-ATPase and vacuolar Na+/H+ antiporter as a response to NaCl treatment in a C3 halophyte Suaeda salsa. Plant Sci. 2007, 172, 1218–1225. [Google Scholar] [CrossRef]
- Guo, J.; Dong, X.; Han, G.; Wang, B. Salt-enhanced reproductive development of Suaeda salsa L. coincided withion transporter gene upregulation in flowers and increased pollen K+ content. Front. Plant Sci. 2019, 10, 333. [Google Scholar] [CrossRef]
- Gharat, S.A.; Parmar, S.; Tambat, S.; Vasudevan, M.; Shaw, B.P. Transcriptome analysis of the response to NaCl in Suaeda maritima provides an insight into salt tolerance mechanisms in halophytes. PLoS ONE 2016, 119, e0163485. [Google Scholar] [CrossRef]
- Shrikanth, K.S.; Parida, A.K.; Girivasan, K.P. Differentially expressed long-term salinity responsive sequences in halophyte Suaeda maritima L. Dumort. Eur. J. Biol. Biotechnol. 2022, 31, 59–67. [Google Scholar] [CrossRef]
- Diray-Arce, J.; Clement, M.; Gul, B.; Khan, M.A.; Nielsen, B.L. Transcriptome assembly, profiling and differential gene expression analysis of the halophyte Suaeda fruticosa provides insights into salt tolerance. BMC Genom. 2015, 16, 353. [Google Scholar] [CrossRef]
- Wang, B.; Lüttge, U.; Ratajczak, R. Effects of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J. Exp. Bot. 2001, 52, 2355–2365. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Wang, N.; Dong, Y.; Fan, X.; Liu, X.; Yang, J.; Li, H. Cloning of a vacuolar H+-pyrophosphatase gene from the halophyte Suaeda corniculata whose heterologous overexpression improves salt, saline-alkali and drought tolerance in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 731–742. [Google Scholar] [CrossRef]
- Guo, S.; Yin, H.; Zhang, X.; Zhao, F.; Li, P.; Chen, S.; Zhao, Y.; Zhang, H. Molecular cloning and characterization of a Vacuolar H+-pyrophos-phatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Mol. Biol. 2006, 60, 41–50. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Duan, H.; Yin., X.; Cui, Y.; Chai, W.; Song, X.; Flowers, T.; Wang, S. SsHKT1;1 is coordinated with SsSOS1 and SsNHX1 to regulate Na+ homeostasis in Suaeda salsa under saline conditions. Plant Soil 2020, 449, 117–131. [Google Scholar] [CrossRef]
- Hasegawa, P.M. Sodium Na+ homeostasis and salt tolerance of plants. Environ. Exp. Bot. 2013, 92, 19–31. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Li, Y.; Ding, J.; Du, H.; Zhao, Z.; Zhou, L.; Liu, C.; Gao, S.; Cao, M.; et al. Overexpression of the Suaeda salsa SsNHX1 gene confers enhanced salt and drought tolerance to transgenic Zea mays. J. Integr. Agric. 2018, 17, 2612–2623. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, F.; Sun, D.; Wang, N.; Dong, Y.; Liu, W.; Liu, X.; Yao, N.; Chen, H.; Chi, M.; et al. Cloning and characterization of SucNHX1, a novel vacular Na+/H+ antiporter from the halophyte Suaeda corniculata that enhances the saline-alkali tolerance in Arabidopsis by its overexpression. Plant Cell Tissue Organ Cult. 2018, 134, 395–407. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, Z.; Zhang, Q.; Zhao, Y.; Zhang, H. Analysis of the physiological mechanism of salt-tolerant transgenic rice carrying a vacuolar Na+/H+ antiporter gene from Suaeda salsa. J. Plant Res. 2006, 119, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Wang, B. Cloning and expression of subunit H of V-H+-ATPase in vacuole membrane in the leaves of the halophyte Suaeda salsa under salt stress. Acta. Bot. Boreal. Occident. Sin. 2006, 26, 63–67. [Google Scholar]
