Identification and Expression Profile Analysis of the OSCA Gene Family Related to Abiotic and Biotic Stress Response in Cucumber
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of OSCA Family Genes in Cucumber
2.2. Sequence Alignment and Phylogenetic Analysis
2.3. Conserved Motif and Gene Structure Analysis
2.4. Chromosomal Location and Promoter Analysis
2.5. In Silico Expression Analysis Based on RNA-Seq
2.6. Plant Materials and Treatments
2.7. RNA Extraction, cDNA Synthesis, and qRT-PCR
3. Results
3.1. Identification and Characterization of OSCA Family Genes in Cucumber
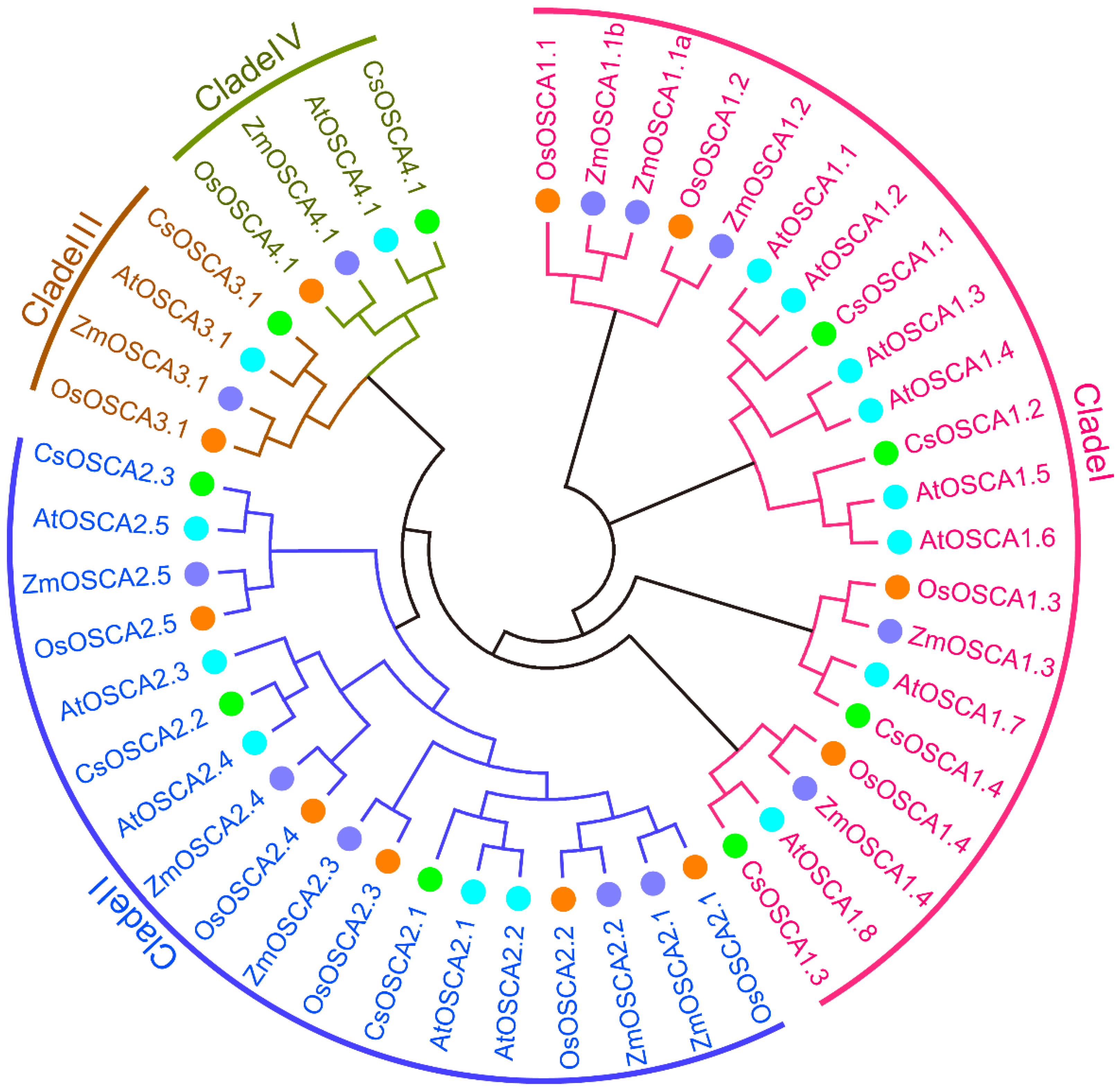
3.2. Phylogenetic Analysis of OSCA Proteins
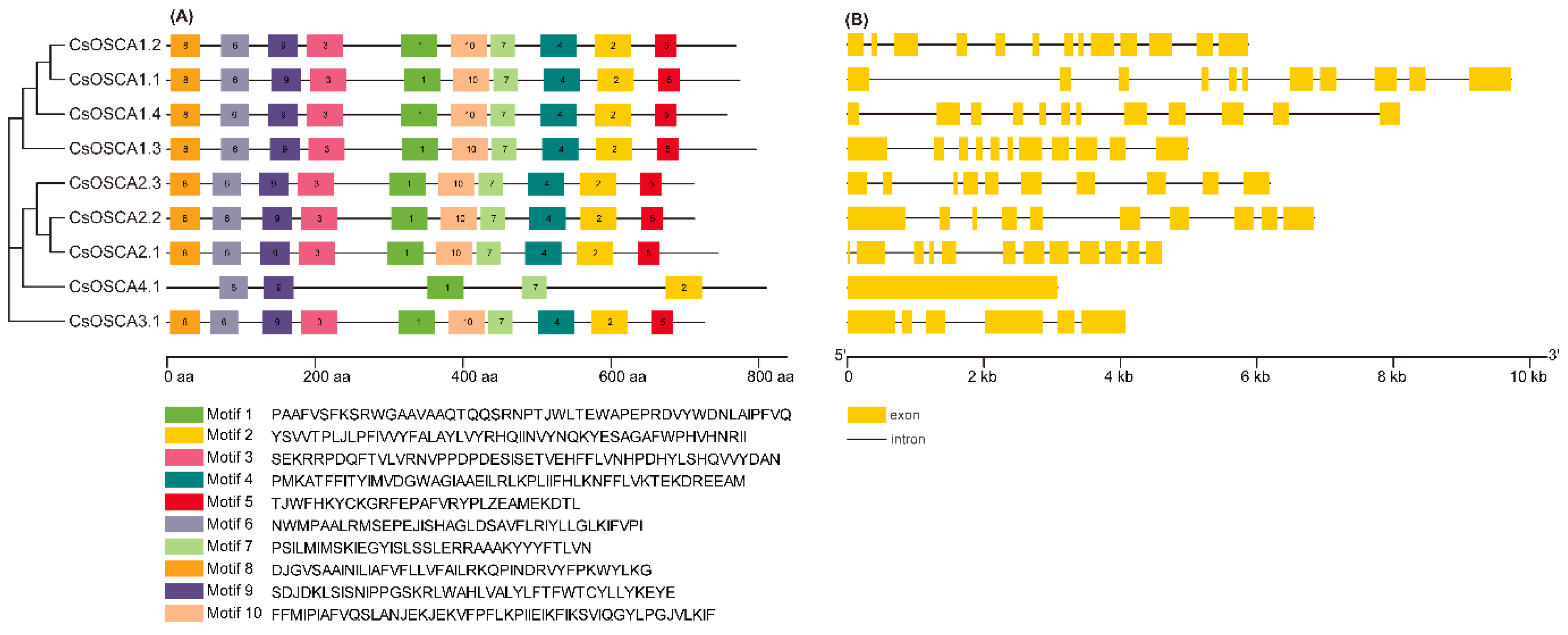
3.3. Comparison of Conserved Motifs and Gene Structures of CsOSCAs
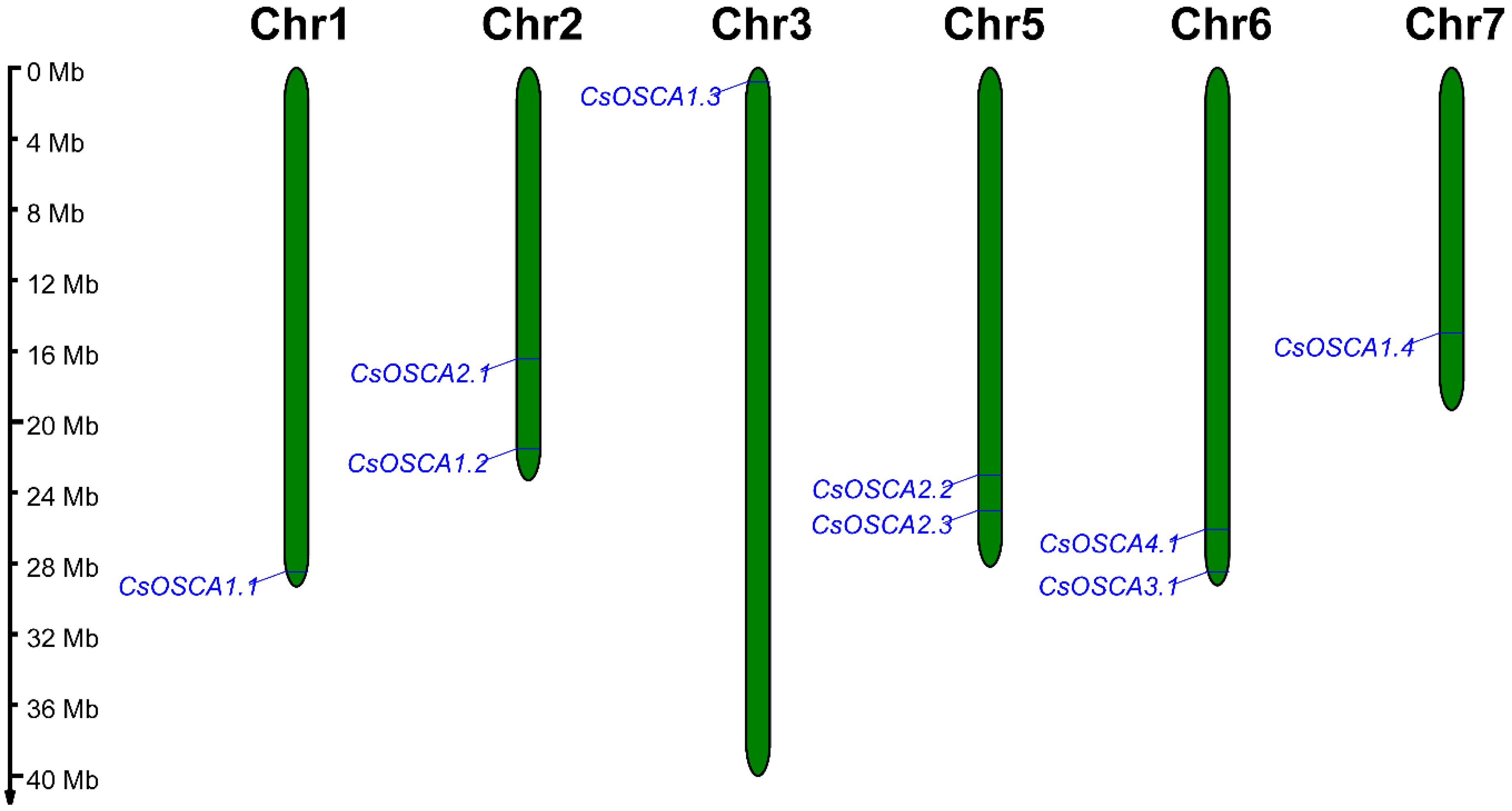
3.4. Chromosomal Location of CsOSCA Genes
3.5. Cis-Elements in the Promoters of CsOSCA Genes
3.6. Tissue Expression Profiles of CsOSCA Genes
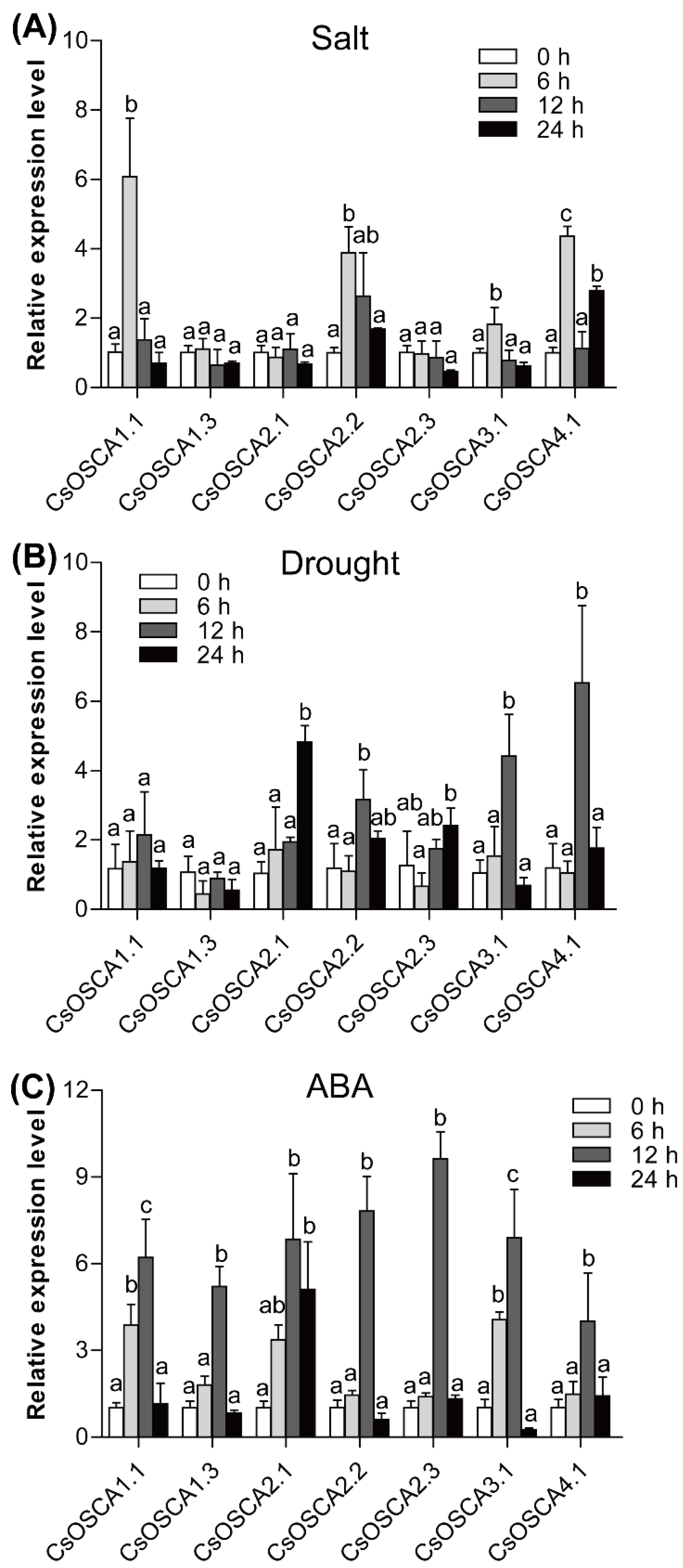
3.7. Expression Patterns of the CsOSCA Genes under Abiotic Stress and ABA Treatment
3.8. Expression Patterns of CsOSCA Genes under Biotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garrido-Gala, J.; Higuera, J.J.; Muñoz-Blanco, J.; Amil-Ruiz, F.; Caballero, J.L. The VQ motif-containing proteins in the diploid and octoploid strawberry. Sci. Rep. 2019, 9, 4942. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Xiao, L.; Hu, Y.; Liu, L.; Liu, H.; Hu, Z.; Liu, S.; Zhou, Y. Genome-wide survey and expression analysis of B-box family genes in cucumber reveal their potential roles in response to diverse abiotic and