Methionine Metabolism Is Down-Regulated in Heart of Long-Lived Mammals
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Samples
2.3. Sample Homogeneization and Quantification
2.4. Sample Processing
2.5. Analysis Conditions
2.6. Equipment
2.7. Statistics
3. Results
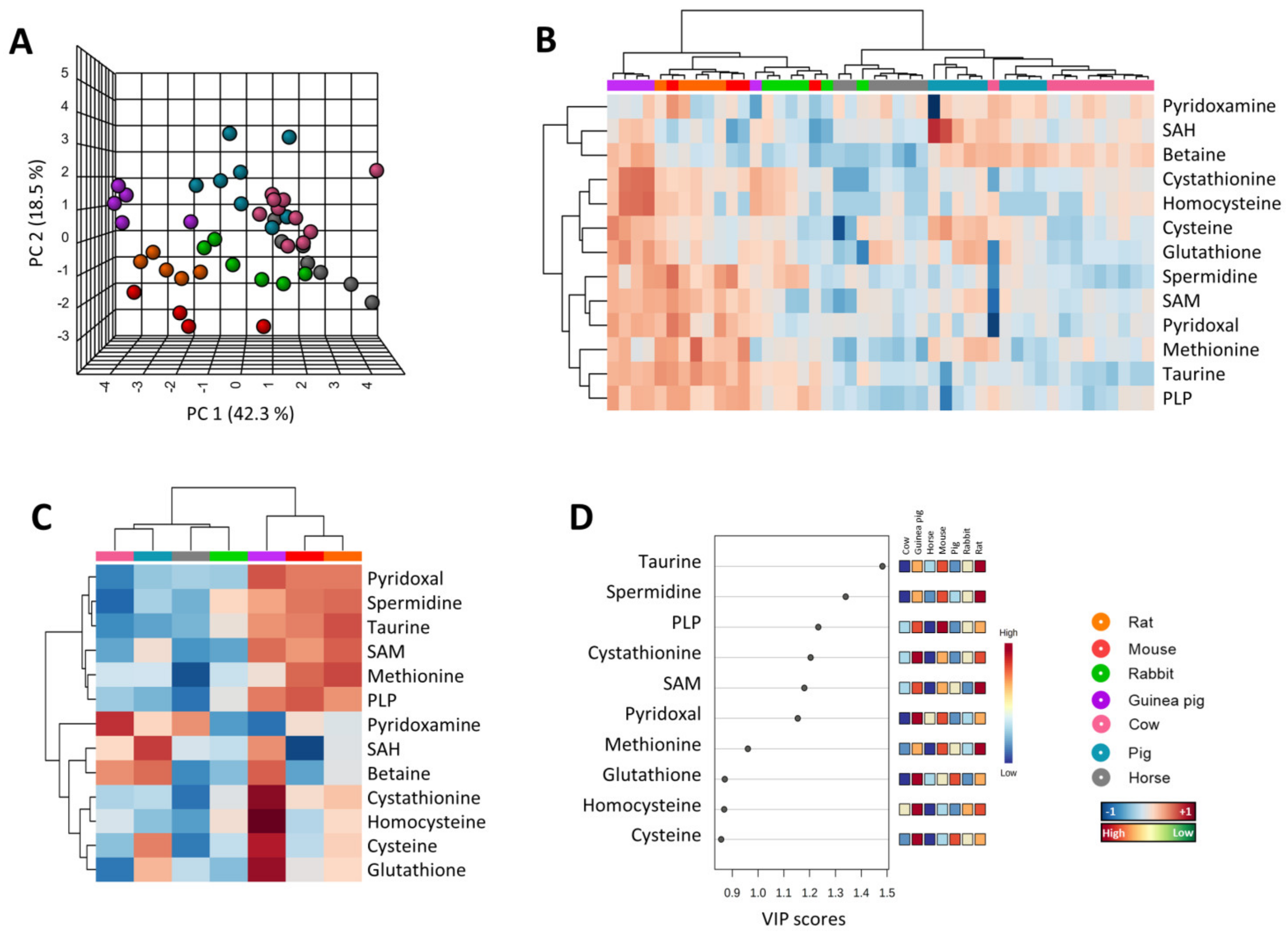
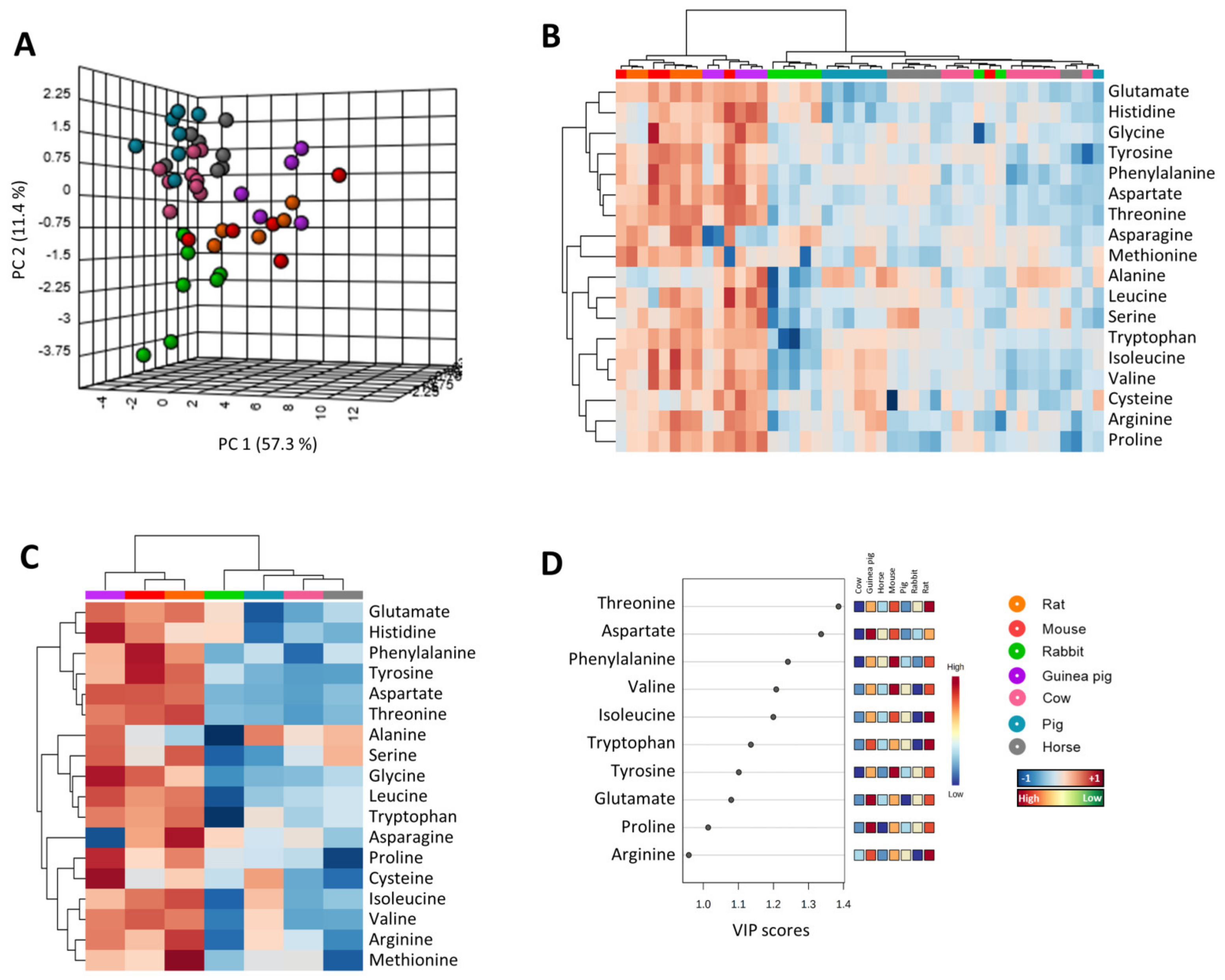
3.1. Multivariate Statistics Reveal a Heart-Species-Specific Methionine-Related Metabolite Profile
3.2. Low Levels of Methionine Related Metabolites in the Heart of Long-Lived Animals
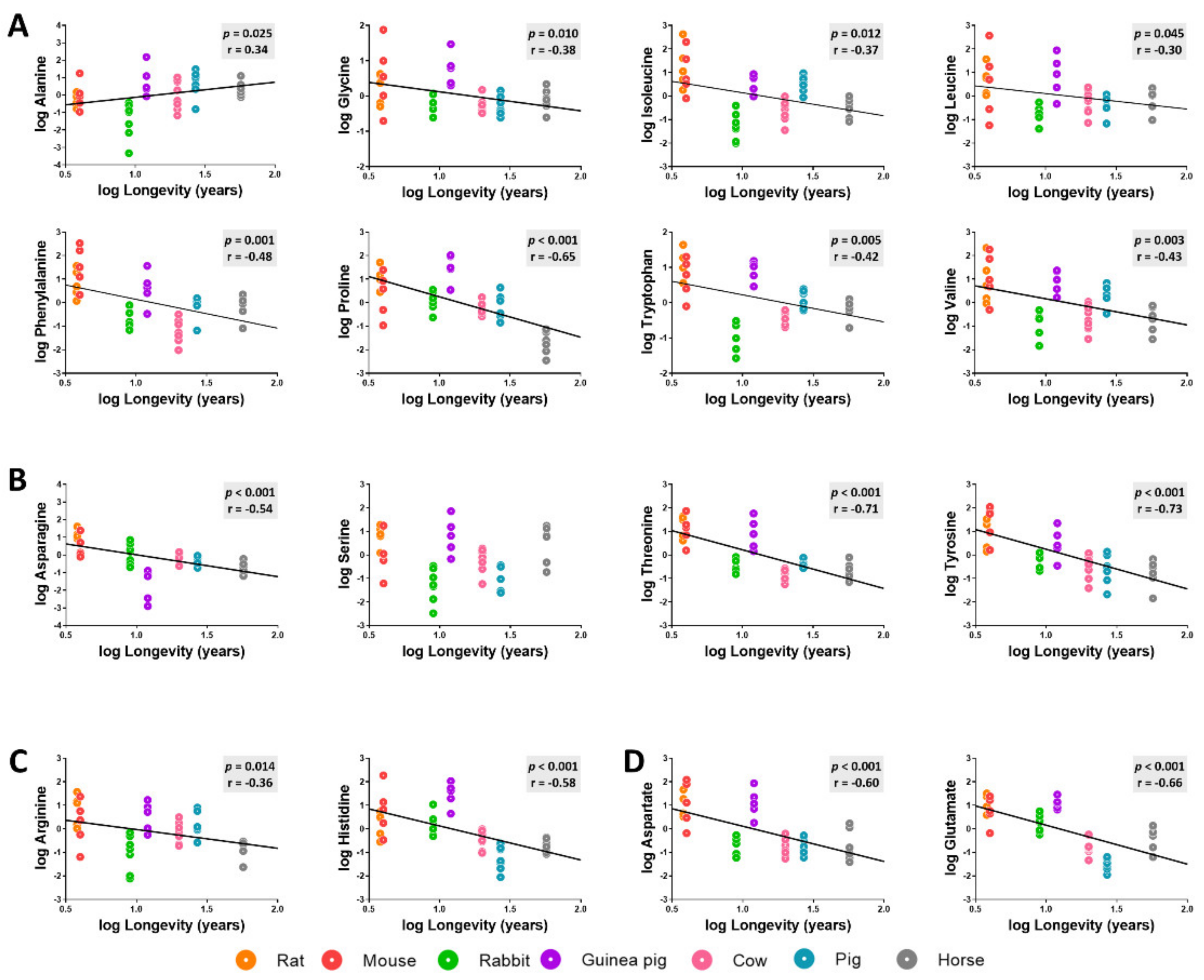
3.3. Amino Acid Content Is Decreased in Heart from Long-Lived Animals
3.4. Heart Metabolome Is Also Related to Longevity concerning Specific Lipid Intermediates
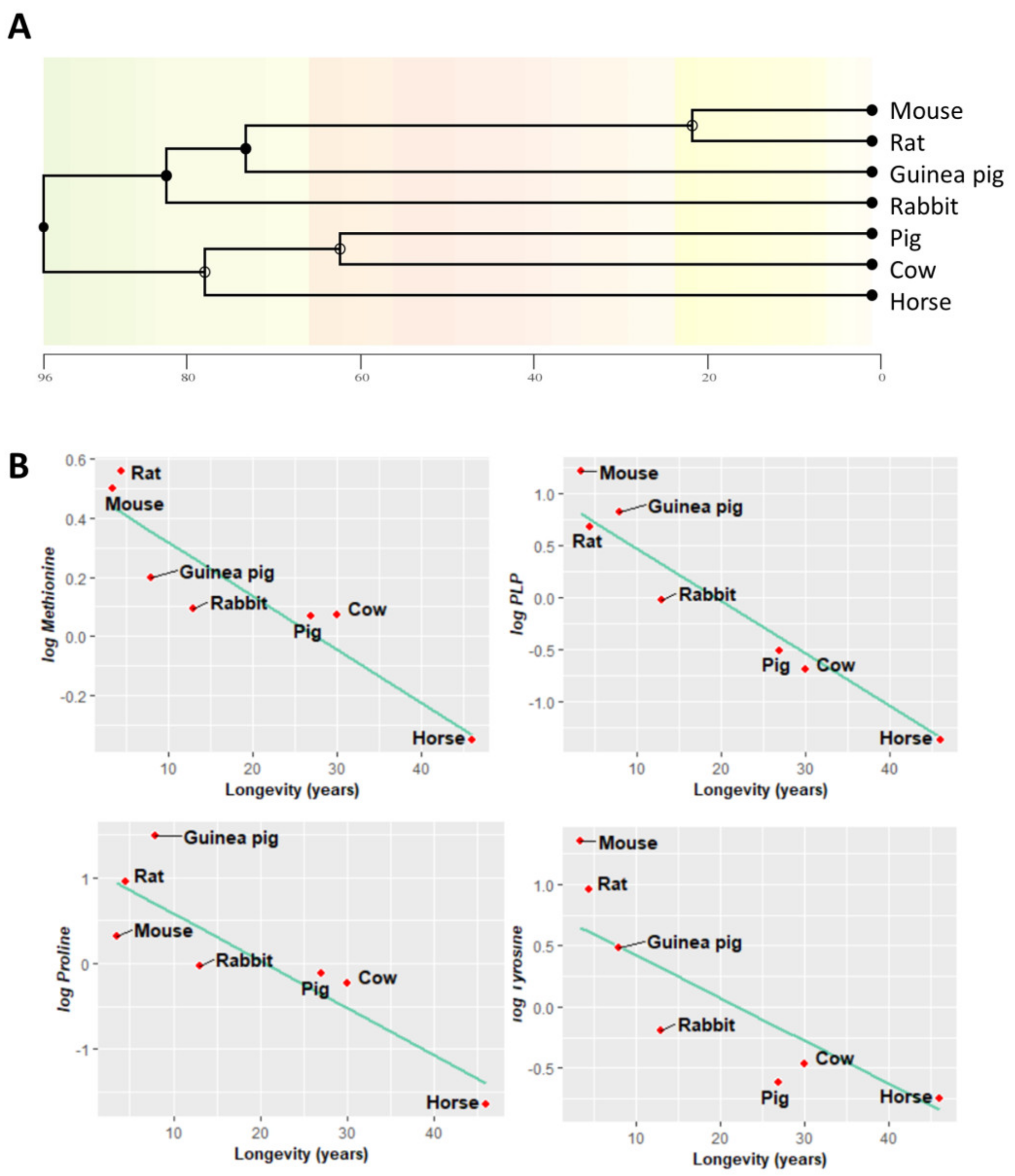
3.5. Methionine-Related Metabolites and Amino Acids Also Correlate with Longevity after Controlling for Phylogenetic Relationships
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Pathway | Metabolite | Rat | Mouse | Rabbit | Guinea Pig | Cow | Pig | Horse |
|---|---|---|---|---|---|---|---|---|
| Methionine metabolism | Betaine * | 6016 ± 726 | 3292 ± 847 | 3355 ± 650 | 19814 ± 3683 | 13995 ± 1173 | 16845 ± 1817 | 2103 ± 301 |
| Cystathionine | 0.12 ± 0.02 | 0.10 ± 0.03 | 0.13 ± 0.03 | 2.10 ± 0.72 | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.01 ± 0.00 | |
| Cysteine | 0.16 ± 0.03 | 0.14 ± 0.04 | 0.11 ± 0.02 | 0.49 ± 0.13 | 0.08 ± 0.02 | 0.30 ± 0.06 | 0.07 ± 0.02 | |
| GSH | 1.45 ± 0.48 | 2.34 ± 1.16 | 1.23 ± 0.30 | 5.00 ± 1.19 | 0.41 ± 0.04 | 2.15 ± 0.43 | 1.21 ± 0.43 | |
| Homocysteine * | 1.42 ± 0.33 | 1.84 ± 0.64 | 1.35 ± 0.41 | 22.5 ± 7.89 | 0.66 ± 0.13 | 0.67 ± 0.13 | 0.45 ± 0.11 | |
| Methionine | 1.08 ± 0.24 | 0.91 ± 0.20 | 0.40 ± 0.02 | 0.65 ± 0.14 | 0.38 ± 0.03 | 0.47 ± 0.08 | 0.22 ± 0.01 | |
| Pyridoxal * | 62.7 ± 11.6 | 66.0 ± 14.0 | 30.2 ± 2.40 | 67.0 ± 6.20 | 26.5 ± 2.00 | 29.1 ± 2.30 | 30.1 ± 2.20 | |
| PLP * | 24.7 ± 5.10 | 44.9 ± 11.6 | 13.0 ± 3.10 | 28.3 ± 5.30 | 7.20 ± 1.40 | 9.90 ± 3.00 | 2.80 ± 0.40 | |
| Pyridoxamine * | 7.50 ± 2.30 | 12.3 ± 4.60 | 5.7 ± 0.8 | 6.10 ± 1.70 | 8.70 ± 0.90 | 8.20 ± 1.30 | 7.90 ± 1.00 | |
| SAH | 0.08 ± 0.01 | 0.02 ± 0.00 | 0.08 ± 0.02 | 0.15 ± 0.02 | 0.11 ± 0.01 | 0.16 ± 0.02 | 0.08 ± 0.01 | |
| SAM | 1.98 ± 0.26 | 2.03 ± 0.65 | 0.32 ± 0.08 | 1.87 ± 0.30 | 0.33 ± 0.04 | 0.74 ± 0.10 | 0.26 ± 0.07 | |
| Spermidine * | 134 ± 34.9 | 232 ± 93.0 | 67.2 ± 15.4 | 98.8 ± 33.8 | 8.40 ± 1.20 | 21.9 ± 2.90 | 15.3 ± 3.60 | |
| Taurine | 362 ± 53.5 | 413 ± 132 | 28.0 ± 11.1 | 79.8 ± 23.2 | 8.32 ± 1.49 | 11.58 ± 2.42 | 14.3 ± 2.54 | |
| Amino acids | Alanine | 1.54 ± 0.12 | 2.16 ± 0.62 | 1.07 ± 0.23 | 2.95 ± 0.47 | 5.58 ± 1.42 | 5.17 ± 1.31 | 5.05 ± 0.68 |
| Arginine | 1.67 ± 0.29 | 1.21 ± 0.30 | 0.54 ± 0.09 | 1.38 ± 0.22 | 0.87 ± 0.08 | 1.01 ± 0.12 | 0.57 ± 0.04 | |
| Asparagine | 0.33 ± 0.06 | 0.14 ± 0.06 | 0.09 ± 0.02 | ND | 0.06 ± 0.00 | 0.07 ± 0.01 | 0.03 ± 0.00 | |
| Aspartate | 10.4 ± 2.38 | 14.3 ± 4.79 | 2.20 ± 0.25 | 12.4 ± 3.23 | 2.05 ± 0.25 | 2.69 ± 0.37 | 2.71 ± 0.63 | |
