Transgenic Rice Plants Expressing Artificial miRNA Targeting the Rice Stripe Virus MP Gene Are Highly Resistant to the Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence and amiRNA Prediction
2.2. Transformation
2.3. Production of T0 Generation Transgenic Plants
2.4. Detection of Foreign Gene Insertion Sites and Homozygotes
2.5. Assessment of Resistance to RSV
2.6. RNA Analysis
2.7. Real-Time Quantitative PCR and Semiquantitative RT-PCR Analysis
2.8. Western Blotting
3. Results
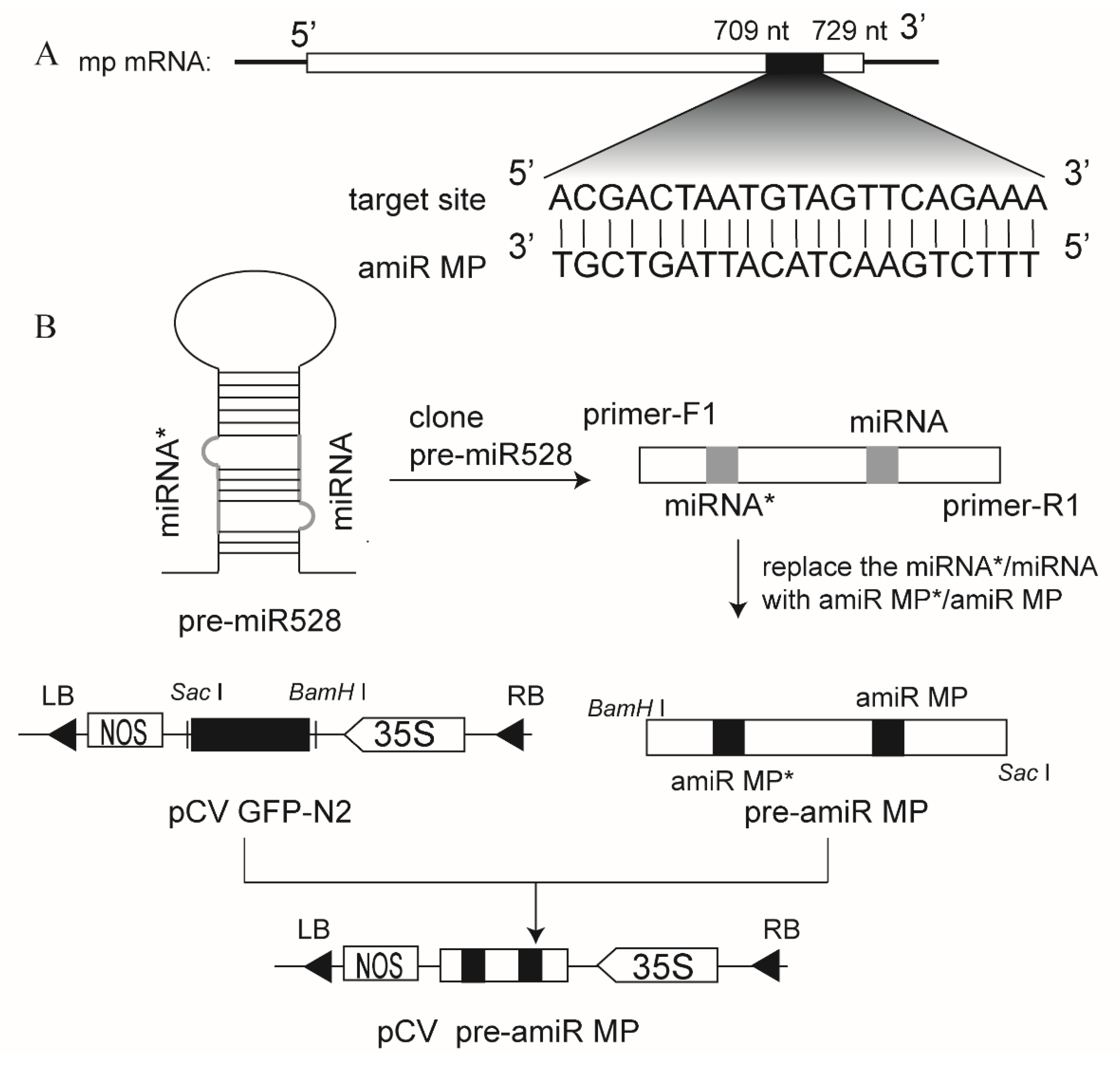
3.1. Construction of the Vector Expressing amiRNA Targeting RSV MP mRNA (amiR MP)
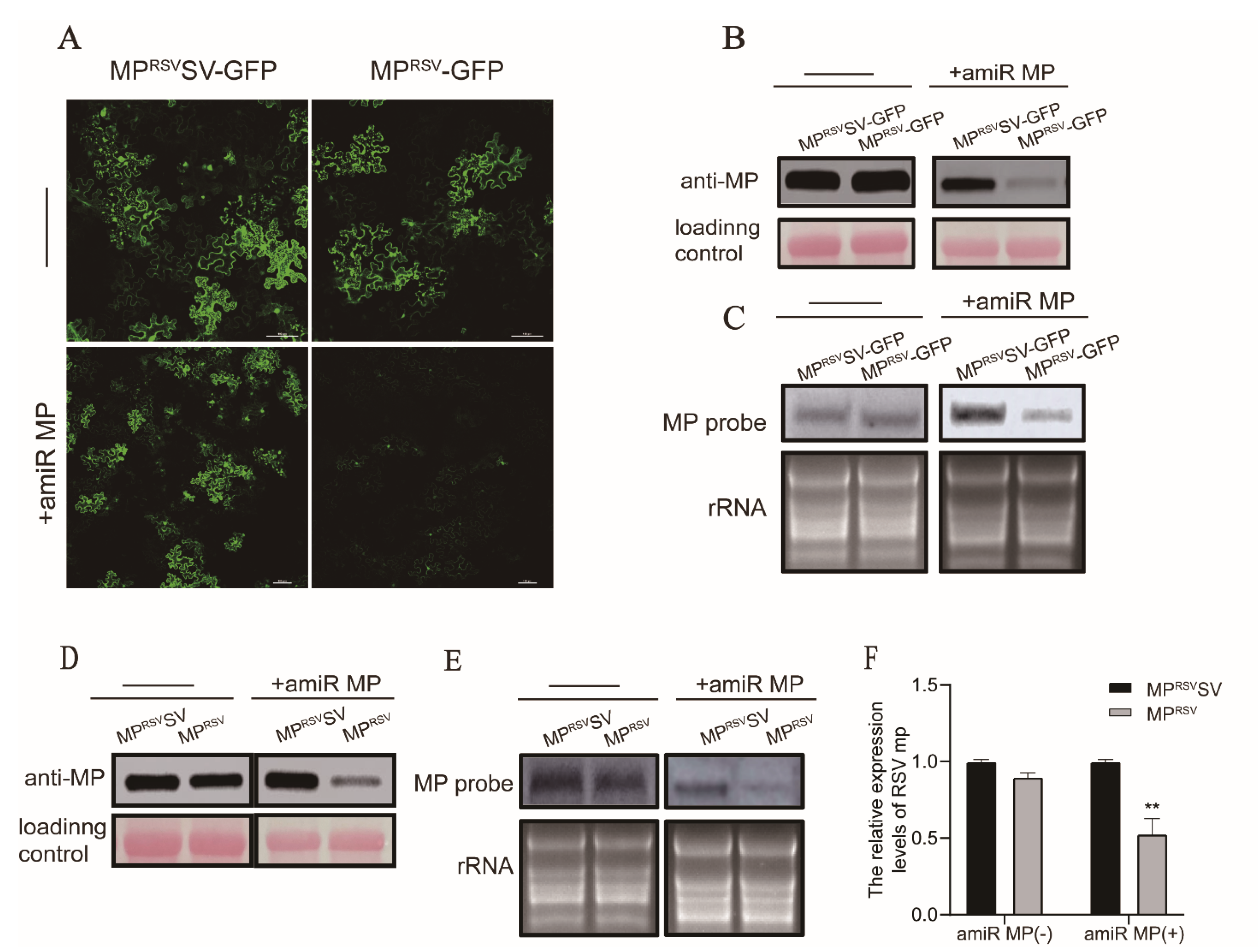
3.2. Expression of amiR MP Decreased Accumulation of MP mRNA in a Transient Expression Assay
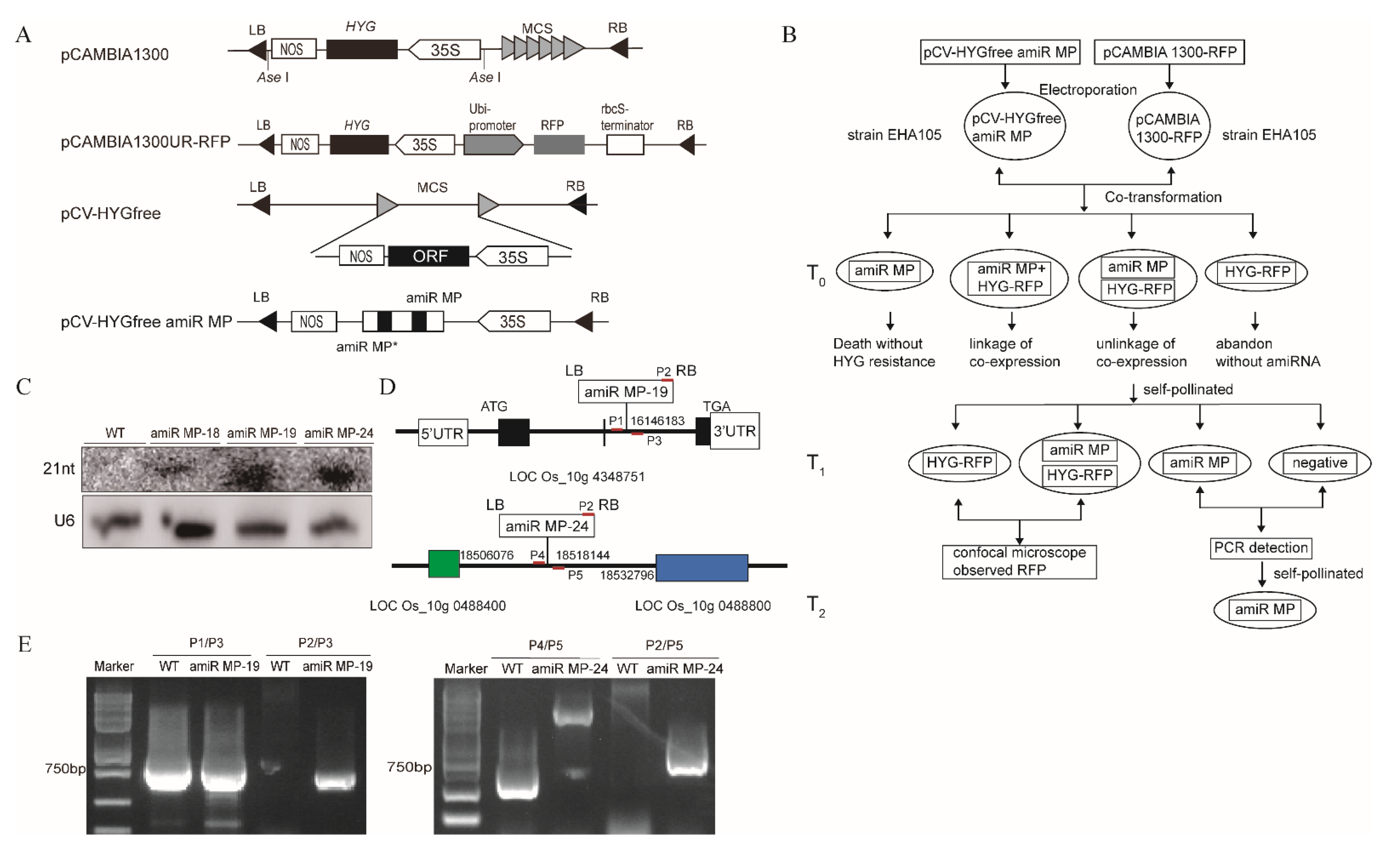
3.3. Generation of Marker-Free Transgenic Rice Expressing amiR MP
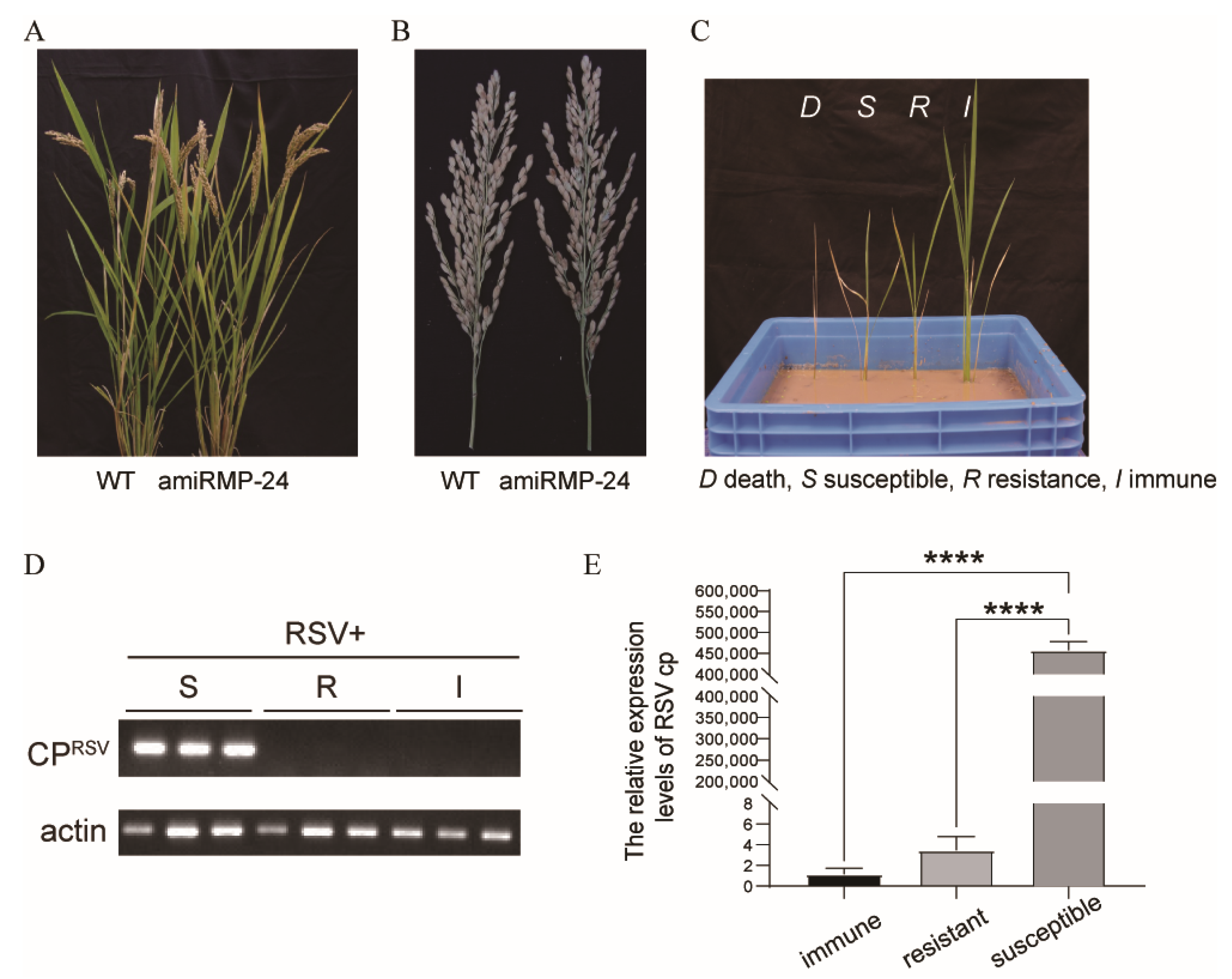
3.4. Transgenic Rice Plants Were Highly Resistant to RSV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, M.; Guan, S.Y.; He, C.Q. Evolution of rice stripe virus. Mol. Phylogenet. Evol. 2017, 109, 343–350. [Google Scholar] [CrossRef]
- Wei, T.Y.; Yang, J.G.; Liao, F.L.; Gao, F.L.; Lu, L.M.; Zhang, X.T.; Li, F.; Wu, Z.J.; Lin, Q.Y.; Xie, L.H.; et al. Genetic diversity and population structure of rice stripe virus in China. J. Gen. Virol. 2009, 90, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Toriyama, S.; Takahashi, M.; Sano, Y.; Shimizu, T.; Ishihama, A. Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 1994, 75, 3569–3579. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Wu, J.; Zhou, Y.; Zhou, X. Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 2008, 82, 12304–12311. [Google Scholar] [CrossRef] [Green Version]