- Liu, Q.; Liu, R.; Ma, Y.; Song, J. Physiological and molecular evidence for Na+ and Cl− exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 2018, 146, 1–7. [Google Scholar] [CrossRef]
- Khan, M.A.; Ungar, I.A.; Showalter, A.M. The effect of salinity on the growth, water status, and ion content of a leaf succulent perennial halophyte, Suaeda fruticosa L. Forssk. J. Arid. Environ. 2000, 451, 73–84. [Google Scholar] [CrossRef]
- Mori, S.; Suzuki, K.; Oda, R.; Higuchi, K.; Maeda, Y.; Yoshiba, M.; Tadano, T. Characteristics of Na+ and K+ absorption in Suaeda salsa L. Pall. Soil Sci. Plant Nutr. 2011, 573, 377–386. [Google Scholar] [CrossRef]
- Mori, S.; Akiya, M.; Yamamura, K.; Murano, H.; Arao, T.; Kawasaki, A.; Higuchi, K.; Maeda, Y.; Yoshiba, M.; Tadano, T. Physiological role of sodium in the growth of the halophyte Suaeda salsa (L.) Pall. under high-sodium conditions. Crop Sci. 2010, 50, 2492–2498. [Google Scholar] [CrossRef]
- Song, J.; Chen, M.; Feng, G.; Jia, Y.H.; Wang, B.S.; Zhang, F.S. Effect of salinity on growth, ion accumulation and the roles of ions in osmotic adjustment of two populations of Suaeda salsa. Plant Soil 2009, 314, 133–141. [Google Scholar] [CrossRef]
- Wang, S.M.; Zhang, J.L.; Flowers, T.J. Low-affinity Na+ uptake in the halophyte Suaeda maritima. Plant Physiol. 2007, 1452, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shi, D.; Wang, D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca Bge. Plant Growth Regul. 2008, 562, 179–190. [Google Scholar] [CrossRef]
- Yeo, A.R. Salt tolerance in the halophyte Suaeda maritima L. Dum.: Intracellular compartmentation of ions. J. Exp. Bot. 1981, 32, 487–497. [Google Scholar] [CrossRef]
- Yeo, A.R.; Flowers, T.J. Salt tolerance in the halophyte Suaeda maritima L. Dum.: Evaluation of the effect of salinity upon growth. J. Exp. Bot. 1980, 31, 1171–1183. [Google Scholar] [CrossRef]
- Shao, Q.; Han, N.; Ding, T.; Zhou, F.; Wang, B. SsHKT1;1 is a potassium transporter of the C3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Biol. 2014, 41, 790–802. [Google Scholar] [CrossRef]
- Lebaudy, A.; Véry, A.A.; Sentenac, H. K+ channel activity in plants: Genes, regulations and functions. FEBS. Lett. 2007, 581, 2357–2366. [Google Scholar] [CrossRef]
- Corratgé-Faillie, C.; Jabnoune, M.; Zimmermann, S.; Véry, A.A.; Fizames, C.; Sentenac, H. Potassium and sodium transport in non-animal cells: The Trk/Ktr/HKT transporter family. Cell Mol. Life Sci. 2010, 67, 2511–2532. [Google Scholar] [CrossRef]
- Cuéllar, T.; Pascaud, F.; Verdeil, J.L.; Torregrosa, L.; Adam-Blondon, A.F.; Thibaud, J.B.; Sentenac, H.; Gaillard, I. A grapevine Shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J. 2010, 51, 58–69. [Google Scholar] [CrossRef]
- Duan, H.; Ma, Q.; Zhang, J.; Hu, J.; Bao, A.; Wei, L.; Wang, Q.; Luan, S.; Wang, S. The inward-rectifying K+ channel SsAKT1 is a candidate involved in K+ uptake in the halophyte Suaeda salsa under saline condition. Plant Soil 2015, 395, 173–187. [Google Scholar] [CrossRef]
- Jin, H.; Dong, D.; Yang, Q.; Zhu, D. Salt responsive transcriptome profiling of Suaeda glauca via RNA sequencing. PLoS ONE 2016, 11, e0150504. [Google Scholar] [CrossRef] [PubMed]