| Glutamate | 4.29 ± 0.67 | 3.66 ± 0.71 | 2.24 ± 0.26 | 4.44 ± 0.42 | 0.92 ± 0.08 | 0.65 ± 0.10 | 1.44 ± 0.19 | |
| Glycine | 2.33 ± 0.47 | 4.82 ± 2.32 | 1.26 ± 0.15 | 4.20 ± 1.14 | 1.12 ± 0.09 | 2.36 ± 0.72 | 1.35 ± 0.17 | |
| Histidine | 0.70 ± 0.11 | 1.37 ± 0.50 | 0.76 ± 0.11 | 1.90 ± 0.30 | 0.47 ± 0.05 | 0.50 ± 0.17 | 0.35 ± 0.03 | |
| Isoleucine | 44.5 ± 12.7 | 35.9 ± 9.77 | 7.81 ± 1.27 | 22.3 ± 2.06 | 13.7 ± 1.35 | 49.0 ± 10.8 | 14.8 ± 1.33 | |
| Leucine | 4.40 ± 0.54 | 4.79 ± 1.42 | 2.12 ± 0.25 | 4.92 ± 0.82 | 2.98 ± 0.2 | 2.12 ± 0.35 | 3.12 ± 0.21 | |
| Phenylalanine | 0.73 ± 0.12 | 1.18 ± 0.26 | 0.29 ± 0.02 | 0.65 ± 0.13 | 0.23 ± 0.02 | 0.67 ± 0.16 | 0.40 ± 0.04 | |
| Proline | 5.77 ± 1.10 | 3.57 ± 1.06 | 2.81 ± 0.39 | 9.25 ± 1.64 | 2.28 ± 0.19 | 4.21 ± 1.22 | 0.51 ± 0.10 | |
| Serine | 1.60 ± 0.20 | 0.61 ± 0.09 | 0.37 ± 0.07 | 1.72 ± 0.46 | 0.85 ± 0.09 | 0.62 ± 0.08 | 1.39 ± 0.27 | |
| Threonine | 3.92 ± 0.63 | 3.50 ± 0.79 | 0.75 ± 0.12 | 3.07 ± 0.81 | 0.52 ± 0.03 | 1.11 ± 0.25 | 0.69 ± 0.09 | |
| Tryptophan | 0.41 ± 0.09 | 0.31 ± 0.07 | 0.04 ± 0.01 | 0.36 ± 0.04 | 0.10 ± 0.01 | 0.22 ± 0.03 | 0.13 ± 0.01 | |
| Tyrosine | 1.15 ± 0.29 | 1.83 ± 0.47 | 0.31 ± 0.03 | 0.67 ± 0.22 | 0.23 ± 0.04 | 0.44 ± 0.13 | 0.17 ± 0.03 | |
| Valine | 2.42 ± 0.61 | 2.66 ± 0.60 | 0.82 ± 0.10 | 2.20 ± 0.25 | 1.04 ± 0.11 | 2.63 ± 0.59 | 0.98 ± 0.10 | |
| Lipid and protein intermediates | Carnitine * | 5418 ± 425 | 3402 ± 872 | 2298 ± 207 | 4589 ± 780 | 3405 ± 327 | 811 ± 211 | 3594 ± 482 |
| Choline * | 1981 ± 721 | 2585 ± 964 | 879.9 ± 130 | 3707 ± 735 | 528 ± 71.3 | 2280 ± 848 | 474 ± 47.9 |
| Pathway | Metabolites | Pearson r Values | PGLS λ |
|---|---|---|---|
| Methionine metabolism | Betaine | 0.06 (p = 0.69) | 0 (p = 0.834) |
| Cystathionine | −0.54 (p < 0.001) | 0.7 (p = 0.070) | |
| Cysteine | −0.19 (p = 0.214) | 1 (p = 0.667) | |
| GSH | −0.2 (p = 0.196) | 0.7 (p = 0.590) | |
| Homocysteine | −0.28 (p = 0.131) | 1 (p = 0.391) | |
| Methionine | −0.75 (p < 0.001) | 0.5 (p = 0.00) | |
| Pyridoxal | −0.56 (p < 0.001) | 1 (p = 0.371) | |
| PLP | −0.8 (p < 0.001) | 1 (p = 0.001) | |
| Pyridoxamine | 0.13 (p = 0.382) | 1 (p = 0.742) | |
| SAH | 0.37 (p = 0.014) | 1 (p = 0.754) | |
| SAM | −0.55 (p < 0.001) | 1 (p = 0.089) | |
| Spermidine | −0.67 (p < 0.001) | 1 (p = 0.287) | |
| Taurine | −0.78 (p < 0.001) | 1 (p = 0.262) | |
| Amino acids | Alanine | 0.34 (p = 0.025) | 0.0 (p = 0.501) |
| Arginine | −0.36 (p = 0.014) | 0.8 (p = 0.217) | |
| Asparagine | −0.54 (p < 0.001) | 0.0 (p = 0.434) | |
| Aspartate | −0.6 (p < 0.001) | 1.2 (p = 0.336) | |
| Glutamate | −0.66 (p < 0.001) | 1.1 (p = 0.492) | |
| Glycine | −0.38 (p = 0.01) | 1.2 (p = 0.556) | |
| Histidine | −0.58 (p < 0.001) | 1.2 (p = 0.177) | |
| Isoleucine | −0.37 (p = 0.012) | 1.2 (p = 0.674) | |
| Leucine | −0.3 (p = 0.045) | 1.2 (p = 0.640) | |
| Phenylalanine | −0.48 (p = 0.001) | 0.7 (p = 0.481) | |
| Proline | −0.65 (p < 0.001) | 0.5 (p = 0.019) | |