- Jian, Y.; Zhang, F.; Li, J.; Chen, J.P.; Zhang, H.M.; Sek-Man, W. Integrative Analysis of the microRNAome and Transcriptome Illuminates the Response of Susceptible Rice Plants to Rice Stripe Virus. PLoS ONE 2016, 11, e0146946. [Google Scholar]
- Falk, B.W.; Tsai, J.H. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998, 36, 139–163. [Google Scholar] [CrossRef]
- Lin, H.A.; Chen, S.Y.; Chang, F.Y.; Tung, C.W.; Chen, Y.C.; Shen, W.C.; Chen, R.S.; Wu, C.W.; Chung, C.L. Genome-wide association study of rice genes and loci conferring resistance to Magnaporthe oryzae isolates from Taiwan. Bot. Stud. 2018, 59, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Hu, T.; Liu, J.; Li, W.; Wang, D.; Chen, S.; Jiang, Y.; et al. Deep Learning: Individual maize segmentation from terrestrial lidar data using faster R-CNN and regional growth algorithms. Front. Recent Dev. Plant Sci. 2018, 9, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, T.; Zhu, Y.; Itoh, K.; Kimura, Y.; Izawa, T.; Shimamoto, K.; Toriyama, S. Genetically engineered rice resistant to rice stripe virus, an insect-transmitted virus. J. Appl. Biol. Sci. 1992, 89, 9865–9869. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Nakazono-Nagaoka, E.; Uehara-Ichiki, T.; Sasaya, T.; Omura, T. Targeting specific genes for RNA interference is crucial to the development of strong resistance to rice stripe virus. Plant Biotechnol. J. 2011, 9, 503–512. [Google Scholar] [CrossRef]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Norman, W.; Hao, C.; Stephan, O.; Detlef, W.; Philippe, H.; Borevitz, J.O. Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 2008, 3, e1829. [Google Scholar]
