The Transcription Factor FgAtrR Regulates Asexual and Sexual Development, Virulence, and DON Production and Contributes to Intrinsic Resistance to Azole Fungicides in Fusarium graminearum
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Construction of FgAtrR Deletion Mutants and Complemented Strains
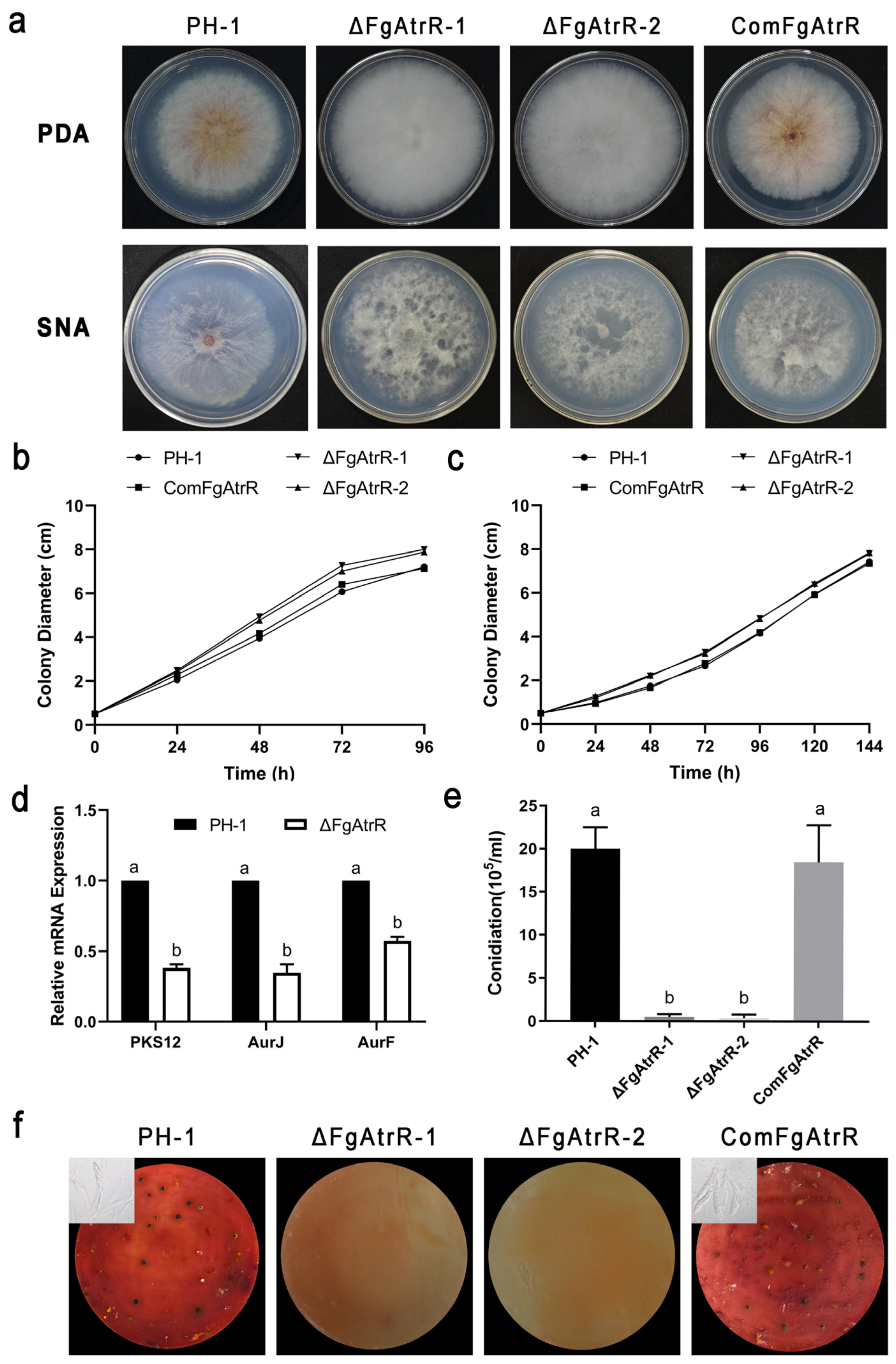
2.3. Assessment of Conidial Production and Sexual Development
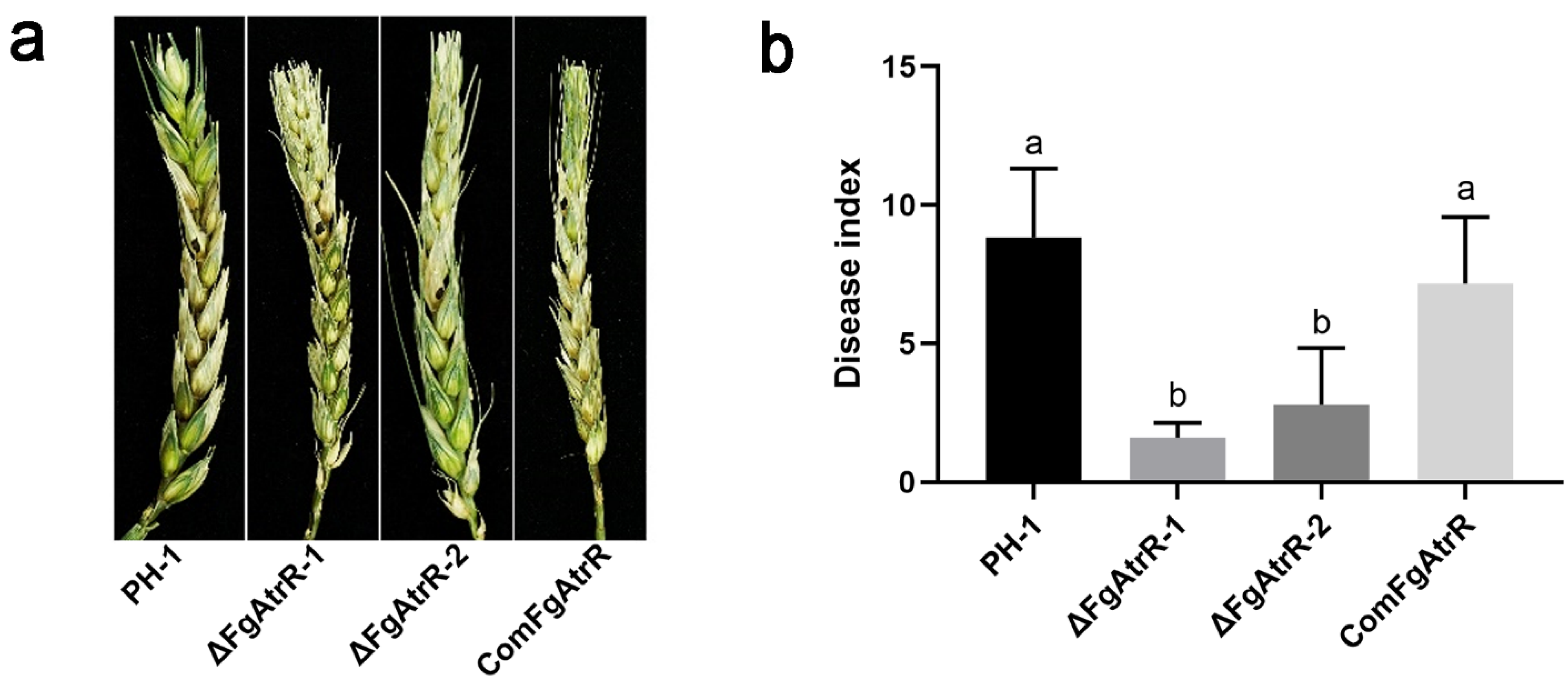
2.4. Pathogenicity Assays
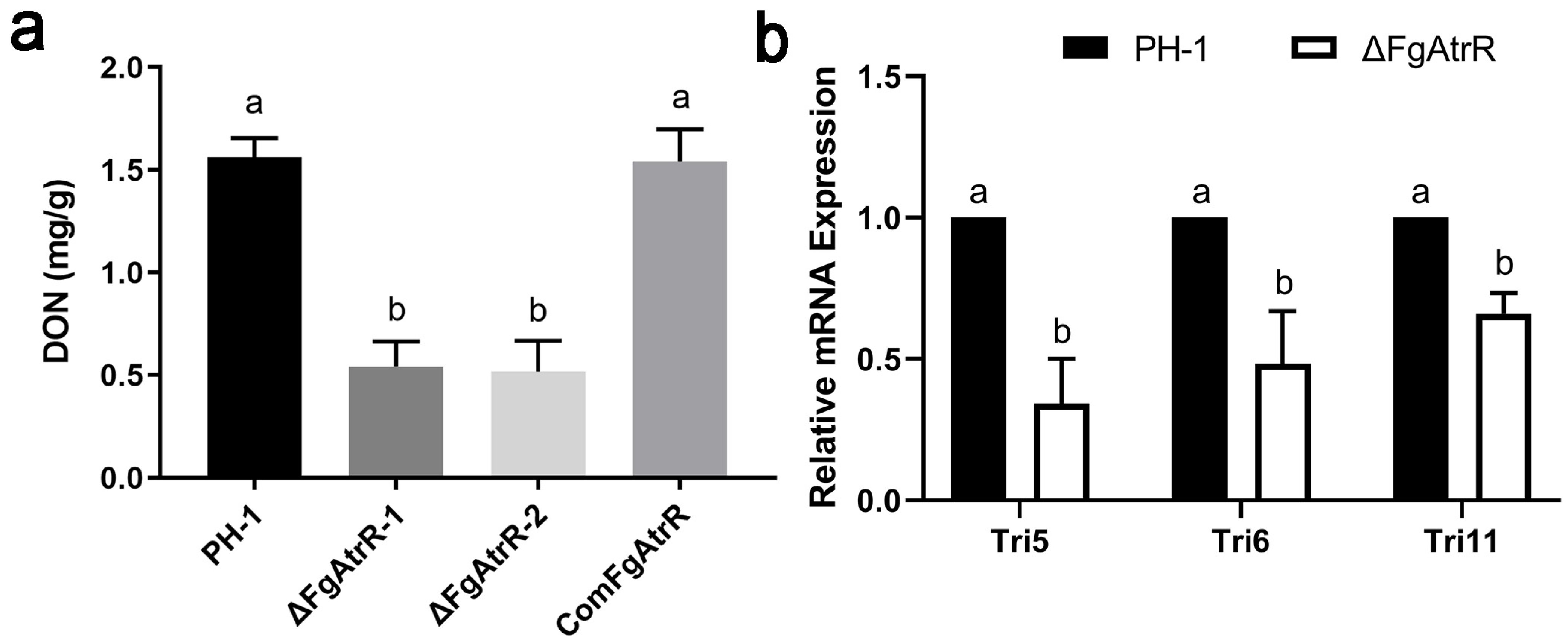
2.5. Analysis of DON Production
2.6. Azole Fungicide Sensitivity Testing
2.7. Microscopic Examinations
2.8. Quantitative Real-Time PCR (qRT-PCR) Assays
2.9. RNA Sequencing and Bioinformatics Analysis
3. Results
3.1. Identification of the AtrR ortholog in F. graminearum
3.2. FgAtrR Is Involved in Vegetative Growth and Pigmentation in F. graminearum
3.3. FgAtrR Is Essential for Asexual and Sexual Development
3.4. FgAtrR Is Necessary for Full Virulence in F. graminearum
3.5. FgAtrR Is Required for DON Biosynthesis
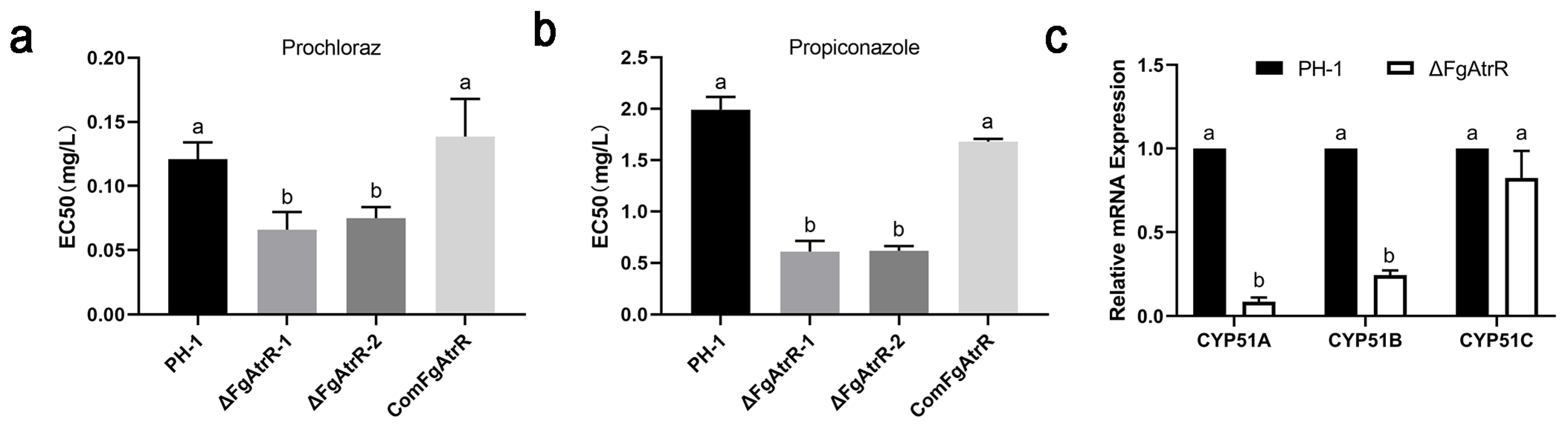
3.6. FgAtrR Contributes to Intrinsic Resistance to Azole Fungicides
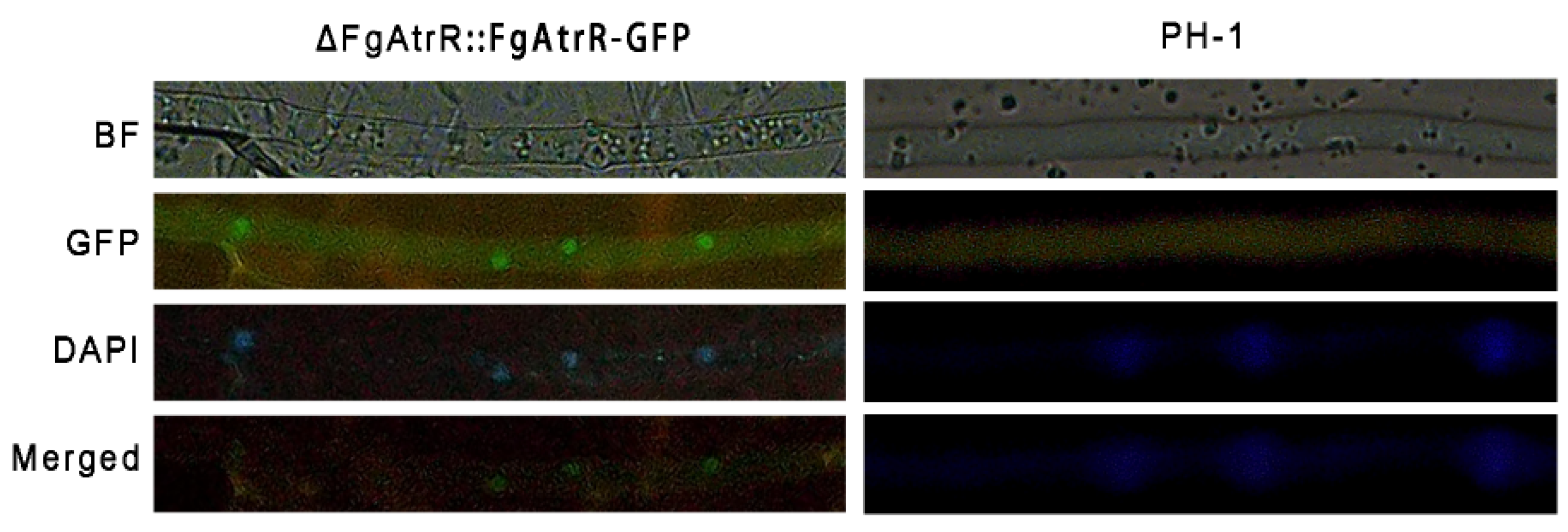
3.7. FgAtrR Mainly Localizes to the Nucleus
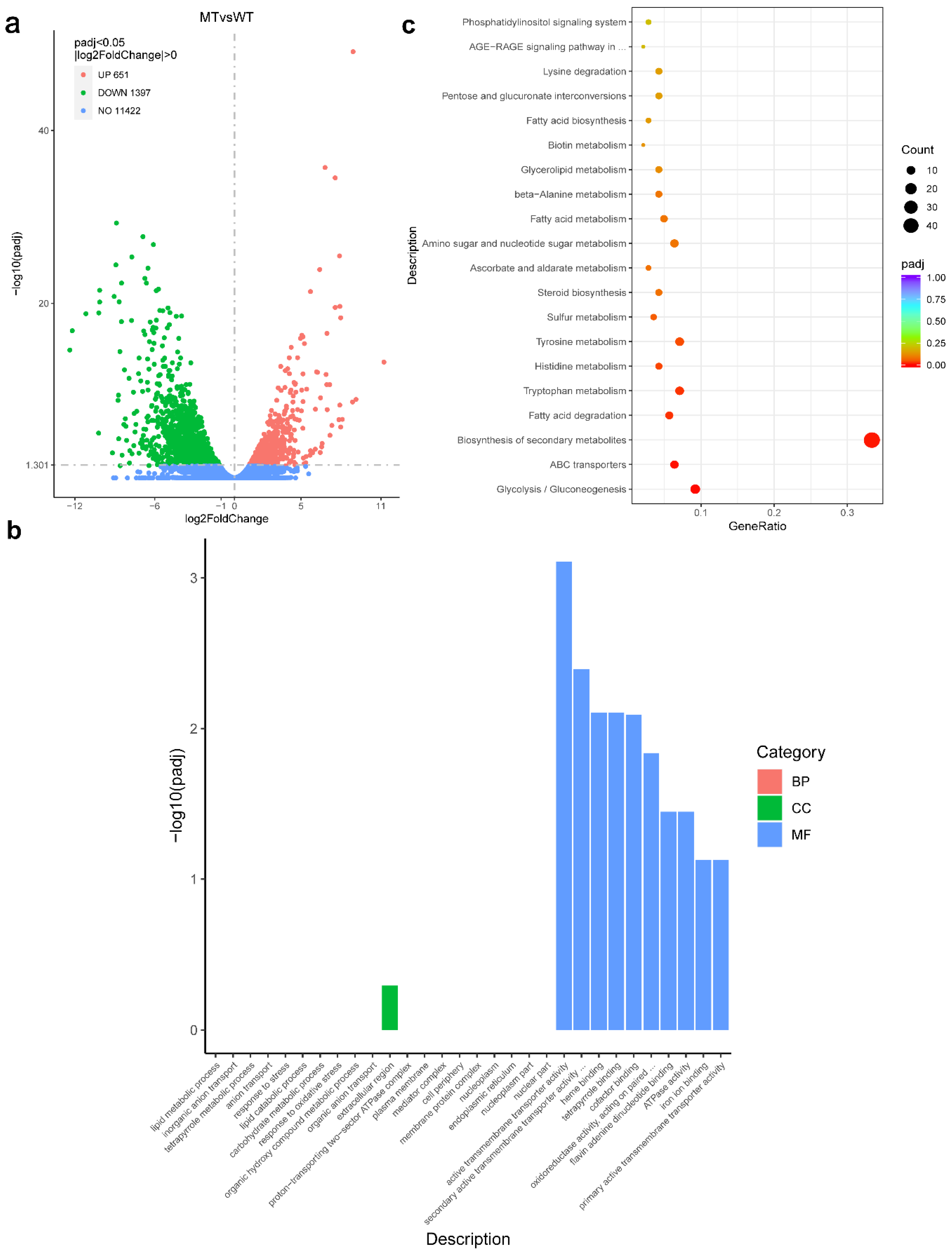
3.8. RNA-seq Analysis with the ΔFgAtrR Mutant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dweba, C.C.; Figlan, S.; Shimelis, H.A.; Motaung, T.E.; Sydenham, S.; Mwadzingeni, L.; Tsilo, T.J. Fusarium Head Blight of Wheat: Pathogenesis and Control Strategies. Crop Prot. 2017, 91, 114–122. [Google Scholar] [CrossRef]
- West, J.S.; Holdgate, S.; Townsend, J.A.; Edwards, S.G.; Jennings, P.; Fitt, B.D.L. Impacts of Changing Climate and Agronomic Factors on Fusarium Ear Blight of Wheat in the UK. Fungal Ecol. 2012, 5, 53–61. [Google Scholar] [CrossRef]
- Chakraborty, S.; Newton, A.C. Climate Change, Plant Diseases and Food Security: An Overview. Plant Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Ehling, G.; Cockburn, A.; Snowdon, P.; Buschhaus, H. The Significance of the Fusarium Toxin Deoxynivalenol (DON) for Human and Animal Health. Cereal Res. Commun. 1997, 25, 443–447. [Google Scholar] [CrossRef]
- Ji, F.; He, D.; Olaniran, A.O.; Mokoena, M.P.; Xu, J.; Shi, J. Occurrence, Toxicity, Production and Detection of Fusarium Mycotoxin: A Review. Food Prod. Process. Nutr. 2019, 1, 6. [Google Scholar] [CrossRef]
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A Unified Effort to Fight an Enemy of Wheat and Barley: Fusarium Head Blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.A.; Lipps, P.E.; Hershman, D.E.; McMullen, M.P.; Draper, M.A.; Madden, L.V. Efficacy of Triazole-Based Fungicides for Fusarium Head Blight and Deoxynivalenol Control in Wheat: A Multivariate Meta-Analysis. Phytopathology 2008, 98, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide Emergence of Resistance to Antifungal Drugs Challenges Human Health and Food Security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef]
- Song, J.; Zhang, S.; Lu, L. Fungal Cytochrome P450 Protein Cyp51: What We Can Learn from Its Evolution, Regulons and Cyp51-Based Azole Resistance. Fungal Biol. Rev. 2018, 32, 131–142. [Google Scholar] [CrossRef]