- Nedelyaeva, O.I.; Popova, L.G.; Volkov, V.S.; Balnokin, Y.V. Molecular cloning and characterization of SaCLCd, SaCLCf, and SaCLCg, novel proteins of the chloride channel family CLC from the halophyte Suaeda altissima L. Pall. Plants 2022, 11, 409. [Google Scholar] [CrossRef] [PubMed]
- Nedelyaeva, O.I.; Shuvalov, A.V.; Mayorova, O.V.; Yurchenko, A.A.; Popova, L.G.; Balnokin, Y.V.; Karpichev, I.V. Cloning and functional analysis of SaCLCc1, a gene belonging to the chloride channel family (CLC), from the halophyte Suaeda altissima (L.) Pall. Dokl. Biochem. Biophys. 2018, 481, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Loescher, W.H. Expression of a celery mannose 6-phosphate reductase in Arabidopsis thaliana enhances salt tolerance and induces biosynthesis of both mannitol and aglucosyl-mannitol dimer. Plant Cell Environ. 2003, 26, 275–283. [Google Scholar]
- Moghaieb, R.E.A.; Saneoka, H.; Fujita, K. Effect of salinity on osmotic adjustment, glycine betaine accumulation and the betaine aldehyde dehydrogenase gene expression in two halophytic plants, Salicornia europaea and Suaeda maritime. Plant Sci. 2004, 166, 1345–1349. [Google Scholar] [CrossRef]
- Liu, J.R.; Yi, Y.J.; Zhao, K.F. Effects of salinity onion contents, betaine level and betaine-aldehyde dehydrogenase activity in seepweed Suaeda salsa seedlings. J. Integr. Plant Biol. 1994, 36, 622–626. [Google Scholar]
- Flowers, T.J.; Hall, J.L. Salt tolerance in the halophyte, Suaeda rnaritima L. Dum.: The influence of the salinity of the culture solution on the content of various organic compounds. Ann. Bot. 1978, 42, 1057–1063. [Google Scholar] [CrossRef]
- Park, J.; Okita, T.W.; Edwards, G.E. Salt tolerant mechanismsin single-cell C4 species Bienertia sinuspersici and Suaeda aralocaspica Chenopodiaceae. Plant Sci. 2009, 176, 616–626. [Google Scholar] [CrossRef]
- Song, J.; Ding, X.D.; Feng, G.; Zhang, F.S. Nutritional and osmotic roles of nitrate in a euhalophyte and xerophyte in saline conditions. New Phytol. 2006, 171, 357–366. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Zhao, K.F. Effects of salt and water stresses on osmotic adjustment of Suaeda salsa seedlings. Acta. Bot. Sin. 1998, 40, 56–61. [Google Scholar]
- Tipirdamaz, R.; Gagneul, D.; Duhaze, C.; Aïnouche, A.; Monnier, C.; Özkum, D.; Larher, F. Clustering of halophytes from an inland salt marsh in Turkey according to their ability to accumulate sodium and nitrogenous osmolytes. Environ. Exp. Bot. 2006, 57, 139–153. [Google Scholar] [CrossRef]
- Anbarasi, G.; Somasundaram, S.T. Growth regulation and proteomic approaches of exogenous abscisic acid induced changes on salt tolerance factors in Suaeda maritima. Plant Physiol. Rep. 2020, 25, 33–50. [Google Scholar] [CrossRef]
- Greenway, H.; Osmond, C.B. Salt responses of enzymes from species differing in salt tolerance. Plant Physiol. 1972, 49, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Dong, D.; Yang, C.; Yuan, F.; Yu, X.; Fu, X.; Zhu, D. Cloning and analysis of PEAMT gene in Suaeda Glauca. Chin. Agric. Sci. Bull. 2015, 31, 178–183. [Google Scholar]
- Li, W.; Zhang, C.Y.; Lu, Q.T.; Wen, X.G.; Lu, C.M. The combined effect of salt stress and heat shock on proteome profiling in Suaeda salsa. J. Plant Physiol. 2011, 168, 1743–1752. [Google Scholar] [CrossRef]