- Liu, S.R.; Zhou, J.J.; Hu, C.G.; Wei, C.L.; Zhang, J.Z. MicroRNA-mediated gene silencing in plant defense and viral counter-defense. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Lu, Y.D.; Gan, Q.H.; Chi, X.Y.; Qin, S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 2008, 27, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.Q.; Hao, J.; Li, J.; Barbara, B.; Laixin, L. Artificial microRNA-mediated resistance to Cucumber Green Mottle Mosaic Virus in Nicotiana benthamiana. Planta 2019, 250, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Simon-Mateo, C.; Garcia, J.A. MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 2006, 80, 2429–2436. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Song, Y.; Han, Q.; Zhu, C.; Wen, F. The choice of target site is crucial in artificial miRNA-mediated virus resistance in transgenic Nicotiana tabacum. Physiol. Mol. Plant Pathol. 2011, 76, 2–8. [Google Scholar]
- Ali, I.; Amin, I.; Briddon, R.W.; Mansoor, S. Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virol. J. 2013, 10, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Lin, C.; Du, J.; Song, Y.; Jiang, M.; Liu, H.; Zhou, S.; Wen, F.; Zhu, C. Dimeric artificial microRNAs mediate high resistance to RSV and RBSDV in transgenic rice plants. Plant Cell Tissue Organ Cult. 2016, 126, 127–139. [Google Scholar] [CrossRef]
- Molnar, A.; Bassett, A.; Thuenemann, E.; Schwach, F.; Karkare, S.; Ossowski, S.; Weigel, D.; Baulcombe, D. Highly specific gene silencing by artificial micrornas in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009, 58, 165–174. [Google Scholar] [CrossRef]
- Yan, F.; Lu, Y.; Wu, G.; Peng, J.; Zheng, H.; Lin, L.; Chen, J. A simplified method for constructing artificial microRNAs based on the osa-MIR528 precursor. J. Biotechnol. 2012, 160, 146–150. [Google Scholar] [CrossRef]