biotic stresses. Agriculture 2022, 12, 827. [Google Scholar] [CrossRef]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997, 12, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Kleist, T.J.; Wudick, M.M. Shaping up: Recent advances in the study of plant calcium channels. Curr. Opin. Cell Biol. 2022, 76, 102080. [Google Scholar] [CrossRef]
- Jammes, F.; Hu, H.C.; Villiers, F.; Bouten, R.; Kwak, J.M. Calcium-permeable channels in plant cells. FEBS J. 2011, 278, 4262–4276. [Google Scholar] [CrossRef]
- Swarbreck, S.M.; Colaço, R.; Davies, J.M. Plant calcium-permeable channels. Plant Physiol. 2013, 163, 514–522. [Google Scholar] [CrossRef] [Green Version]
- Davies, J.M. Annexin-mediated calcium signalling in plants. Plants 2014, 3, 128–140. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, Z.; Kakar, K.U.; Ullah, R.; Yu, S.; Zhang, J.; Shu, Q.Y.; Ren, X.L. Genome-wide identification, evolution and expression analysis of cyclic nucleotide-gated channels in tobacco (Nicotiana tabacum L.). Genomics 2019, 111, 142–158. [Google Scholar] [CrossRef]
- Zeng, H.; Zhao, B.; Wu, H.; Zhu, Y.; Chen, H. Comprehensive in silico characterization and expression profiling of nine gene families associated with calcium transport in soybean. Agronomy 2020, 10, 1539. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Thor, K.; Jiang, S.; Michard, E.; George, J.; Scherzer, S.; Huang, S.; Dindas, J.; Derbyshire, P.; Leitão, N.; DeFalco, T.A.; et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 2020, 585, 569–573. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Sun, L. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018, 9, 5060. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Kang, Y.; Wu, J.X.; Yao, F.; Pan, C.; Yan, Z.; Song, C.; Chen, L. Structure of the mechanosensitive OSCA channels. Nat. Struct. Mol. Biol. 2018, 25, 850–858. [Google Scholar] [CrossRef]
- Wu, X.; Yuan, F.; Wang, X.; Zhu, S.; Pei, Z.M. Evolution of osmosensing OSCA1 Ca2+ channel family coincident with plant transition from water to land. Plant. Genome 2022, 15, e20198. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015, 15, 261. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Xu, Y.; Yang, F.; Magwanga, R.O.; Cai, X.; Wang, X.; Wang, Y.; Hou, Y.; Wang, K.; Liu, F.; et al. Genome-wide identification of OSCA gene family and their potential function in the regulation of dehydration and salt stress in Gossypium hirsutum. J. Cotton Res. 2019, 2, 11. [Google Scholar] [CrossRef]
- Tong, K.; Wu, X.; He, L.; Qiu, S.; Liu, S.; Cai, L.; Rao, S.; Chen, J. Genome-wide identification and expression profile of OSCA gene family members in Triticum aestivum L. Int. J. Mol. Sci. 2022, 23, 469. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, P.; Lu, X.; Wang, G.; Wang, Z.; Zhang, Q.; Zhang, X.; Wei, X.; Mei, F.; Wei, L.; et al. Systematic analysis of the maize OSCA Genes revealing ZmOSCA family members involved in osmotic stress and ZmOSCA2.4 confers enhanced drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 351. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Wen, Z.; Fang, W.; Wang, Y.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Han, S. Functional analysis of rice OSCA genes overexpressed in the Arabidopsis osca1 mutant due to drought and salt stresses. Transgenic Res. 2021, 30, 811–820. [Google Scholar] [CrossRef]