- Becher, R.; Wirsel, S.G.R. Fungal Cytochrome P450 Sterol 14α-Demethylase (CYP51) and Azole Resistance in Plant and Human Pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef]
- Wang, J.; Yu, J.; Liu, J.; Yuan, Y.; Li, N.; He, M.; Qi, T.; Hui, G.; Xiong, L.; Liu, D. Novel Mutations in CYP51B from Penicillium digitatum Involved in Prochloraz Resistance. J. Microbiol. 2014, 52, 762–770. [Google Scholar] [CrossRef]
- Pereira, D.A.; McDonald, B.A.; Brunner, P.C. Mutations in the CYP51 Gene Reduce DMI Sensitivity in Parastagonospora nodorum Populations in Europe and China. Pest Manag. Sci. 2017, 73, 1503–1510. [Google Scholar] [CrossRef]
- Gonzalez-Jimenez, I.; Lucio, J.; Amich, J.; Cuesta, I.; Sanchez Arroyo, R.; Alcazar-Fuoli, L.; Mellado, E. A Cyp51B Mutation Contributes to Azole Resistance in Aspergillus fumigatus. J. Fungi 2020, 6, 315. [Google Scholar] [CrossRef]
- Franco, C.H.; Warhurst, D.C.; Bhattacharyya, T.; Au, H.Y.A.; Le, H.; Giardini, M.A.; Pascoalino, B.S.; Torrecilhas, A.C.; Romera, L.M.D.; Madeira, R.P.; et al. Novel Structural CYP51 Mutation in Trypanosoma cruzi Associated with Multidrug Resistance to CYP51 Inhibitors and Reduced Infectivity. Int. J. Parasitol. Drugs Drug Resist. 2020, 13, 107–120. [Google Scholar] [CrossRef]
- Toyotome, T.; Onishi, K.; Sato, M.; Kusuya, Y.; Hagiwara, D.; Watanabe, A.; Takahashi, H. Identification of Novel Mutations Contributing to Azole Tolerance of Aspergillus fumigatus through in vitro Exposure to Tebuconazole. Antimicrob. Agents Chemother. 2021, 65, e02657-20. [Google Scholar] [CrossRef]
- Hamamoto, H.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. Tandem Repeat of a Transcriptional Enhancer Upstream of the Sterol 14α-Demethylase Gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 2000, 66, 3421–3426. [Google Scholar] [CrossRef]
- Ma, Z.; Proffer, T.J.; Jacobs, J.L.; Sundin, G.W. Overexpression of the 14α-Demethylase Target Gene (CYP51) Mediates Fungicide Resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 2006, 72, 2581–2585. [Google Scholar] [CrossRef]
- Cools, H.J.; Bayon, C.; Atkins, S.; Lucas, J.A.; Fraaije, B.A. Overexpression of the Sterol 14α-Demethylase Gene (MgCYP51) in Mycosphaerella graminicola Isolates Confers a Novel Azole Fungicide Sensitivity Phenotype. Pest. Manag. Sci. 2012, 68, 1034–1040. [Google Scholar] [CrossRef]
- Villani, S.M.; Hulvey, J.; Hily, J.-M.; Cox, K.D. Overexpression of the CYP51A1 Gene and Repeated Elements Are Associated with Differential Sensitivity to DMI Fungicides in Venturia inaequalis. Phytopathology 2016, 106, 562–571. [Google Scholar] [CrossRef]
- Chen, S.; Yuan, N.; Schnabel, G.; Luo, C. Function of the Genetic Element ‘Mona’ Associated with Fungicide Resistance in Monilinia fructicola. Mol. Plant Pathol. 2017, 18, 90–97. [Google Scholar] [CrossRef]
- Nakaune, R.; Adachi, K.; Nawata, O.; Tomiyama, M.; Akutsu, K.; Hibi, T. A Novel ATP-Binding Cassette Transporter Involved in Multidrug Resistance in the Phytopathogenic Fungus Penicillium digitatum. Appl. Environ. Microbiol. 1998, 64, 3983–3988. [Google Scholar] [CrossRef]
