Nutrient Digestibility, Microbial Fermentation, and Response in Bacterial Composition to Methionine Dipeptide: An In Vitro Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Design and In Vitro Batch Culture
2.3. DNA Extraction and Sequencing
2.4. Statistical Analysis
2.5. Sequencing Data Analysis
3. Results
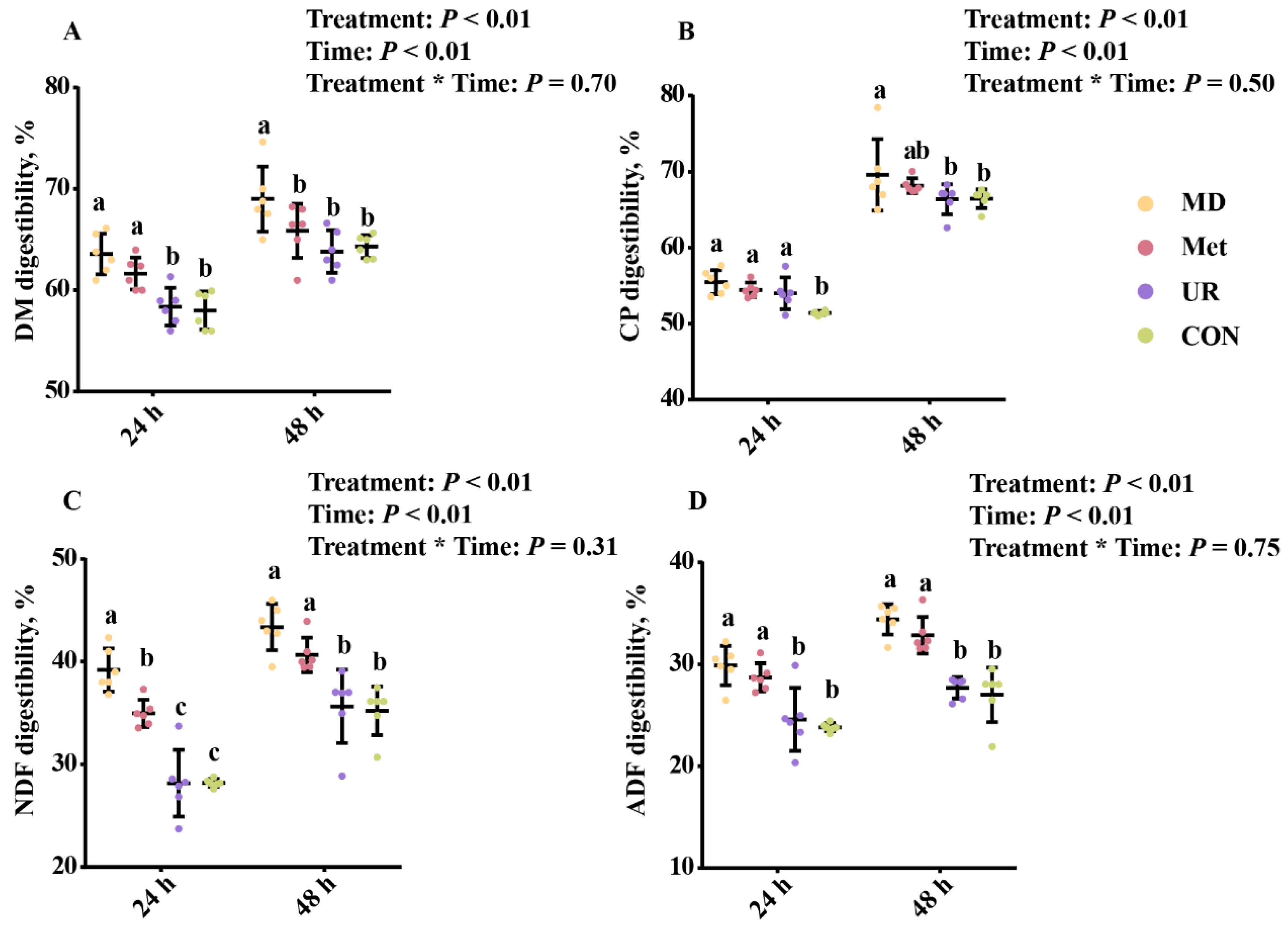
3.1. Nutrient Digestibility
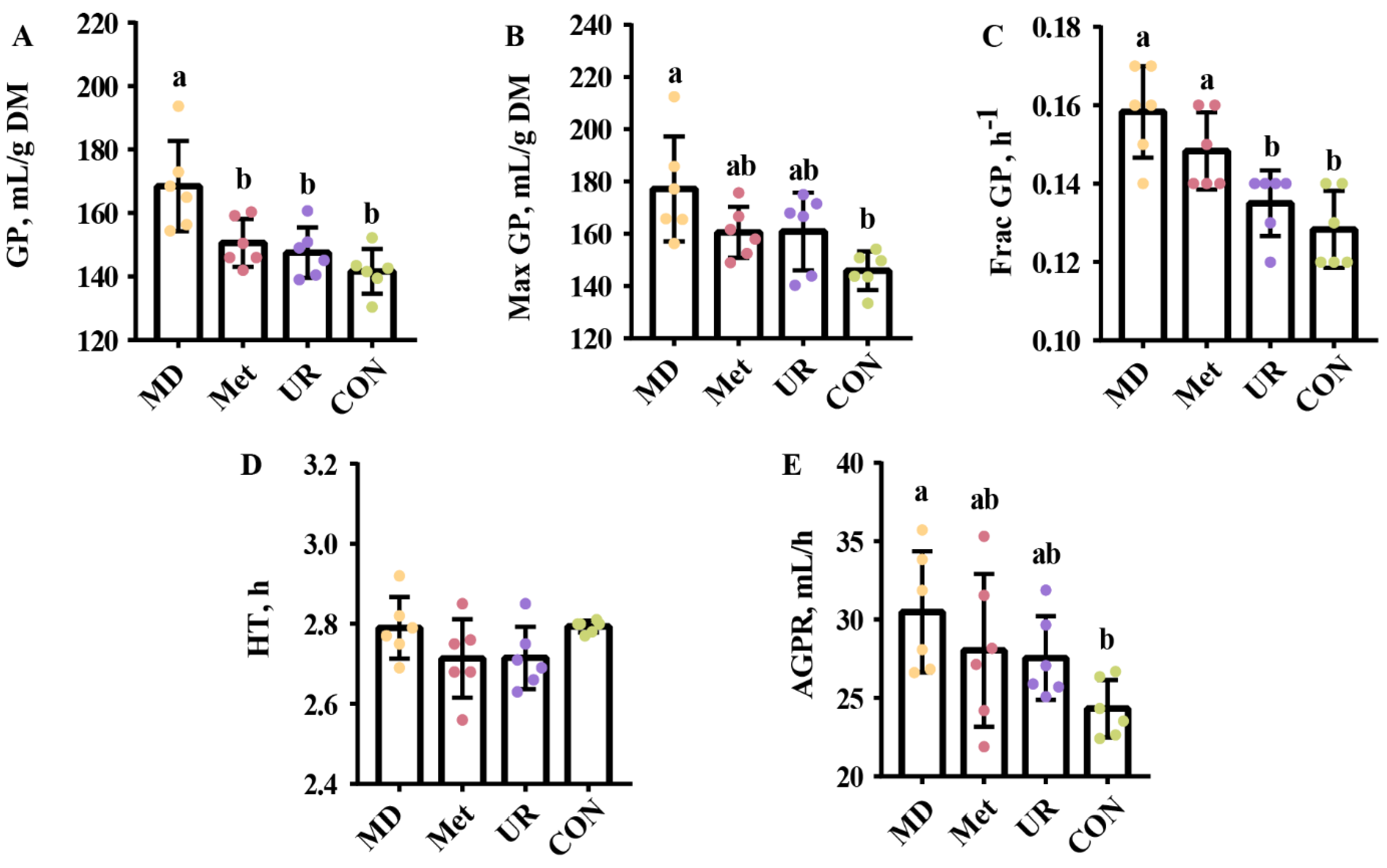
3.2. Fermentation Kinetics Parameters
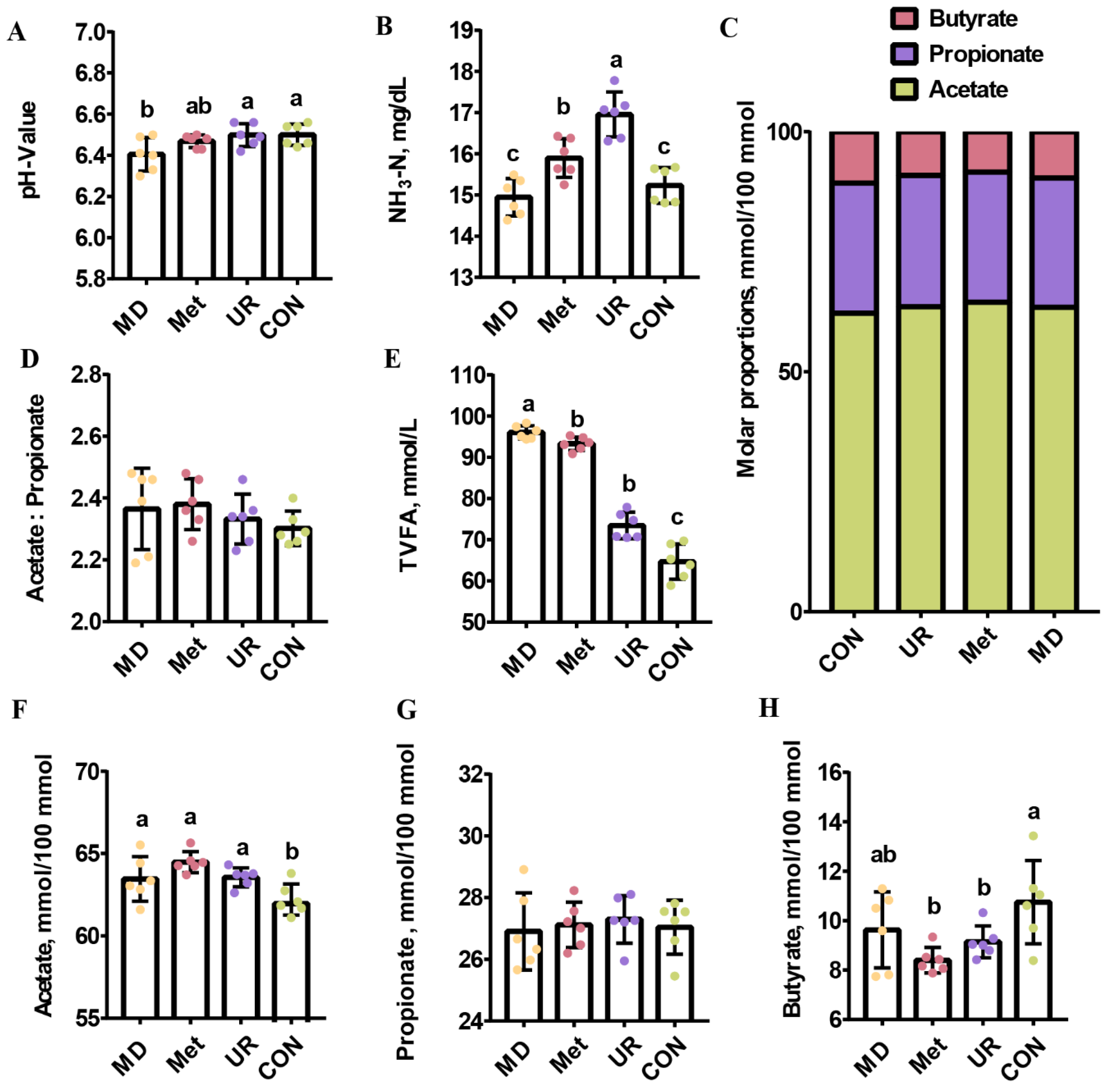
3.3. pH Value, NH3-N Concentration, and VFA Proportion
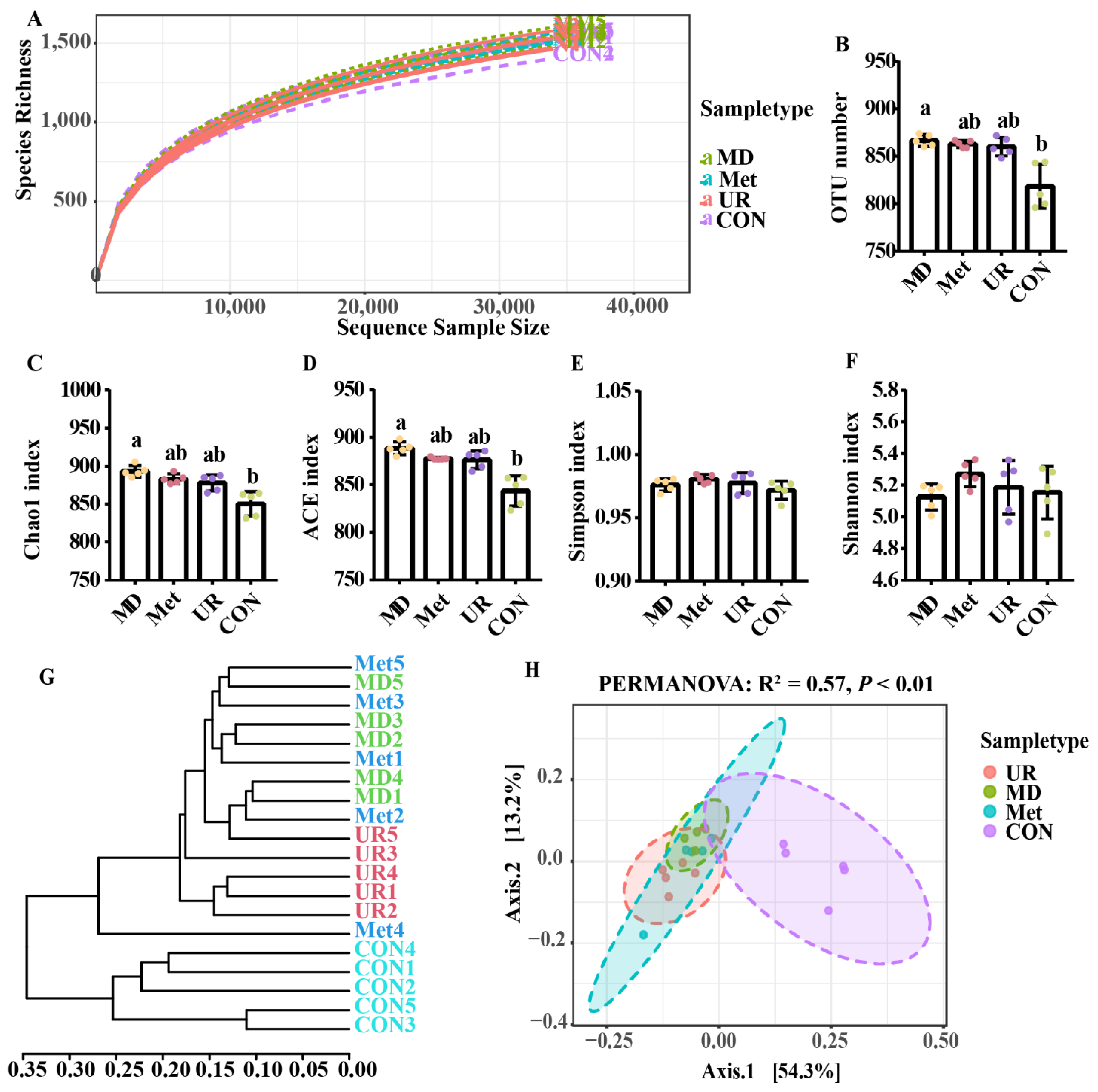
3.4. Diversity of Microbiota
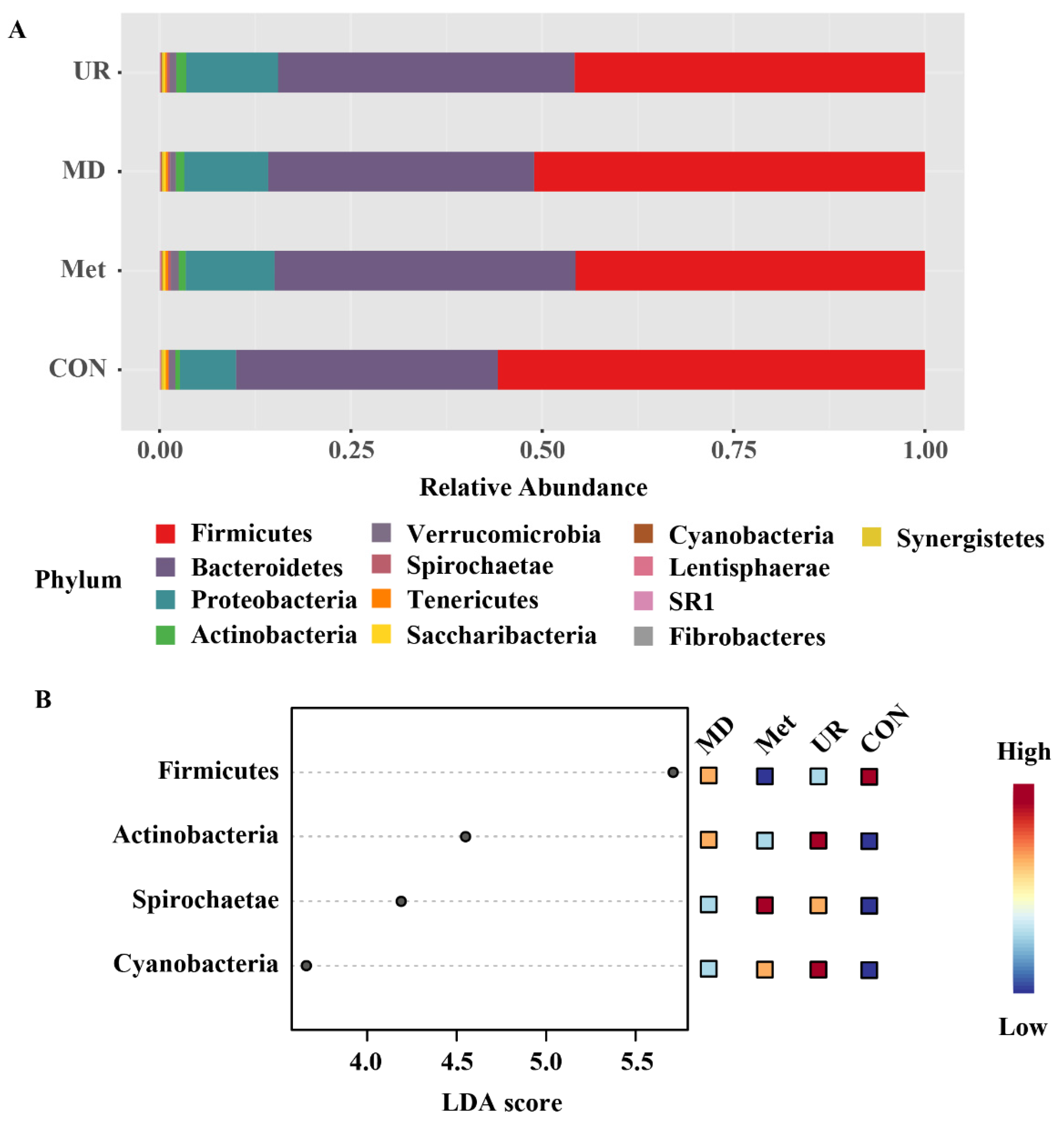
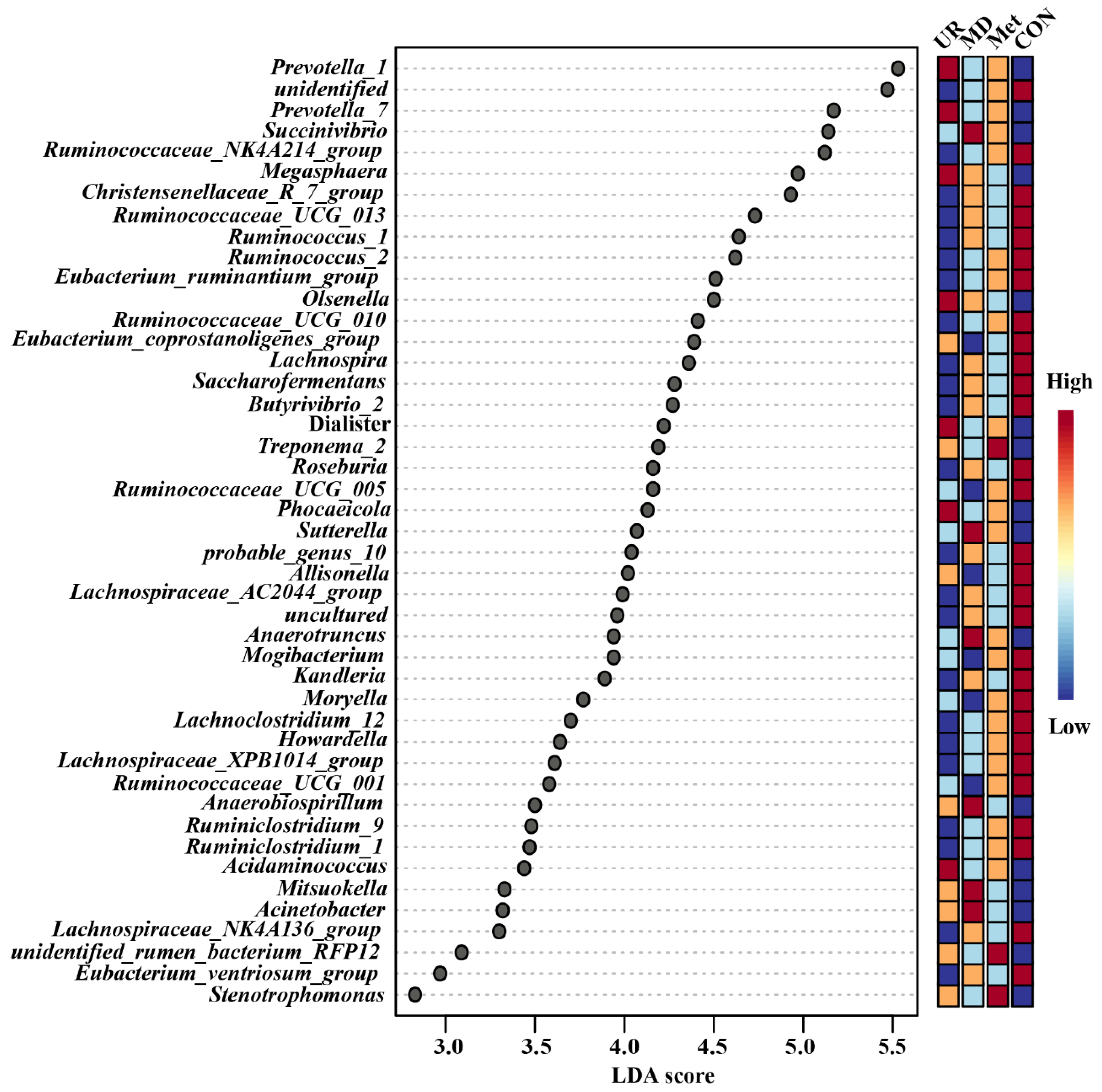
3.5. Microbial Composition and Its Comparison in Response to Urea, Methionine, and Methionine Dipeptide
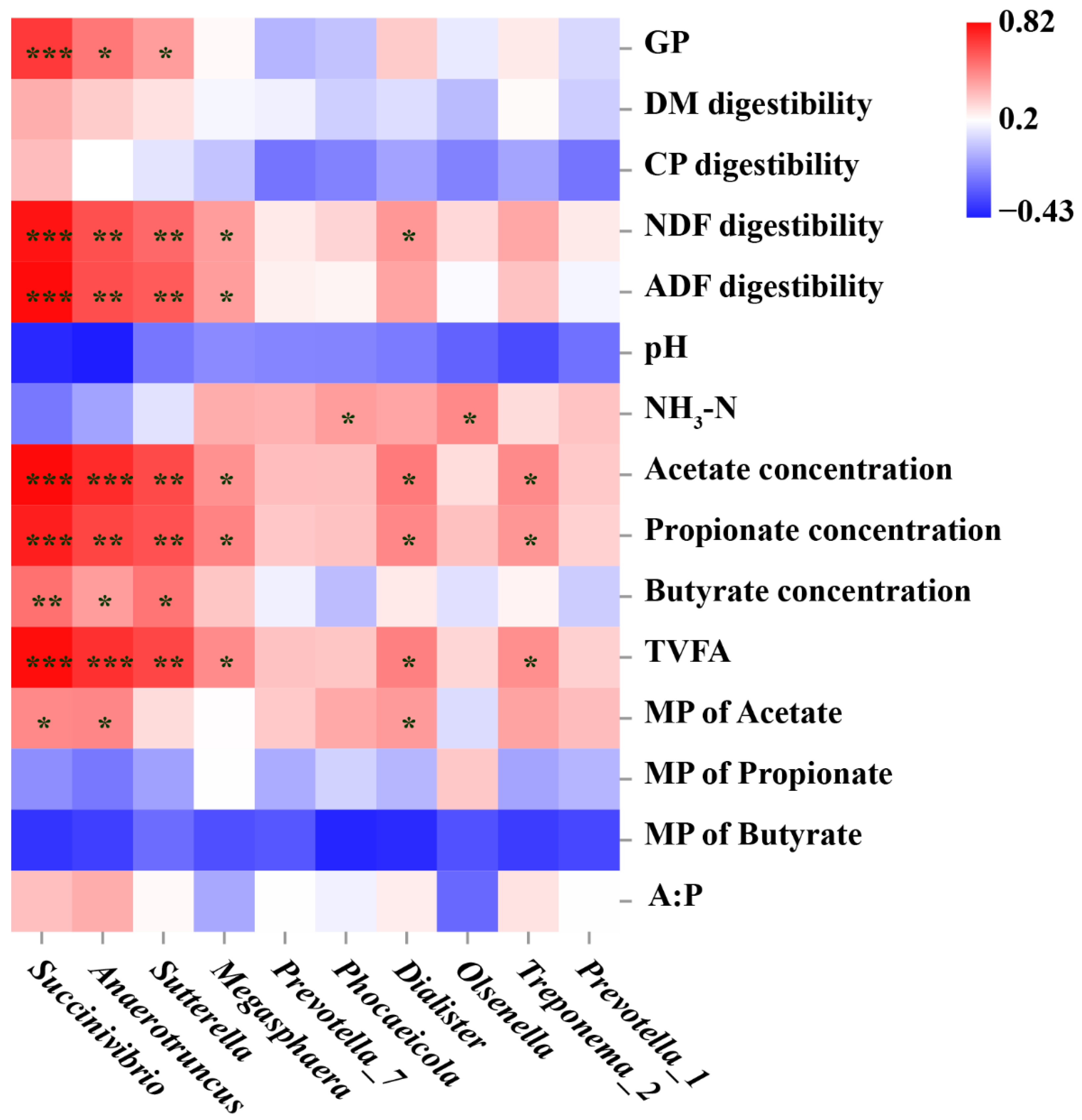