- Zhai, Y.; Wen, Z.; Han, Y.; Zhuo, W.; Wang, F.; Xi, C.; Liu, J.; Gao, P.; Zhao, H.; Wang, Y.; et al. Heterogeneous expression of plasma-membrane-localised OsOSCA1.4 complements osmotic sensing based on hyperosmolality and salt stress in Arabidopsis osca1 mutant. Cell Calcium 2020, 91, 102261. [Google Scholar] [CrossRef]
- Akita, K.; Miyazawa, Y. The mechanosensitive Ca2+ channel, OSCA1.1, modulates root hydrotropic bending in Arabidopsis thaliana. Environ. Exp. Bot. 2022, 197, 104825. [Google Scholar] [CrossRef]
- Liao, L.; Hu, Z.; Liu, S.; Yang, Y.; Zhou, Y. Characterization of germin-like proteins (GLPs) and their expression in response to abiotic and biotic stresses in cucumber. Horticulturae 2021, 7, 412. [Google Scholar] [CrossRef]
- Liu, X.; Lu, H.; Liu, P.; Miao, H.; Bai, Y.; Gu, X.; Zhang, S. Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants 2020, 9, 1659. [Google Scholar] [CrossRef]
- Ding, S.; Feng, X.; Du, H.; Wang, H. Genome-wide analysis of maize OSCA family members and their involvement in drought stress. PeerJ 2019, 7, e6765. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [Green Version]
- Waseem, M.; Aslam, M.M.; Shaheen, I. The DUF221 domain-containing (DDP) genes identification and expression analysis in tomato under abiotic and phytohormone stress. GM Croops Food 2021, 12, 586–599. [Google Scholar] [CrossRef]