- Hayashi, K.; Schoonbeek, H.; De Waard, M.A. Expression of the ABC Transporter BcatrD from Botrytis cinerea Reduces Sensitivity to Sterol Demethylation Inhibitor Fungicides. Pestic. Biochem. Physiol. 2002, 73, 110–121. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Zhang, X.; Zhang, H.; Sun, X.; Hu, C.; Li, S. CDR4 Is the Major Contributor to Azole Resistance among Four Pdr5p-like ABC Transporters in Neurospora crassa. Fungal Biol. 2012, 116, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Omrane, S.; Sghyer, H.; Audéon, C.; Lanen, C.; Duplaix, C.; Walker, A.-S.; Fillinger, S. Fungicide Efflux and the MgMFS1 Transporter Contribute to the Multidrug Resistance Phenotype in Zymoseptoria tritici Field Isolates. Environ. Microbiol. 2015, 17, 2805–2823. [Google Scholar] [CrossRef]
- Whaley, S.G.; Zhang, Q.; Caudle, K.E.; Rogers, P.D. Relative Contribution of the ABC Transporters Cdr1, Pdh1, and Snq2 to Azole Resistance in Candida glabrata. Antimicrob. Agents Chemother. 2018, 62, e01070-18. [Google Scholar] [CrossRef]
- Dunkel, N.; Liu, T.T.; Barker, K.S.; Homayouni, R.; Morschhäuser, J.; Rogers, P.D. A Gain-of-Function Mutation in the Transcription Factor Upc2p Causes Upregulation of Ergosterol Biosynthesis Genes and Increased Fluconazole Resistance in a Clinical Candida albicans Isolate. Eukaryot. Cell 2008, 7, 1180–1190. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, Y.; Wu, Z.; Li, N.; Chen, Y.; Qin, T.; Geng, H.; Xiong, L.; Liu, D. A Novel Sterol Regulatory Element-Binding Protein Gene (sreA) Identified in Penicillium digitatum Is Required for Prochloraz Resistance, Full Virulence and Erg11 (Cyp51) Regulation. PLoS ONE 2015, 10, e0117115. [Google Scholar] [CrossRef]
- Du, W.; Zhai, P.; Wang, T.; Bromley, M.J.; Zhang, Y.; Lu, L. The C2H2 Transcription Factor SltA Contributes to Azole Resistance by Coregulating the Expression of the Drug Target Erg11A and the Drug Efflux Pump Mdr1 in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2021, 65, e01839-20. [Google Scholar] [CrossRef]
- Hagiwara, D.; Miura, D.; Shimizu, K.; Paul, S.; Ohba, A.; Gonoi, T.; Watanabe, A.; Kamei, K.; Shintani, T.; Moye-Rowley, W.S.; et al. A Novel Zn2-Cys6 Transcription Factor AtrR Plays a Key Role in an Azole Resistance Mechanism of Aspergillus fumigatus by Co-Regulating cyp51A and cdr1B Expressions. PLoS Pathog. 2017, 13, e1006096. [Google Scholar] [CrossRef]
- Liu, Z.; Jian, Y.; Chen, Y.; Kistler, H.C.; He, P.; Ma, Z.; Yin, Y. A Phosphorylated Transcription Factor Regulates Sterol Biosynthesis in Fusarium graminearum. Nat. Commun. 2019, 10, 1228. [Google Scholar] [CrossRef]
- Vik, Å.; Rine, J. Upc2p and Ecm22p, Dual Regulators of Sterol Biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 6395–6405. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, S.; Akache, B.; Weber, S.; Deken, X.D.; Raymond, M.; Turcotte, B. Candida albicans Zinc Cluster Protein Upc2p Confers Resistance to Antifungal Drugs and Is an Activator of Ergosterol Biosynthetic Genes. Antimicrob. Agents Chemother. 2005, 49, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.G.; Stamnes, M.A.; Li, Y.; Rogers, P.D.; Moye-Rowley, W.S. The Candida glabrata Upc2A Transcription Factor Is a Global Regulator of Antifungal Drug Resistance Pathways. PLoS Genet. 2021, 17, e1009582. [Google Scholar] [CrossRef] [PubMed]