- Chen, B.; Lin, L.; Lu, Y.; Peng, J.; Zheng, H.; Yang, Q.; Rao, S.; Wu, G.; Li, J.; Chen, Z.; et al. Ubiquitin-Like protein 5 interacts with the silencing suppressor p3 of rice stripe virus and mediates its degradation through the 26S proteasome pathway. PLoS Pathog. 2020, 16, e1008780. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2010, 47, 969–976. [Google Scholar] [CrossRef]
- Jia, X.; Lin, X.; Chen, J. Linear and exponential TAIL-PCR: A method for efficient and quick amplification of flanking sequences adjacent to Tn5 transposon insertion sites. AMB Express 2017, 7, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Jiang, L.; Yang, J.; Peng, J.; Lu, Y.; Zheng, H.; Lin, L.; Chen, J.; Yan, F. Over-expression of Oryza sativa Xrn4 confers plant resistance to virus infection. Gene 2018, 639, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lu, Y.; Zheng, X.; Yang, X.; Chen, Y.; Zhang, T.; Zhao, X.; Wang, S.; Zhao, X.; Song, X.; et al. The plant protein NbP3IP directs degradation of Rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8. New Phytol. 2021, 229, 1036–1051. [Google Scholar] [CrossRef]
- Dong, R.X.; Chen, J.; Wang, X.M.; Li, J.S.; Jie, Z.; Yang, Y.; Yu, C.L.; Cheng, Y.; Yan, C.Q.; Chen, J.P. Agrobacterium -mediated transformation efficiency is altered in a novel rice bacterial blight resistance cultivar and is influenced by environmental temperature. Physiol. Mol. Plant Pathol. 2012, 77, 33–40. [Google Scholar] [CrossRef]
- Ban, L.; Zhang, S.; Huang, Z.; He, Y.; Peng, Y.; Gao, C. Resistance monitoring and assessment of resistance risk to pymetrozine in Laodelphax striatellus (Hemiptera: Delphacidae). J. Econ. Entomol. 2012, 105, 2129–2135. [Google Scholar] [CrossRef]
- He, Y.; Zhang, J.; Chen, J.; Shen, J. Using synergists to detect multiple insecticide resistance in field populations of rice stem borer. Pestic. Biochem. Physiol. 2012, 103, 121–126. [Google Scholar] [CrossRef]