- Wu, H.; Liu, X.; You, L.; Zhang, L.; Zhou, D.; Feng, J.; Zhao, J.; Yu, J. Effects of salinity on metabolic profiles, gene expressions, and antioxidant enzymes in halophyte Suaeda salsa. J. Plant Growth Regul. 2012, 31, 332–341. [Google Scholar] [CrossRef]
- Askari, H.; Edqvist, J.; Hajheidari, M.; Kafi, M.; Salekdeh, G.H. Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics 2006, 6, 2542–2554. [Google Scholar] [CrossRef]
- Li, Q.; Gao, X.; Yu, X.; Wang, X.; An, L. Molecular cloning and characterization of betaine aldehyde dehydrogenase gene from Suaeda liaotungensis and its use in improved tolerance to salinity in transgenic tobacco. Biotechnol. Lett. 2003, 25, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, D.; Gao, X.; Su, Q.; Jia, A. Cloning of cDNA encoding choline monooxygenase from Suaeda liaotungensis and salt tolerance of transgenic tobacco. J. Integr. Plant Biol. 2003, 45, 242–247. [Google Scholar]
- Wang, F.; Wang, M.; Guo, C.; Wang, N.; Li, X.; Chen, H.; Dong, Y.; Chen, X.; Wang, Z.; Li, H. Cloning and characterization of a novel betaine aldehyde dehydrogenase gene from Suaeda corniculata. Genet. Mol. Res. 2016, 15, gmr15027848. [Google Scholar] [CrossRef]
- Wang, P.; Ma, C.; Zhao, K.; Zhao, Y.; Zhang, H. Isolation and characterizing of a Δ1-pyrroline-5-carboxylate synthase gene in Suaeda salsa under salinity stress. J. Shandong Norm. Univ. Nat. Sci. 2002, 17, 59–62. [Google Scholar]
- Wang, P.; Ma, C.; Cao, Z.; Zhao, Y.; Zhang, H. Molecular cloning and differential expression of amyo-inositol-1-phosphate synthase gene in Suaeda salsa under salinity stress. J. Plant Physiol. Mol. Biol. 2002, 28, 175–180. [Google Scholar]
- Li, Q.; Song, J. Analysis of widely targeted metabolites of the euhalophyte Suaeda salsa under saline conditions provides new insights into salt tolerance and nutritional value in halophytic species. BMC Plant Biol. 2019, 19, 388. [Google Scholar] [CrossRef]
- Pang, Q.; Zhang, A.; Zang, W.; Wei, L.; Yan, X. Integrated proteomics and metabolomics for dissecting the mechanism of global responses to salt and alkali stress in Suaeda corniculata. Plant Soil 2016, 402, 379–394. [Google Scholar] [CrossRef]
- Zang, W.; Miao, R.; Zhang, Y.; Yuan, Y.; Pang, Q.; Zhou, Z. Metabolic and molecular basis for the salt and alkali responses of Suaeda corniculate. Environ. Exp. Bot. 2021, 192, 104643. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, R.; Sui, N.; Shi, W.; Wang, L.; Tian, C.; Song, J. Changes in endogenous hormones and seed-coat phenolics during seed storage of two Suaeda salsa populations. Aust. J. Bot. 2016, 64, 325–332. [Google Scholar] [CrossRef]
- Wang, B.; Luttge, U.; Ratajczak, R. Specific regulation of SOD isoforms by NaCl and osmotic stress in leaves of the C3 halophyte Suaeda salsa L. J. Plant Physiol. 2004, 161, 285–293. [Google Scholar] [CrossRef]
- Guan, B.; Yu, J.; Lu, Z.; Zhang, Y.; Wang, X. Effects of water-salt stresses on seedling growth and activities of antioxidative enzyme of Suaeda salsa in coastal wetlands of the Yellow River Delta. Environ. Sci. 2011, 32, 2422–2429. [Google Scholar]
- Pang, C.; Zhang, S.; Gong, Z.; Wang, B. NaCl treatment markedly enhances H2O2-scavenging system in leaves of halophyte Suaeda salsa. Physiol. Plantarum 2005, 125, 490–499. [Google Scholar]