- Silver, P.M.; Oliver, B.G.; White, T.C. Role of Candida albicans Transcription Factor Upc2p in Drug Resistance and Sterol Metabolism. Eukaryot. Cell 2004, 3, 1391–1397. [Google Scholar] [CrossRef]
- Yokoyama, C.; Wang, X.; Briggs, M.R.; Admon, A.; Wu, J.; Hua, X.; Goldstein, J.L.; Brown, M.S. SREBP-1, a Basic-Helix-Loop-Helix-Leucine Zipper Protein That Controls Transcription of the Low Density Lipoprotein Receptor Gene. Cell 1993, 75, 187–197. [Google Scholar] [CrossRef]
- Hua, X.; Yokoyama, C.; Wu, J.; Briggs, M.R.; Brown, M.S.; Goldstein, J.L.; Wang, X. SREBP-2, a Second Basic-Helix-Loop-Helix-Leucine Zipper Protein That Stimulates Transcription by Binding to a Sterol Regulatory Element. Proc. Natl. Acad. Sci. USA 1993, 90, 11603–11607. [Google Scholar] [CrossRef]
- Hughes, A.L.; Todd, B.L.; Espenshade, P.J. SREBP Pathway Responds to Sterols and Functions as an Oxygen Sensor in Fission Yeast. Cell 2005, 120, 831–842. [Google Scholar] [CrossRef]
- Chang, Y.C.; Bien, C.M.; Lee, H.; Espenshade, P.J.; Kwon-Chung, K.J. Sre1p, a Regulator of Oxygen Sensing and Sterol Homeostasis, Is Required for Virulence in Cryptococcus neoformans. Mol. Microbiol. 2007, 64, 614–629. [Google Scholar] [CrossRef]
- DuBois, J.C.; Smulian, A.G. Sterol Regulatory Element Binding Protein (Srb1) Is Required for Hypoxic Adaptation and Virulence in the Dimorphic Fungus Histoplasma capsulatum. PLoS ONE 2016, 11, e0163849. [Google Scholar] [CrossRef]
- Qin, L.; Wu, V.W.; Glass, N.L. Deciphering the Regulatory Network between the SREBP Pathway and Protein Secretion in Neurospora crassa. mBio 2017, 8, e00233-17. [Google Scholar] [CrossRef]
- Blosser, S.J.; Cramer, R.A. SREBP-Dependent Triazole Susceptibility in Aspergillus fumigatus Is Mediated through Direct Transcriptional Regulation of Erg11A (Cyp51A). Antimicrob. Agents Chemother. 2012, 56, 248–257. [Google Scholar] [CrossRef]
- Carvajal, E.; van den Hazel, H.B.; Cybularz-Kolaczkowska, A.; Balzi, E.; Goffeau, A. Molecular and Phenotypic Characterization of Yeast PDR1 Mutants That Show Hyperactive Transcription of Various ABC Multidrug Transporter Genes. Mol. Gen. Genet. 1997, 256, 406–415. [Google Scholar] [CrossRef][Green Version]
- Katzmann, D.J.; Hallstrom, T.C.; Mahé, Y.; Moye-Rowley, W.S. Multiple Pdr1p/Pdr3p Binding Sites Are Essential for Normal Expression of the ATP Binding Cassette Transporter Protein-Encoding Gene PDR5. J. Biol. Chem. 1996, 271, 23049–23054. [Google Scholar] [CrossRef]
- Coste, A.T.; Karababa, M.; Ischer, F.; Bille, J.; Sanglard, D. TAC1, Transcriptional Activator of CDR Genes, Is a New Transcription Factor Involved in the Regulation of Candida albicans ABC Transporters CDR1 and CDR2. Eukaryot. Cell 2004, 3, 1639–1652. [Google Scholar] [CrossRef]