| Serine | −0.07 (p = 0.648) | 1.0 (p = 0.639) | |
| Threonine | −0.71 (p < 0.001) | 1.2 (p = 0.265) | |
| Tryptophan | −0.42 (p = 0.005) | 1.2 (p = 0.803) | |
| Tyrosine | −0.73 (p < 0.001) | 1.2 (p = 0.041) | |
| Valine | −0.43 (p = 0.003) | 1.3 (p = 0.401) | |
| Lipid and protein intermediates | Carnitine | −0.28 (p = 0.06) | 1 (p = 0.825) |
| Choline | −0.51 (p < 0.001) | 1 (p = 0.221) |
| Pathway | Compound | Precursor | Product | Frag | CE | CAV | RT | ΔRT | Polarity | Extraction | Method |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Methionine metabolism | Betaine | 118.09 | 59.2 | 136 | 16 | 7 | 0.425 | 2 | Positive | Methanol | 1 |
| Betaine | 118.09 | 58.2 | 136 | 32 | 7 | 0.425 | 2 | Positive | Methanol | 1 | |
| Cysteine | 122.02 | 76 | 64 | 12 | 7 | 6.312 | 4 | Positive | ACN | 2 | |
| Cysteine | 122.02 | 59 | 64 | 24 | 7 | 6.312 | 4 | Positive | ACN | 2 | |
| Cystathionine | 223.07 | 134 | 88 | 8 | 7 | 6.818 | 4 | Positive | ACN | 2 | |
| Cystathionine | 223.07 | 88 | 88 | 28 | 7 | 6.818 | 4 | Positive | ACN | 2 | |
| GSH | 308.09 | 179 | 88 | 8 | 7 | 0.5 | 2 | Positive | ACN | 1 | |
| GSH | 308.09 | 76 | 88 | 24 | 7 | 0.5 | 2 | Positive | ACN | 1 | |
| Homocysteine | 136.18 | 90.1 | 135 | 15 | 7 | 7.225 | 4 | Positive | ACN | 2 | |
| Homocysteine | 136.18 | 56.2 | 135 | 15 | 7 | 7.225 | 4 | Positive | ACN | 2 | |
| Methionine | 150.05 | 104 | 64 | 4 | 7 | 0.48 | 2 | Positive | ACN | 1 | |
| Pyridoxal | 168.05 | 150 | 64 | 8 | 7 | 0.522 | 2 | Positive | Methanol | 1 | |
| Pyridoxal | 168.05 | 94 | 64 | 24 | 7 | 0.522 | 2 | Positive | Methanol | 1 | |
| PLP | 248.03 | 150 | 112 | 12 | 7 | 0.7 | 2 | Positive | Methanol | 1 | |
| PLP | 248.03 | 67 | 112 | 32 | 7 | 0.7 | 2 | Positive | Methanol | 1 | |
| Pyridoxamine | 169.09 | 152 | 64 | 8 | 7 | 0.366 | 2 | Positive | Methanol | 1 | |
| Pyridoxamine | 169.09 | 134 | 64 | 20 | 7 | 0.366 | 2 | Positive | Methanol | 1 | |
| SAH | 385.1 | 136 | 112 | 20 | 7 | 1.13 | 2 | Positive | Methanol | 1 | |
| SAH | 385.1 | 88 | 112 | 48 | 7 | 1.13 | 2 | Positive | Methanol | 1 | |
| SAM | 399.1 | 250 | 112 | 12 | 7 | 0.396 | 2 | Positive | ACN | 1 | |
| SAM | 399.1 | 136 | 112 | 28 | 7 | 0.396 | 2 | Positive | ACN | 1 | |
| Spermidine | 146.1 | 84 | 88 | 24 | 7 | 0.3 | 2 | Positive | Methanol | 1 | |
| Spermidine | 146.1 | 72 | 88 | 12 | 7 | 0.3 | 2 | Positive | Methanol | 1 | |
| Taurine | 126.02 | 108 | 88 | 8 | 7 | 0.38 | 2 | Positive | ACN | 1 | |
| Taurine | 124 | 80 | 112 | 20 | 7 | 0.38 | 2 | Negative | ACN | 1 | |
| Amino acids | Alanine | 90.06 | 44.2 | 40 | 8 | 7 | 0.376 | 2 | Positive | Methanol | 1 |
| Arginine | 175.1 | 70.2 | 60 | 20 | 7 | 0.32 | 2 | Positive | Methanol | 1 | |
| Arginine | 175.1 | 60.2 | 60 | 15 | 7 | 0.32 | 2 | Positive | Methanol | 1 | |
| Asparagine | 133 | 74.1 | 60 | 15 | 7 | 0.376 | 2 | Positive | Methanol | 1 | |
| Aspartate | 134 | 43.2 | 60 | 15 | 7 | 0.362 | 2 | Positive | Methanol | 1 | |
| Aspartate | 132 | 88.1 | 60 | 15 | 7 | 0.362 | 2 | Negative | Methanol | 1 | |