- Mohamed, E.; Matsuda, R.; El-Khatib, A.A.; Takechi, K.; Takano, H.; Takio, S. Characterization of the superoxide dismutase genes of the halophyte Suaeda maritima in Japan and Egypt. Plant Cell Rep. 2015, 34, 2099–2110. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Wen, W.; Yang, G. The antioxidant system in Suaeda salsa under salt stress. Plant Signal Behav. 2020, 15, 1771939. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.B.; Shaw, B.P. Isolation, identification and expression analysis of salt induced genes in Suaeda maritima, a natural halophyte, using PCR-based suppression subtractive hybridization. BMC Plant Biol. 2009, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Hussain, T.; Gulzar, S.; Aziz, I.; Gul, B.; Khan, M.A. Salt tolerance of a cash crop halophyte Suaeda fruticosa: Biochemical responses to salt and exogenous chemical treatments. Acta Physiol. Plant. 2012, 34, 2331–2340. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, S.; Wang, L.; Wang, M.; Zhang, H. Overexpression of GST gene accelerates the growth of transgenic Arabidopsis under salt stress. J. Plant Physiol. Mol. Biol. 2004, 30, 517–522. [Google Scholar]
- Hara, M.; Fujinaga, M.; Kuboi, T. Metal binding by citrus dehydrin with histidine–rich domains. J. Exp. Bot. 2005, 56, 2695–2703. [Google Scholar] [CrossRef]
- Tiwari, P.; Chakrabarty, D. Dehydrin in the past four decades: From chaperones to transcription co-regulators in regulating abiotic stress response. Curr. Res. Biotech. 2021, 3, 249–259. [Google Scholar] [CrossRef]
- Ma, H.; Sun, H.; Li, H.; Deng, S.; Wang, X.; Chen, S.; Chen, L.; Zhang, L.; Zhong, M. Functional analysis of the SsDHN of Suaeda salsa L. Pall. Mol. Plant Breed. 2021, 19, 827–832. [Google Scholar]
- Ayarpadikannan, S.; Chung, E.; Cho, C.W.; So, H.A.; Kim, S.O.; Jeon, J.M.; Kwak, M.H.; Lee, S.W.; Lee, J.H. Exploration for the salt stress tolerance genes from a salt-treated halophyte, Suaeda asparagoides. Plant Cell Rep. 2012, 31, 35–48. [Google Scholar] [CrossRef]
- Ma, C.; Wang, P.; Cao, Z.; Zhao, Y.; Zhang, H. cDNA cloning and gene expression of APX in Suaeda salsa in response to salt stress. J. Plant Physiol. Mol. Biol. 2002, 28, 261–266. [Google Scholar]
- Ma, C.; Wang, P.; Cao, Z.; Zhao, Y.; Zhang, H. Cloning and differential gene expression of two catalases in Suaeda salsa in response to salt stress. J. Integr. Plant Biol. 2003, 45, 93–97. [Google Scholar]
- Li, K.; Pang, C.H.; Ding, F.; Sui, N.; Feng, Z.T.; Wang, B.S. Overexpression of Suaeda salsa stroma ascorbate peroxidase in Arabidopsis. S. Afr. J. Bot. 2012, 78, 235–245. [Google Scholar] [CrossRef]
- Behera, L.M.; Hembram, P. Advances on plant salinity stress responses in the post-genomic era: A review. J. Crop Sci. Biotech. 2021, 24, 117–126. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Li, W.; Liu, X.; Ajmal Khan, M.; Yamaguchi, S. The effect of plant growth regulators, nitric oxide, nitrate, nitrite and light on the germination of dimorphic seeds of Suaeda salsa under saline conditions. J. Plant Res. 2005, 118, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yamaguchi, S.; Khan, M.; An, P.; Liu, X.; Tran, L.S. Roles of gibberellins and abscisic acid in regulating germination of Suaeda salsa dimorphic seeds under salt stress. Front. Plant Sci. 2016, 6, 1235. [Google Scholar] [CrossRef] [PubMed]
- Boucaud, J.; Ungar, I.A. Hormonal control of germination under saline conditions of three halophytic taxa in the genus Suaeda. Physiol. Plant. 1976, 37, 143–148. [Google Scholar] [CrossRef]