- Coste, A.; Turner, V.; Ischer, F.; Morschhäuser, J.; Forche, A.; Selmecki, A.; Berman, J.; Bille, J.; Sanglard, D. A Mutation in Tac1p, a Transcription Factor Regulating CDR1 and CDR2, Is Coupled with Loss of Heterozygosity at Chromosome 5 to Mediate Antifungal Resistance in Candida albicans. Genetics 2006, 172, 2139–2156. [Google Scholar] [CrossRef]
- Morschhäuser, J.; Barker, K.S.; Liu, T.T.; Blaß-Warmuth, J.; Homayouni, R.; Rogers, P.D. The Transcription Factor Mrr1p Controls Expression of the MDR1 Efflux Pump and Mediates Multidrug Resistance in Candida albicans. PLoS Pathog. 2007, 3, e164. [Google Scholar] [CrossRef]
- Yin, Y.; Zhang, H.; Zhang, Y.; Hu, C.; Sun, X.; Liu, W.; Li, S. Fungal Zn(II)2Cys6 Transcription Factor ADS-1 Regulates Drug Efflux and Ergosterol Metabolism under Antifungal Azole Stress. Antimicrob. Agents Chemother. 2021, 65, e01316-20. [Google Scholar] [CrossRef]
- Furukawa, T.; van Rhijn, N.; Fraczek, M.; Gsaller, F.; Davies, E.; Carr, P.; Gago, S.; Fortune-Grant, R.; Rahman, S.; Gilsenan, J.M.; et al. The Negative Cofactor 2 Complex Is a Key Regulator of Drug Resistance in Aspergillus fumigatus. Nat. Commun. 2020, 11, 427. [Google Scholar] [CrossRef]
- Paul, S.; Stamnes, M.; Thomas, G.H.; Liu, H.; Hagiwara, D.; Gomi, K.; Filler, S.G.; Moye-Rowley, W.S. AtrR Is an Essential Determinant of Azole Resistance in Aspergillus fumigatus. mBio 2019, 10, e02563-18. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, S.; Li, W.; Sun, H.; Deng, Y.; Zhang, A.; Chen, H. Functional Characterization of Calcineurin-Responsive Transcription Factors Fg01341 and Fg01350 in Fusarium graminearum. Front. Microbiol. 2020, 11, 597998. [Google Scholar] [CrossRef]
- Zhao, Y.; Chi, M.; Sun, H.; Qian, H.; Yang, J.; Huang, J. The FgCYP51B Y123H Mutation Confers Reduced Sensitivity to Prochloraz and Is Important for Conidiation and Ascospore Development in Fusarium graminearum. Phytopathology 2021, 111, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Catlett, N.L.; Lee, B.-N.; Yoder, O.C.; Turgeon, B.G. Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced Virulence of Gibberella zeae Caused by Disruption of a Trichothecene Toxin Biosynthetic Gene. Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, G.; Xu, J.-R. Efficient approaches for generating GFP fusion and epitope-tagging constructs in Filamentous fungi. In Fungal Genomics: Methods and Protocols; Xu, J.-R., Bluhm, B.H., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; pp. 199–212. ISBN 978-1-61779-040-9. [Google Scholar]
- Cavinder, B.; Sikhakolli, U.; Fellows, K.M.; Trail, F. Sexual Development and Ascospore Discharge in Fusarium graminearum. JoVE 2012, 61, e3895. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An Orphan Protein of Fusarium graminearum Modulates Host Immunity by Mediating Proteasomal Degradation of TaSnRK1α. Nat. Commun. 2020, 11, 4382. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.; Hou, Z.; Tracy, M.; Kistler, H.C.; Xu, J.-R. Random Insertional Mutagenesis Identifies Genes Associated with Virulence in the Wheat Scab Fungus Fusarium graminearum. Phytopathology 2005, 95, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Menke, J.; Dong, Y.; Kistler, H.C. Fusarium graminearum Tri12p Influences Virulence to Wheat and Trichothecene Accumulation. Mol. Plant Microbe Interact. 2012, 25, 1408–1418. [Google Scholar] [CrossRef]