3.6. Correlation Analysis between Microbial Features and Nutrient Digestibility, Gas Production Parameters, and Fermentation Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Xu, L.; Zhao, F.; Liu, H. Regulation of Milk Protein Synthesis by Free and Peptide-Bound Amino Acids in Dairy Cows. Biology 2021, 10, 1044. [Google Scholar] [CrossRef]
- Pan, Y.; Bender, P.K.; Akers, R.M.; Webb, K.E., Jr. Methionine-containing peptides can be used as methionine sources for protein accretion in cultured C2C12 and MAC-T cells. J. Nutr. 1996, 126, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Yang, J.; Zhao, K.; Liu, H.; Wu, Y.; Liu, J. Effects of methionine-containing dipeptides on αs1casein expression in bovine mammary epithelial cells. J. Anim. Feed Sci. 2007, 16, 325–329. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Liu, Z.; Liu, H.; Zhao, F.; Huang, X.; Wu, Y.; Liu, J. Functional characterization of oligopeptide transporter 1 of dairy cows. J. Anim. Sci. Biotechnol. 2018, 9, 7. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.B.; Mertens, D.R. A 100-Year Review: Carbohydrates-Characterization, digestion, and utilization. J. Dairy Sci. 2017, 100, 10078–10093. [Google Scholar] [CrossRef] [Green Version]