| Glutamate | 146 | 102.1 | 60 | 15 | 7 | 0.363 | 2 | Negative | Methanol | 1 | |
| Glutamate | 146 | 41 | 60 | 15 | 7 | 0.363 | 2 | Negative | Methanol | 1 | |
| Glycine | 76.04 | 48 | 40 | 0 | 7 | 0.34 | 2 | Positive | Methanol | 1 | |
| Glycine | 76.04 | 30 | 40 | 4 | 7 | 0.34 | 2 | Positive | Methanol | 1 | |
| Histidine | 156 | 110.1 | 60 | 15 | 7 | 0.32 | 2 | Positive | Methanol | 1 | |
| Histidine | 156 | 56.2 | 60 | 25 | 7 | 0.32 | 2 | Positive | Methanol | 1 | |
| Isoleucine | 132.1 | 86 | 64 | 8 | 7 | 0.591 | 2 | Positive | Methanol | 1 | |
| Isoleucine | 132.1 | 69 | 64 | 16 | 7 | 0.591 | 2 | Positive | Methanol | 1 | |
| Leucine | 132.1 | 86 | 64 | 8 | 7 | 0.591 | 2 | Positive | Methanol | 1 | |
| Leucine | 132.1 | 69 | 64 | 16 | 7 | 0.591 | 2 | Positive | Methanol | 1 | |
| Phenylalanine | 164 | 147 | 100 | 15 | 7 | 0.841 | 2 | Negative | Methanol | 1 | |
| Phenylalanine | 164 | 103.1 | 100 | 15 | 7 | 0.841 | 2 | Negative | Methanol | 1 | |
| Proline | 116 | 70.2 | 60 | 15 | 7 | 0.392 | 2 | Positive | Methanol | 1 | |
| Serine | 106.05 | 60 | 64 | 8 | 7 | 0.35 | 2 | Positive | Methanol | 1 | |
| Serine | 106.05 | 42 | 64 | 24 | 7 | 0.35 | 2 | Positive | Methanol | 1 | |
| Serine | 104.03 | 74 | 64 | 8 | 7 | 0.35 | 2 | Negative | Methanol | 1 | |
| Threonine | 120 | 74.2 | 60 | 15 | 7 | 0.358 | 2 | Positive | Methanol | 1 | |
| Threonine | 120 | 56.2 | 60 | 15 | 7 | 0.358 | 2 | Positive | Methanol | 1 | |
| Tryptophan | 205 | 188.1 | 60 | 15 | 7 | 1.23 | 2 | Positive | Methanol | 1 | |
| Tryptophan | 205 | 146.1 | 60 | 15 | 7 | 1.23 | 2 | Positive | Methanol | 1 | |
| Tyrosine | 180.1 | 163.1 | 100 | 15 | 7 | 0.548 | 2 | Negative | Methanol | 1 | |
| Tyrosine | 180.1 | 119.1 | 100 | 15 | 7 | 0.548 | 2 | Negative | Methanol | 1 | |
| Valine | 118.08 | 72 | 64 | 8 | 7 | 0.43 | 2 | Positive | Methanol | 1 | |
| Valine | 118.08 | 55 | 64 | 20 | 7 | 0.43 | 2 | Positive | Methanol | 1 | |
| Lipid and protein intermediates | Carnitine | 162.12 | 103.1 | 88 | 16 | 7 | 0.393 | 2 | Positive | Methanol | 1 |
| Choline | 104.11 | 60.2 | 112 | 16 | 7 | 0.39 | 2 | Positive | Methanol | 1 | |
| ISTD | PheC13 | 167.09 | 120.1 | 70 | 8 | 7 | 0.87 | 2 | Positive | Methanol/ACN | 1/2 |
| PheC13 | 167.09 | 77 | 70 | 44 | 7 | 0.87 | 2 | Positive | Methanol/ACN | 1/2 | |
| PheC13 | 167.09 | 103 | 70 | 28 | 7 | 0.87 | 2 | Positive | Methanol/ACN | 1/2 | |
| PheC13 | 167.09 | 51.1 | 70 | 60 | 7 | 0.87 | 2 | Positive | Methanol/ACN | 1/2 |
References
- Pamplona, R.; Barja, G. Highly resistant macromolecular components and low rate of generation of endogenous damage: Two key traits of longevity. Ageing Res. Rev. 2007, 6, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Barja, G. Updating the mitochondrial free radical theory of aging: An integrated view, key aspects, and confounding concepts. Antioxid. Redox Signal. 2013, 19, 1420–1445. [Google Scholar] [CrossRef] [PubMed]
- Jové, M.; Mota-Martorell, N.; Pradas, I.; Galo-Licona, J.D.; Martín-Gari, M.; Obis, È.; Sol, J.; Pamplona, R. The lipidome fingerprint of longevity. Molecules 2020, 25, 4343. [Google Scholar] [CrossRef] [PubMed]