- Qian, H.; Du, J.; Chi, M.; Sun, X.; Liang, W.; Huang, J.; Li, B. The Y137H Mutation in the Cytochrome P450 FgCYP51B Protein Confers Reduced Sensitivity to Tebuconazole in Fusarium graminearum. Pest. Manag. Sci. 2018, 74, 1472–1477. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Becher, R.; Weihmann, F.; Deising, H.B.; Wirsel, S.G. Development of a Novel Multiplex DNA Microarray for Fusarium graminearum and Analysis of Azole Fungicide Responses. BMC Genom. 2011, 12, 52. [Google Scholar] [CrossRef]
- Dufresne, M.; van der Lee, T.; M’Barek, S.B.; Xu, X.; Zhang, X.; Liu, T.; Waalwijk, C.; Zhang, W.; Kema, G.H.J.; Daboussi, M.-J. Transposon-Tagging Identifies Novel Pathogenicity Genes in Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 1552–1561. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Lee, W.; Wong, P.; Münsterkötter, M.; Mewes, H.-W.; Schmeitzl, C.; Varga, E.; Berthiller, F.; Adam, G.; Güldener, U. The Fusarium graminearum Genome Reveals More Secondary Metabolite Gene Clusters and Hints of Horizontal Gene Transfer. PLoS ONE 2014, 9, e110311. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, J.C.; Jin, J.M.; Yun, S.H.; Lee, Y.W. Functional characterization of genes located at the aurofusarin biosynthesis gene cluster in Gibberella zeae. Plant Pathol. J. 2008, 24, 8–16. [Google Scholar] [CrossRef]
- Frandsen, R.J.N.; Nielsen, N.J.; Maolanon, N.; Sørensen, J.C.; Olsson, S.; Nielsen, J.; Giese, H. The Biosynthetic Pathway for Aurofusarin in Fusarium graminearum Reveals a Close Link between the Naphthoquinones and Naphthopyrones. Mol. Microbiol. 2006, 61, 1069–1080. [Google Scholar] [CrossRef]
- Yun, Y.; Yin, D.; Dawood, D.H.; Liu, X.; Chen, Y.; Ma, Z. Functional Characterization of FgERG3 and FgERG5 Associated with Ergosterol Biosynthesis, Vegetative Differentiation and Virulence of Fusarium graminearum. Fungal Genet. Biol. 2014, 68, 60–70. [Google Scholar] [CrossRef]
- Fan, J.; Urban, M.; Parker, J.E.; Brewer, H.C.; Kelly, S.L.; Hammond-Kosack, K.E.; Fraaije, B.A.; Liu, X.; Cools, H.J. Characterization of the Sterol 14α-Demethylases of Fusarium graminearum Identifies a Novel Genus-Specific CYP51 Function. New Phytol. 2013, 198, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, F.; Schnabel, G.; Wu, J.; Wang, Z.; Ma, Z. Paralogous Cyp51 Genes in Fusarium graminearum Mediate Differential Sensitivity to Sterol Demethylation Inhibitors. Fungal Genet. Biol. 2011, 48, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, R.J.N.; Schütt, C.; Lund, B.W.; Staerk, D.; Nielsen, J.; Olsson, S.; Giese, H. Two Novel Classes of Enzymes Are Required for the Biosynthesis of Aurofusarin in Fusarium graminearum. J. Biol. Chem. 2011, 286, 10419–10428. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-E.; Han, K.-H.; Jin, J.; Kim, H.; Kim, J.-C.; Yun, S.-H.; Lee, Y.-W. Putative Polyketide Synthase and Laccase Genes for Biosynthesis of Aurofusarin in Gibberella zeae. Appl. Environ. Microbiol. 2005, 71, 1701–1708. [Google Scholar] [CrossRef]