- Gu, X.; Wang, P.; Liu, Z.; Wang, L.; Huang, Z.; Zhang, S.; Wu, J. Genome-wide identification and expression analysis of the OSCA gene family in Pyrus bretschneideri. Can. J. Plant Sci. 2018, 98, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Zaynab, M.; Peng, J.; Sharif, Y.; Albaqami, M.; Al-Yahyai, R.; Fatima, M.; Nadeem, M.A.; Khan, K.A.; Alotaibi, S.S.; Alaraidh, I.A.; et al. Genome-wide identification and expression profiling of DUF221 gene family provides new insights into abiotic stress responses in potato. Front. Plant Sci. 2022, 12, 804600. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, Y.; Zhai, Y.; Wen, Z.; Liu, J.; Xi, C.; Zhao, H.; Wang, Y.; Han, S. OsOSCA1.1 mediates hyperosmolality and salt stress sensing in Oryza sativa. Biology 2022, 11, 678. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, M.; Wu, R.; Chen, X.; Liu, F.; Xing, B. Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response. BMC Plant Biol. 2021, 21, 408. [Google Scholar] [CrossRef] [PubMed]
- Moeder, W.; Phan, V.; Yoshioka, K. Ca2+ to the rescue—Ca2+ channels and signaling in plant immunity. Plant Sci. 2019, 279, 19–26. [Google Scholar] [CrossRef]



| Gene | Accession No. (v2) | Chromosome: Position | gDNA/bp | CDS/bp | Protein | |||
|---|---|---|---|---|---|---|---|---|
| AA | MW/kDa | pI | GRAVY | |||||
| CsOSCA1.1 | Csa1G701320.1 | Chr1: 28,326,385–28,336,117 | 9733 | 2322 | 773 | 87.97 | 9.16 | 0.186 |
| CsOSCA1.2 | Csa2G416110.1 | Chr2: 21,427,024–21,432,896 | 5873 | 2307 | 768 | 87.75 | 8.93 | 0.168 |
| CsOSCA1.3 | Csa3G006670.1 | Chr3: 814,253–819,247 | 4995 | 2388 | 795 | 91.15 | 9.32 | 0.134 |
| CsOSCA1.4 | Csa7G392390.1 | Chr7: 14,860,224–14,868,316 | 8093 | 2271 | 756 | 86.89 | 8.78 | 0.156 |