- Mota-Martorell, N.; Jové, M.; Pradas, I.; Sanchez, I.; Gómez, J.; Naudí, A.; Barja, G.; Pamplona, R. Low abundance of NDUFV2 and NDUFS4 subunits of the hydrophilic complex I domain and VDAC1 predicts mammalian longevity. Redox Biol. 2020, 34, 101539. [Google Scholar] [CrossRef]
- Pamplona, R.; Barja, G. Mitochondrial oxidative stress, aging and caloric restriction: The protein and methionine connection. Biochim. Biophys. Acta-Bioenerg. 2006, 1757, 496–508. [Google Scholar] [CrossRef]
- Lambert, A.J.; Buckingham, J.A.; Boysen, H.M.; Brand, M.D. Low complex I content explains the low hydrogen peroxide production rate of heart mitochondria from the long-lived pigeon, Columba livia. Aging Cell 2010, 9, 78–91. [Google Scholar] [CrossRef]
- Miwa, S.; Jow, H.; Baty, K.; Johnson, A.; Czapiewski, R.; Saretzki, G.; Treumann, A.; Von Zglinicki, T. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat. Commun. 2014, 5, 3837. [Google Scholar] [CrossRef]
- Barja, G. Towards a unified mechanistic theory of aging. Exp. Gerontol. 2019, 124, 110627. [Google Scholar] [CrossRef]
- Mota-Martorell, N.; Jove, M.; Pradas, I.; Berdún, R.; Sanchez, I.; Naudi, A.; Gari, E.; Barja, G.; Pamplona, R. Gene expression and regulatory factors of the mechanistic target of rapamycin (mTOR) complex 1 predict mammalian longevity. GeroScience 2020, 42, 1157–1173. [Google Scholar] [CrossRef]
- Tyshkovskiy, A.; Bozaykut, P.; Borodinova, A.A.; Gerashchenko, M.V.; Ables, G.P.; Garratt, M.; Khaitovich, P.; Clish, C.B.; Miller, R.A.; Gladyshev, V.N. Identification and application of gene expression signatures associated with lifespan extension. Cell Metab. 2019, 30, 573–593.e8. [Google Scholar] [CrossRef]
- Swovick, K.; Welle, K.A.; Hryhorenko, J.R.; Seluanov, A.; Gorbunova, V.; Ghaemmaghami, S. Cross-species comparison of proteome turnover kinetics. Mol. Cell. Proteomics 2018, 17, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Faulkes, C.G.; Eykyn, T.R.; Aksentijevic, D. Cardiac metabolomic profile of the naked mole-rat—Glycogen to the rescue. Biol. Lett. 2019, 15, 20190710. [Google Scholar] [CrossRef]
- Ma, S.; Gladyshev, V.N. Molecular signatures of longevity: Insights from cross-species comparative studies. Semin. Cell Dev. Biol. 2017, 70, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Moosmann, B. Redox biochemistry of the genetic code. Trends Biochem. Sci. 2021, 46, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Moosmann, B.; Schindeldecker, M.; Hajieva, P. Cysteine, glutathione and a new genetic code: Biochemical adaptations of the primordial cells that spread into open water and survived biospheric oxygenation. Biol. Chem. 2020, 401, 213–231. [Google Scholar] [CrossRef]
- Aledo, J.C.; Li, Y.; de Magalhães, J.P.; Ruíz-Camacho, M.; Pérez-Claros, J.A. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell 2011, 10, 198–207. [Google Scholar] [CrossRef]