- Malz, S.; Grell, M.N.; Thrane, C.; Maier, F.J.; Rosager, P.; Felk, A.; Albertsen, K.S.; Salomon, S.; Bohn, L.; Schäfer, W.; et al. Identification of a Gene Cluster Responsible for the Biosynthesis of Aurofusarin in the Fusarium graminearum Species Complex. Fungal Genet. Biol. 2005, 42, 420–433. [Google Scholar] [CrossRef]
- Lee, S.; Son, H.; Lee, J.; Lee, Y.-R.; Lee, Y.-W. A Putative ABC Transporter Gene, ZRA1, Is Required for Zearalenone Production in Gibberella zeae. Curr. Genet. 2011, 57, 343–351. [Google Scholar] [CrossRef]
- Ammar, G.A.; Tryono, R.; Döll, K.; Karlovsky, P.; Deising, H.B.; Wirsel, S.G.R. Identification of ABC Transporter Genes of Fusarium graminearum with Roles in Azole Tolerance and/or Virulence. PLoS ONE 2013, 8, e79042. [Google Scholar] [CrossRef]
- Son, H.; Seo, Y.-S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.-M.; Lin, Y.; Cao, P.; Hong, S.-Y.; Kim, E.-K.; et al. A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef]
- Shin, J.; Bui, D.-C.; Kim, S.; Jung, S.Y.; Nam, H.J.; Lim, J.Y.; Choi, G.J.; Lee, Y.-W.; Kim, J.-E.; Son, H. The Novel BZIP Transcription Factor Fpo1 Negatively Regulates Perithecial Development by Modulating Carbon Metabolism in the Ascomycete Fungus Fusarium graminearum. Environ. Microbiol. 2020, 22, 2596–2612. [Google Scholar] [CrossRef]
- Lin, Y.; Son, H.; Min, K.; Lee, J.; Choi, G.J.; Kim, J.-C.; Lee, Y.-W. A Putative Transcription Factor MYT2 Regulates Perithecium Size in the Ascomycete Gibberella zeae. PLoS ONE 2012, 7, e37859. [Google Scholar] [CrossRef][Green Version]
- Son, H.; Min, K.; Lee, J.; Choi, G.J.; Kim, J.-C.; Lee, Y.-W. Differential Roles of Pyruvate Decarboxylase in Aerial and Embedded Mycelia of the Ascomycete Gibberella zeae. FEMS Microbiol. Lett. 2012, 329, 123–130. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Chen, Q.; Liu, C.-H.; Liu, Y.-B.; Yi, P.; Niu, K.-X.; Wang, Y.-Q.; Wang, A.-Q.; Yu, H.-Y.; Pu, Z.-E.; et al. Chitin Synthase Gene FgCHS8 Affects Virulence and Fungal Cell Wall Sensitivity to Environmental Stress in Fusarium graminearum. Fungal Biol. 2016, 120, 764–774. [Google Scholar] [CrossRef]
- Kim, D.-W.; Shin, Y.-K.; Lee, S.-W.; Wimonmuang, K.; Kang, K.B.; Lee, Y.-S.; Yun, S.-H. FgPKS7 Is an Essential Player in Mating-Type-Mediated Regulatory Pathway Required for Completing Sexual Cycle in Fusarium graminearum. Environ. Microbiol. 2021, 23, 1972–1990. [Google Scholar] [CrossRef]
- Luo, X.; Mao, H.; Wei, Y.; Cai, J.; Xie, C.; Sui, A.; Yang, X.; Dong, J. The Fungal-Specific Transcription Factor Vdpf Influences Conidia Production, Melanized Microsclerotia Formation and Pathogenicity in Verticillium dahliae: The Function of Vdpf in Verticillium dahliae. Mol. Plant Pathol. 2016, 17, 1364–1381. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Wenner, N.; Kuldau, G.A.; Trail, F. Deoxynivalenol Biosynthesis-Related Gene Expression during Wheat Kernel Colonization by Fusarium graminearum. Phytopathology 2011, 101, 1091–1096. [Google Scholar] [CrossRef]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.-H.; Felk, A.; Maier, F.J. Infection Patterns in Barley and Wheat Spikes Inoculated with Wild-Type and Trichodiene Synthase Gene Disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Zhang, X.-W.; Jia, L.-J.; Zhang, Y.; Jiang, G.; Li, X.; Zhang, D.; Tang, W.-H. In Planta Stage-Specific Fungal Gene Profiling Elucidates the Molecular Strategies of Fusarium graminearum Growing inside Wheat Coleoptiles. Plant Cell 2012, 24, 5159–5176. [Google Scholar] [CrossRef]