- Sasaya, T.; Nakazono-Nagaoka, E.; Saika, H.; Aoki, H.; Hiraguri, A.; Netsu, O.; Uehara-Ichiki, T.; Onuki, M.; Toki, S.; Saito, K.; et al. Transgenic strategies to confer resistance against viruses in rice plants. Front. Virol. 2014, 4, 1–11. [Google Scholar] [CrossRef]
- Han, S.; Wu, Z.; Yang, H.; Rong, W.; Yin, Y.; Xie, L.; Tien, P. Ribozyme-mediated resistance to rice dwarf virus and the transgene silencing in the progeny of transgenic rice plants. Transgenic Res. 2000, 9, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Yoshii, M.; Wei, T.; Hirochika, H.; Omura, T. Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of Rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 2010, 7, 24–32. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakazono-Nagaoka, E.; Akita, F.; Uehara-Ichiki, T.; Omura, T.; Sasaya, T. Immunity to Rice black streaked dwarf virus, a plant reovirus, can be achieved in rice plants by RNA silencing against the gene for the viroplasm component protein. Virus Res. 2011, 160, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Mengistu, A.A.; Tenkegna, T.A. The role of miRNA in plant-virus interaction: A review. Mol. Biol. Rep. 2021, 48, 2853–2861. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A Signaling Cascade from miR444 to RDR1 in Rice Antiviral RNA Silencing Pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [Green Version]
- Akhter, Y.; Khan, J.A. Genome wide identification of cotton (Gossypium hirsutum)—Encoded microRNA targets against Cotton leaf curl Burewala virus. Gene 2018, 638, 60–65. [Google Scholar]