| CsOSCA2.1 | Csa2G352960.1 | Chr2: 16,330,484–16,335,086 | 4603 | 2232 | 743 | 84.16 | 8.97 | 0.261 |
| CsOSCA2.2 | Csa5G606420.1 | Chr5: 22,846,868–22,853,705 | 6838 | 2139 | 712 | 80.73 | 8.81 | 0.364 |
| CsOSCA2.3 | Csa5G623630.1 | Chr5: 24,915,913–24,922,103 | 6191 | 2136 | 771 | 80.66 | 8.63 | 0.269 |
| CsOSCA3.1 | Csa6G525220.1 | Chr6: 28,314,457–28,318,525 | 4069 | 2178 | 725 | 81.87 | 9.37 | 0.310 |
| CsOSCA4.1 | Csa6G507280.1 | Chr6: 25,978,314–25,981,392 | 3079 | 2430 | 809 | 92.01 | 6.42 | 0.178 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Zhu, C.; Chen, J.; Zhao, J.; Hu, Z.; Liu, S.; Zhou, Y. Identification and Expression Profile Analysis of the OSCA Gene Family Related to Abiotic and Biotic Stress Response in Cucumber. Biology 2022, 11, 1134. https://doi.org/10.3390/biology11081134
Yang S, Zhu C, Chen J, Zhao J, Hu Z, Liu S, Zhou Y. Identification and Expression Profile Analysis of the OSCA Gene Family Related to Abiotic and Biotic Stress Response in Cucumber. Biology. 2022; 11(8):1134. https://doi.org/10.3390/biology11081134
Chicago/Turabian StyleYang, Shuting, Chuxia Zhu, Jingju Chen, Jindong Zhao, Zhaoyang Hu, Shiqiang Liu, and Yong Zhou. 2022. "Identification and Expression Profile Analysis of the OSCA Gene Family Related to Abiotic and Biotic Stress Response in Cucumber" Biology 11, no. 8: 1134. https://doi.org/10.3390/biology11081134
APA StyleYang, S., Zhu, C., Chen, J., Zhao, J., Hu, Z., Liu, S., & Zhou, Y. (2022). Identification and Expression Profile Analysis of the OSCA Gene Family Related to Abiotic and Biotic Stress Response in Cucumber. Biology, 11(8), 1134. https://doi.org/10.3390/biology11081134