| Protein Name | Gene Locus | Log2(Mutant/Wild-Type) | Reference |
|---|---|---|---|
| Sterol-biosynthesis-related genes | |||
| FgERG6B | FGSG_05740 | −5.52 | [71] |
| FgERG5B | FGSG_03686 | −4.87 | [71] |
| FgCYP51A | FGSG_04092 | −4.25 | [72,73] |
| Aurofusarin biosynthesis gene cluster | |||
| GIP3/AurO | FGSG_02321 | −5.00 | [69,74] |
| GIP4/AurT | FGSG_02322 | −3.21 | [69,74] |
| GIP5/AurR2 | FGSG_02323 | −3.03 | [70] |
| PKS12 | FGSG_02324 | −5.88 | [75,76] |
| GIP6/AurZ | FGSG_02325 | −6.62 | [74] |
| GIP7/AurJ | FGSG_02326 | −5.62 | [69,70] |
| GIP8/AurF | FGSG_02327 | −5.88 | [69,76] |
| GIP1 | FGSG_02328 | −5.05 | [70,75] |
| ABC transporter | |||
| ZRA1 | FGSG_02139 | −4.11 | [77] |
| FgABC1 | FGSG_10995 | −3.15 | [78] |
| FGSG_08312 | −2.16 | [66] | |
| FGSG_05076 | −3.63 | [66] | |
| FGSG_02847 | −2.49 | [66] | |
| FGSG_08373 | −1.89 | [66] | |
| Transcription factors | |||
| FGSG_10470 | −3.41 | [79] | |
| FGSG_00404 | −1.36 | [79] | |
| Fpo1 | FGSG_06651 | 1.26 | [80] |
| MYT2 | FGSG_07546 | 2.39 | [81] |
| Others | |||
| PDC1 | FGSG_09834 | −3.16 | [82] |
| Chs8 | FGSG_06550 | −4.98 | [83] |
| FgPKS7 | FGSG_08795 | −6.34 | [84] |
| FGSG_10608 | −3.98 | [68] | |
| FGSG_10609 | −6.40 | [68] | |
| FGSG_10611 | −8.49 | [68] | |
| FGSG_10612 | −10.14 | [68] | |
| FGSG_10613 | −12.20 | [68] | |
| FGSG_10614 | −8.88 | [68] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Sun, H.; Li, J.; Ju, C.; Huang, J. The Transcription Factor FgAtrR Regulates Asexual and Sexual Development, Virulence, and DON Production and Contributes to Intrinsic Resistance to Azole Fungicides in Fusarium graminearum. Biology 2022, 11, 326. https://doi.org/10.3390/biology11020326
Zhao Y, Sun H, Li J, Ju C, Huang J. The Transcription Factor FgAtrR Regulates Asexual and Sexual Development, Virulence, and DON Production and Contributes to Intrinsic Resistance to Azole Fungicides in Fusarium graminearum. Biology. 2022; 11(2):326. https://doi.org/10.3390/biology11020326
Chicago/Turabian StyleZhao, Yanxiang, Huilin Sun, Jingwen Li, Chao Ju, and Jinguang Huang. 2022. "The Transcription Factor FgAtrR Regulates Asexual and Sexual Development, Virulence, and DON Production and Contributes to Intrinsic Resistance to Azole Fungicides in Fusarium graminearum" Biology 11, no. 2: 326. https://doi.org/10.3390/biology11020326
APA StyleZhao, Y., Sun, H., Li, J., Ju, C., & Huang, J. (2022). The Transcription Factor FgAtrR Regulates Asexual and Sexual Development, Virulence, and DON Production and Contributes to Intrinsic Resistance to Azole Fungicides in Fusarium graminearum. Biology, 11(2), 326. https://doi.org/10.3390/biology11020326