- Guo, J.; Lu, C.; Zhao, F.; Gao, S.; Wang, B. Improved reproductive growth of euhalophyte Suaeda salsa under salinity is correlated with altered phytohormone biosynthesis and signal transduction. Funct. Plant Biol. 2020, 47, 170–183. [Google Scholar] [CrossRef]
- Fisher, D.D.; Schenk, H.J.; Thorsch, J.A.; Ferren, W.R., Jr. Leaf anatomy and subgeneric affiliations of C3 and C4 species of Suaeda Chenopodiaceae in North America. Am. J. Bot. 1997, 849, 1198–1210. [Google Scholar] [CrossRef]
- Li, Q.; Liu, R.; Li, Z.; Fan, H.; Song, J. Positive effects of NaCl on the photoreaction and carbon assimilation efficiency in Suaeda salsa. Plant Physiol. Bioch. 2022, 177, 32–37. [Google Scholar] [CrossRef]
- Ibraheem, F.; Al-Zahrani, A.; Mosa, A. Physiological adaptation of three wild halophytic Suaeda species: Salt tolerance strategies and metal accumulation capacity. Plants 2022, 11, 537. [Google Scholar] [CrossRef]
- Wei, L.; Pang, Q.; Zhang, A.; Guo, J.; Yan, X. Effects of salt and alkali stresses on photosynthetic characteristics of Suaeda corniculata seedlings. J. Northeast. For. Univ. 2012, 40, 32–35. [Google Scholar]
- Lu, C.; Qiu, N.; Wang, B.; Zhang, J. Salinity treatment shows no effects on photosystem II photochemistry, but increases the resistance of photosystem II to heat stress in halophyte Suaeda salsa. J. Exp. Bot. 2003, 54, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Li, J.; Gao, S.; Tuerxun, Z.; Chang, X.; Hu, W.; Chen, G.; Huang, Q. SsPsaH, a H subunit of the photosystem I reaction center of Suaeda salsa, confers the capacity of osmotic adjustment in tobacco. Genes Genom. 2020, 42, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Bohra, A.; Jha, R.; Parida, S.K. Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 2019, 38, 255–277. [Google Scholar] [CrossRef]
- Lawlor, D. Abiotic stress adaptation in plants. Physiological, Molecular and Genomic Foundation. Ann. Bot. 2011, 107, vii–ix. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Y.; Li, X.; Zhang, L.; Fan, S. Transcriptomic analysis identifies novel genes and pathways for salt stress responses in Suaeda salsa leaves. Sci. Rep. 2020, 10, 4236. [Google Scholar] [CrossRef]
- Guo, S.; Tan, Y.; Chu, H.; Sun, M.; Xing, J. Transcriptome sequencing revealed molecular mechanisms underlying tolerance of Suaeda salsa to saline stress. PLoS ONE 2019, 14, e0219979. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.J.; Sun, Y.; Zhang, M.; Zhai, J.T. Transcriptomic profile analysis of the halophyte Suaeda rigida response and tolerance under NaCl stress. Sci. Rep. 2020, 10, 15148. [Google Scholar] [CrossRef]
- Wang, M.; Ren, T.; Marowa, P.; Du, H.; Xu, Z. Identification and selection of reference genes for gene expression analysis by quantitative real-time PCR in Suaeda glauca’s response to salinity. Sci. Rep. 2021, 11, 8569. [Google Scholar] [CrossRef]
- Diray-Arce, J.; Knowles, A.; Suvorov, A.; O’Brien, J.; Hansen, C.; Bybee, S.M.; Gul, B.; Ajmal Khan, M.; Nielsen, B.L. Identification and evolutionary characterization of salt-responsive transcription factors in the succulent halophyte Suaeda fruticosa. PLoS ONE 2019, 14, e0222940. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, X.; Hu, Y.; Yu, X.; Li, Q. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hu, Y.; Li, X.; Yu, X.; Li, Q. Molecular characterization and function analysis of SlNAC2 in Suaeda liaotungensis K. Gene 2014, 543, 190–197. [Google Scholar] [CrossRef]