- Schwab, C.G.; Broderick, G.A. A 100-Year Review: Protein and amino acid nutrition in dairy cows. J. Dairy Sci. 2017, 100, 10094–10112. [Google Scholar] [CrossRef] [Green Version]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Mabjeesh, S.J.; Gal-Garber, O.; Milgram, J.; Feuermann, Y.; Cohen-Zinder, M.; Shamay, A. Aminopeptidase N gene expression and abundance in caprine mammary gland is influenced by circulating plasma peptide. J. Dairy Sci. 2005, 88, 2055–2064. [Google Scholar] [CrossRef] [Green Version]
- Shennan, D.B. Peptide Transport and Metabolism by the Lactating Mammary Gland. J. Nutr. 2016, 146, 384. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.X.; Wang, C.H.; Xu, Q.B.; Zhao, F.Q.; Liu, J.X.; Liu, H.Y. Methionyl-Methionine Promotes α-s1 Casein Synthesis in Bovine Mammary Gland Explants by Enhancing Intracellular Substrate Availability and Activating JAK2-STAT5 and mTOR-Mediated Signaling Pathways. J. Nutr. 2015, 145, 1748–1753. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, F.; Liu, J.; Liu, H. Dipeptide (Methionyl-Methionine) Transport and Its Effect on β-Casein Synthesis in Bovine Mammary Epithelial Cells. Cell Physiol. Biochem. 2018, 49, 479–488. [Google Scholar] [CrossRef]
- Pitta, D.W.; Indugu, N.; Vecchiarelli, B.; Hennessy, M.; Baldin, M.; Harvatine, K.J. Effect of 2-hydroxy-4-(methylthio) butanoate (HMTBa) supplementation on rumen bacterial populations in dairy cows when exposed to diets with risk for milk fat depression. J. Dairy Sci. 2020, 103, 2718–2730. [Google Scholar] [CrossRef]
- Vyas, D.; Erdman, R.A. Meta-analysis of milk protein yield responses to lysine and methionine supplementation. J. Dairy Sci. 2009, 92, 5011–5018. [Google Scholar] [CrossRef] [Green Version]
- Zanton, G.I.; Bowman, G.R.; Vázquez-Añón, M.; Rode, L.M. Meta-analysis of lactation performance in dairy cows receiving supplemental dietary methionine sources or postruminal infusion of methionine. J. Dairy Sci. 2014, 97, 7085–7101. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, I.H.R.; Abbasi, F.; Liu, L.; Bodinga, B.M.; Abdel-Latif, M.A.; Swelum, A.A.; Mohamed, M.A.E.; Cao, Y. Rumen-protected methionine a feed supplement to low dietary protein: Effects on microbial population, gases production and fermentation characteristics. AMB Express 2019, 9, 93. [Google Scholar] [CrossRef]
- Russi, J.P.; Wallace, R.J.; Newbold, C.J. Influence of the pattern of peptide supply on microbial activity in the rumen simulating fermenter (RUSITEC). Br. J. Nutr. 2002, 88, 73–80. [Google Scholar] [CrossRef]
- Fu, C.J.; Felton, E.E.; Lehmkuhler, J.W.; Kerley, M.S. Ruminal peptide concentration required to optimize microbial growth and efficiency. J. Anim. Sci. 2001, 79, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, P.; Xiao, Y.; Liu, J.; Chen, Y.; Yang, Y. Alterations in Rumen Bacterial Community and Metabolome Characteristics of Cashmere Goats in Response to Dietary Nutrient Density. Animals 2020, 10, 1193. [Google Scholar] [CrossRef]
- Argyle, J.L.; Baldwin, R.L. Effects of amino acids and peptides on rumen microbial growth yields. J. Dairy Sci. 1989, 72, 2017–2027. [Google Scholar] [CrossRef]
- Broderick, G.; Craig, W.M. Metabolism of peptides and amino acids during in vitro protein degradation by mixed rumen organisms. J. Dairy Sci. 1989, 72, 2540–2548. [Google Scholar] [CrossRef]
- Cotta, M.A.; Russell, J.B. Effect of Peptides and Amino Acids on Efficiency of Rumen Bacterial Protein Synthesis in Continuous Culture. J. Dairy Sci. 1982, 65, 226–234. [Google Scholar] [CrossRef]
- Russell, J.B.; O’Connor, J.D.; Fox, D.G.; Van Soest, P.J.; Sniffen, C.J. A net carbohydrate and protein system for evaluating cattle diets: I. Ruminal fermentation. J. Anim. Sci. 1992, 70, 3551–3561. [Google Scholar] [CrossRef]
- Russell, J.B.; Sniffen, C.J.; Van Soest, P.J. Effect of carbohydrate limitation on degradation and utilization of casein by mixed rumen bacteria. J. Dairy Sci. 1983, 66, 763–775. [Google Scholar] [CrossRef]