- Jiang, Y.; Lin, S.; Jiang, M.; Li, K.; Zhu, C. Production of marker-free and RSV-resistant transgenic rice using a twin T-DNA system and RNAi. J. Biosci. 2013, 38, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Mccormac, A.C.; Fowler, M.R.; Chen, D.F.; Elliott, M.C. Efficient co-transformation of Nicotiana tabacum by two independent T-DNAs, the effect of T-DNA size and implications for genetic separation. Transgenic Res. 2001, 10, 143–155. [Google Scholar] [CrossRef]
- Komari, T.; Hiei, Y.; Saito, Y. Vectors carrying two separate t-DNAs for co- transformation of higher plants mediated by agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 1996, 10, 165–174. [Google Scholar] [CrossRef]
- Poirier, Y.; Ventre, G.; Nawrath, C. High-frequency linkage of co-expressing T-DNA in transgenic Arabidopsis thaliana transformed by vacuum-infiltration of Agrobacterium tumefaciens. Theor. Appl. Genet. 2000, 100, 487–493. [Google Scholar] [CrossRef]
- Daley, M.; Knauf, V.C.; Summerfelt, K.R.; Turner, J.C. Co-transformation with one Agrobacterium tumefaciens strain containing two binary plasmids as a method for producing marker-free transgenic plants. Plant Cell Rep. 1998, 17, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, X.; Liu, X.; Xu, H.; Meng, K.; Xiao, G.; Wei, X.; Feng, W.; Zhu, Z. Green fluorescent protein as a vital elimination marker to easily screen marker-free transgenic progeny derived from plants co-transformed with a double T-DNA binary vector system. Plant Cell Rep. 2005, 23, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Woodbury, P.B.; DiTommaso, A.; Thies, J.; Ryan, M.; Losey, J. Effects of Transgenic Crops on the Environment, In Environmental Pest Management: Challenges for Agronomists, Ecologists, Economists and Policymakers; Coll, M., Wajnberg, E., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 131–150. [Google Scholar]