- Moosmann, B. Respiratory chain cysteine and methionine usage indicate a causal role for thiyl radicals in aging. Exp. Gerontol. 2011, 46, 164–169. [Google Scholar] [CrossRef]
- Lewis, K.N.; Rubinstein, N.D.; Buffenstein, R. A window into extreme longevity; the circulating metabolomic signature of the naked mole-rat, a mammal that shows negligible senescence. GeroScience 2018, 40, 105–121. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, 18, e13034. [Google Scholar] [CrossRef]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine metabolism in health and cancer: A nexus of diet and precision medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- McIsaac, R.S.; Lewis, K.N.; Gibney, P.A.; Buffenstein, R. From yeast to human: Exploring the comparative biology of methionine restriction in extending eukaryotic life span. Ann. N. Y. Acad. Sci. 2016, 1363, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Mota-Martorell, N.; Jové, M.; Berdún, R.; Pamplona, R. Plasma methionine metabolic profile is associated with longevity in mammals. Commun. Biol. 2021, 4, 725. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, D.; Xu, B.; Li, H.; Dai, X.; Chen, B. Development of a matrix-based candidate reference material of total homocysteine in human serum. Anal. Bioanal. Chem. 2017, 409, 3329–3335. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinforma. 2019, 68, e86. [Google Scholar] [CrossRef]
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Pearse, W. Comparative Analyses of Phylogenetics and Evolution in R. R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Sahm, A.; Bens, M.; Henning, Y.; Vole, C.; Groth, M.; Schwab, M.; Hoffmann, S.; Platzer, M.; Szafranski, K.; Dammann, P. Higher gene expression stability during aging in long-lived giant mole-rats than in short-lived rats. Aging 2018, 10, 3938–3956. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, S.; Yan, P.; Ren, J.; Song, M.; Li, J.; Lei, J.; Pan, H.; Wang, S.; Ma, X.; et al. A single-cell transcriptomic landscape of primate arterial aging. Nat. Commun. 2020, 11, 2202. [Google Scholar] [CrossRef]
- Seim, I.; Ma, S.; Gladyshev, V.N. Gene expression signatures of human cell and tissue longevity. npj Aging Mech. Dis. 2016, 2, 16014. [Google Scholar] [CrossRef]
- Fushan, A.A.; Turanov, A.A.; Lee, S.-G.; Kim, E.B.; Lobanov, A.V.; Yim, S.H.; Buffenstein, R.; Lee, S.-R.; Chang, K.-T.; Rhee, H.; et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell 2015, 14, 352–365. [Google Scholar] [CrossRef]
- Lu, J.Y.; Simon, M.; Zhao, Y.; Ablaeva, J.; Corson, N.; Choi, Y.; Yamada, K.Y.H.; Schork, N.J.; Hood, W.R.; Hill, G.E.; et al. Comparative transcriptomics reveals circadian and pluripotency networks as two pillars of longevity regulation. Cell Metab. 2022, 34, 836–856.e5. [Google Scholar] [CrossRef] [PubMed]