- Wu, D.; Sun, Y.; Wang, H.; Shi, H.; Su, M.; Shan, H.; Li, T.; Li, Q. The SlNAC8 gene of the halophyte Suaeda liaotungensis enhances drought and salt stress tolerance in transgenic Arabidopsis thaliana. Gene 2018, 662, 10–20. [Google Scholar] [CrossRef]
- Wang, H.F.; Shan, H.Y.; Shi, H.; Wu, D.D.; Li, T.T.; Li, Q.L. Characterization of a transcription factor SlNAC7 gene from Suaeda liaotungensis and its role in stress tolerance. J. Plant Res. 2021, 134, 1105–1120. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sun, X.; Wang, X.; MA, H. Cloning and expression analysis of SsDREB gene from Suaeda salsa L. J. Nucl. Agric. Sci. 2011, 25, 684–691. [Google Scholar]
- Sun, X.; Ma, H.; Jia, X.; Chen, Y.; Ye, X. Molecular cloning and characterization of two novel DREB genes encoding dehydration-responsive element binding proteins in halophyte Suaeda salsa. Genes Genom. 2015, 37, 199–212. [Google Scholar] [CrossRef]
- Sun, X.; Su, J.; Jia, X.; Liang, L.; Xiao, Z.; Deng, Y. Cloning and expression analysis of two DREB1/CBF genes in Suaeda salsa L. Sci. Agric. Sin. 2016, 49, 2418–2429. [Google Scholar]
- Liu, X.; Wu, H.; Ji, C.; Wei, L.; Zhao, J.; Yu, J. An integrated proteomic and metabolomic study on the chronic effects of mercury in Suaeda salsa under an environmentally relevant salinity. PLoS ONE 2013, 8, e64041. [Google Scholar]
- Fernandez-Garcia, N.; Hernandez, M.; Casado-vela, J.; Bru, R.; Elortza, F.; Hedden, P.; Olmos, E. Changes to the proteome and targeted metabolites of xylem sap in Brassica oleracea in response to salt stress. Plant Cell Environ. 2011, 34, 821–836. [Google Scholar] [CrossRef]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.X.; Li, H.Y. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.J.; Miras-Moreno, B.; Araniti, F.; Salehi, H.; Bernardo, L.; Parida, A.; Lucini, L. Proteomics revealed distinct responses to salinity between the halophytes Suaeda maritima L. Dumort and Salicornia brachiata Roxb. Plants 2020, 9, 227. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.H.; Chen, M.; Song, J.; Wang, B.S. Increase in aquaporin activity is involved in leaf succulence of the euhalophyte Suaeda salsa, under salinity. Plant Sci. 2009, 176, 200–205. [Google Scholar] [CrossRef]
- Tao, J.; Chen, H.; Ma, B.; Zhang, W.; Chen, S.; Zhang, J. The role of ethylene in plants under salinity stress. Front. Plant Sci. 2015, 6, 1059. [Google Scholar] [CrossRef]
- Benjamin, J.J.; Lucini, L.; Jothiramshekar, S.; Parida, A. Metabolomic insights into the mechanisms underlying tolerance to salinity in different halophytes. Plant Physiol. Biochem. 2019, 135, 528–545. [Google Scholar] [CrossRef]
- Kumari, A.; Parida, A.K. Metabolomics and network analysis reveal the potential metabolites and biological pathways involved in salinity tolerance of the halophyte Salvadora persica. Environ. Exp. Bot. 2018, 148, 85–99. [Google Scholar] [CrossRef]
- Yadav, S.; Elansary, H.O.; Mattar, M.A.; Elhindi, K.M.; Alotaibi, M.A.; Mishra, A. Differential accumulation of metabolites in Suaeda species provides new insights into abiotic stress tolerance in C4-halophytic species in elevated CO2 conditions. Agronomy 2021, 11, 131. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).