- He, M.; Zhang, J.; Shen, L.; Xu, L.; Luo, W.; Li, D.; Zhai, N.; Zhao, J.; Long, Y.; Pei, X.; et al. High-throughput sequencing analysis of microbial community diversity in response to indica and japonica bar-transgenic rice paddy soils. PLoS ONE 2019, 14, e0222191. [Google Scholar] [CrossRef] [PubMed]
| Transgenic Line | Numbers of Plants with Different Resistance Levels | Resistance Ratio (%) a | Resistance Status b | |||
|---|---|---|---|---|---|---|
| I | R | S | D | |||
| amiR MP-7 | 16 | 6 | 6 | 2 | 73 | + |
| amiR MP-8 | 21 | 3 | 4 | 2 | 80 | + |
| amiR MP-10 | 19 | 1 | 8 | 2 | 67 | − |
| amiR MP-12 | 18 | 6 | 3 | 3 | 80 | + |
| amiR MP-18 | 18 | 8 | 3 | 1 | 87 | ++ |
| amiR MP-19 | 17 | 9 | 2 | 2 | 87 | ++ |
| amiR MP-24 | 26 | 1 | 2 | 1 | 90 | ++ |
| WT | 6 | 5 | 18 | 1 | 37 | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Yuan, Q.; Ai, X.; Chen, J.; Lu, Y.; Yan, F. Transgenic Rice Plants Expressing Artificial miRNA Targeting the Rice Stripe Virus MP Gene Are Highly Resistant to the Virus. Biology 2022, 11, 332. https://doi.org/10.3390/biology11020332
Zhou L, Yuan Q, Ai X, Chen J, Lu Y, Yan F. Transgenic Rice Plants Expressing Artificial miRNA Targeting the Rice Stripe Virus MP Gene Are Highly Resistant to the Virus. Biology. 2022; 11(2):332. https://doi.org/10.3390/biology11020332
Chicago/Turabian StyleZhou, Liya, Quan Yuan, Xuhong Ai, Jianping Chen, Yuwen Lu, and Fei Yan. 2022. "Transgenic Rice Plants Expressing Artificial miRNA Targeting the Rice Stripe Virus MP Gene Are Highly Resistant to the Virus" Biology 11, no. 2: 332. https://doi.org/10.3390/biology11020332
APA StyleZhou, L., Yuan, Q., Ai, X., Chen, J., Lu, Y., & Yan, F. (2022). Transgenic Rice Plants Expressing Artificial miRNA Targeting the Rice Stripe Virus MP Gene Are Highly Resistant to the Virus. Biology, 11(2), 332. https://doi.org/10.3390/biology11020332