- Heinze, I.; Bens, M.; Calzia, E.; Holtze, S.; Dakhovnik, O.; Sahm, A.; Kirkpatrick, J.M.; Szafranski, K.; Romanov, N.; Sama, S.N.; et al. Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol. 2018, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Bozek, K.; Khrameeva, E.E.; Reznick, J.; Omerbašić, D.; Bennett, N.C.; Lewin, G.R.; Azpurua, J.; Gorbunova, V.; Seluanov, A.; Regnard, P.; et al. Lipidome determinants of maximal lifespan in mammals. Sci. Rep. 2017, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Viltard, M.; Durand, S.; Pérez-Lanzón, M.; Aprahamian, F.; Lefevre, D.; Leroy, C.; Madeo, F.; Kroemer, G.; Friedlander, G. The metabolomic signature of extreme longevity: Naked mole rats versus mice. Aging 2019, 11, 4783–4800. [Google Scholar] [CrossRef]
- Walters, R.O.; Arias, E.; Diaz, A.; Burgos, E.S.; Guan, F.; Tiano, S.; Mao, K.; Green, C.L.; Qiu, Y.; Shah, H.; et al. Sarcosine is uniquely modulated by aging and dietary restriction in rodents and humans. Cell Rep. 2018, 25, 663–676.e6. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Poonawalla, A.; Icyuz, M.; Swindell, W.R.; Wilson, L.; Barnes, S.; Sun, L.Y. Transcriptomic and metabolomic profiling of long-lived growth hormone releasing hormone knock-out mice: Evidence for altered mitochondrial function and amino acid metabolism. Aging 2020, 12, 3473–3485. [Google Scholar] [CrossRef]
- Ma, S.; Yim, S.H.; Lee, S.-G.; Kim, E.B.; Lee, S.-R.; Chang, K.-T.; Buffenstein, R.; Lewis, K.N.; Park, T.J.; Miller, R.A.; et al. Organization of the mammalian metabolome according to organ function, lineage specialization and longevity. Cell Metab. 2015, 22, 332–343. [Google Scholar] [CrossRef]
- Granold, M.; Hajieva, P.; Toşa, M.I.; Irimie, F.-D.; Moosmann, B. Modern diversification of the amino acid repertoire driven by oxygen. Proc. Natl. Acad. Sci. USA 2018, 115, 41–46. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Binari, R.; Zhang, N.; Asara, J.M.; Demontis, F.; Perrimon, N. Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev. 2016, 30, 1409–1422. [Google Scholar] [CrossRef]
- Annibal, A.; Tharyan, R.G.; Schonewolff, M.F.; Tam, H.; Latza, C.; Auler, M.M.K.; Grönke, S.; Partridge, L.; Antebi, A. Regulation of the one carbon folate cycle as a shared metabolic signature of longevity. Nat. Commun. 2021, 12, 3486. [Google Scholar] [CrossRef]
- Wijeyesekera, A.; Selman, C.; Barton, R.H.; Holmes, E.; Nicholson, J.K.; Withers, D.J. Metabotyping of long-lived mice using 1H NMR spectroscopy. J. Proteome Res. 2012, 11, 2224–2235. [Google Scholar] [CrossRef] [PubMed]
- Swovick, K.; Firsanov, D.; Welle, K.A.; Hryhorenko, J.R.; Wise, J.P.; George, C.; Sformo, T.L.; Seluanov, A.; Gorbunova, V.; Ghaemmaghami, S. Interspecies Differences in Proteome Turnover Kinetics Are Correlated With Life Spans and Energetic Demands. Mol. Cell. Proteomics 2021, 20, 100041. [Google Scholar] [CrossRef] [PubMed]
- Parkhitko, A.A.; Wang, L.; Filine, E.; Jouandin, P.; Leshchiner, D.; Binari, R.; Asara, J.M.; Rabinowitz, J.D.; Perrimon, N. A genetic model of methionine restriction extends Drosophila health and lifespan. Proc. Natl. Acad. Sci. USA 2021, 118, e2110387118. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Li, W.; Kremer, D.M.; Sajjakulnukit, P.; Li, S.; Crespo, J.; Nwosu, Z.C.; Zhang, L.; Czerwonka, A.; Pawłowska, A.; et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 2020, 585, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Lowe, R.; Barton, C.; Jenkins, C.A.; Ernst, C.; Forman, O.; Fernandez-Twinn, D.S.; Bock, C.; Rossiter, S.J.; Faulkes, C.G.; Ozanne, S.E.; et al. Ageing-associated DNA methylation dynamics are a molecular readout of lifespan variation among mammalian species. Genome Biol. 2018, 19, 22. [Google Scholar] [CrossRef]
- Wilkinson, G.S.; Adams, D.M. Recurrent evolution of extreme longevity in bats. Biol. Lett. 2019, 15, 20180860. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Wang, W.-J.; Liu, J.-Q.; Song, Y.-H.; Li, P.; Sun, X.-F.; Cai, G.-Y.; Chen, X.-M. Methionine restriction delays senescence and suppresses the senescence-associated secretory phenotype in the kidney through endogenous hydrogen sulfide. Cell Cycle 2019, 18, 1573–1587. [Google Scholar] [CrossRef]
- Hine, C.; Harputlugil, E.; Zhang, Y.; Ruckenstuhl, C.; Lee, B.C.; Brace, L.; Longchamp, A.; Treviño-Villarreal, J.H.; Mejia, P.; Ozaki, C.K.; et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 2015, 160, 132–144. [Google Scholar] [CrossRef]
- Castro, C.; Krumsiek, J.; Lehrbach, N.J.; Murfitt, S.A.; Miska, E.A.; Griffin, J.L. A study of Caenorhabditis elegans DAF-2 mutants by metabolomics and differential correlation networks. Mol. Biosyst. 2013, 9, 1632. [Google Scholar] [CrossRef]
- Martinez-Miguel, V.E.; Lujan, C.; Espie--Caullet, T.; Martinez-Martinez, D.; Moore, S.; Backes, C.; Gonzalez, S.; Galimov, E.R.; Brown, A.E.X.; Halic, M.; et al. Increased fidelity of protein synthesis extends lifespan. Cell Metab. 2021, 33, 2288–2300.e12. [Google Scholar] [CrossRef] [PubMed]
- Sarabhai, T.; Roden, M. Hungry for your alanine: When liver depends on muscle proteolysis. J. Clin. Investig. 2019, 129, 4563–4566. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Ussher, J.R. Evolving Concepts of Myocardial Energy Metabolism. Circ. Res. 2016, 119, 1173–1176. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mota-Martorell, N.; Jové, M.; Berdún, R.; Òbis, È.; Barja, G.; Pamplona, R. Methionine Metabolism Is Down-Regulated in Heart of Long-Lived Mammals. Biology 2022, 11, 1821. https://doi.org/10.3390/biology11121821
Mota-Martorell N, Jové M, Berdún R, Òbis È, Barja G, Pamplona R. Methionine Metabolism Is Down-Regulated in Heart of Long-Lived Mammals. Biology. 2022; 11(12):1821. https://doi.org/10.3390/biology11121821
Chicago/Turabian StyleMota-Martorell, Natalia, Mariona Jové, Rebeca Berdún, Èlia Òbis, Gustavo Barja, and Reinald Pamplona. 2022. "Methionine Metabolism Is Down-Regulated in Heart of Long-Lived Mammals" Biology 11, no. 12: 1821. https://doi.org/10.3390/biology11121821
APA StyleMota-Martorell, N., Jové, M., Berdún, R., Òbis, È., Barja, G., & Pamplona, R. (2022). Methionine Metabolism Is Down-Regulated in Heart of Long-Lived Mammals. Biology, 11(12), 1821. https://doi.org/10.3390/biology11121821