- Brooks, M.A.; Harvey, R.M.; Johnson, N.F.; Kerley, M.S. Rumen degradable protein supply affects microbial efficiency in continuous culture and growth in steers. J. Anim. Sci. 2012, 90, 4985–4994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.F.; Yang, H.J. Combination effects of nitrocompounds, pyromellitic diimide, and 2-bromoethanesulfonate on in vitro ruminal methane production and fermentation of a grain-rich feed. J. Agric. Food Chem. 2012, 60, 364–371. [Google Scholar] [CrossRef]
- Yáñez-Ruiz, D.R.; Bannink, A.; Dijkstra, J.; Kebreab, E.; Morgavi, D.P.; O’Kiely, P.; Reynolds, C.K.; Schwarm, A.; Shingfield, K.J.; Yu, Z.; et al. Design, implementation and interpretation of in vitro batch culture experiments to assess enteric methane mitigation in ruminants—A review. Anim. Feed Sci. Tech. 2016, 216, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Benchaar, C.; Calsamiglia, S.; Chaves, A.V.; Fraser, G.R.; Colombatto, D.; McAllister, T.A.; Beauchemin, K.A. A review of plant-derived essential oils in ruminant nutrition and production. Anim. Feed Sci. Tech. 2008, 145, 209–228. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Dairy Cattle; National Academies Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Hao, Y.; Huang, S.; Liu, G.; Zhang, J.; Liu, G.; Cao, Z.; Wang, Y.; Wang, W.; Li, S. Effects of Different Parts on the Chemical Composition, Silage Fermentation Profile, In Vitro and In Situ Digestibility of Paper Mulberry. Animals 2021, 11, 413. [Google Scholar] [CrossRef]
- Kong, F.; Lu, N.; Liu, Y.; Zhang, S.; Jiang, H.; Wang, H.; Wang, W.; Li, S. Aspergillus oryzae and Aspergillus niger Co-Cultivation Extract Affects In Vitro Degradation, Fermentation Characteristics, and Bacterial Composition in a Diet-Specific Manner. Animals 2021, 11, 1248. [Google Scholar] [CrossRef]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.; McGarrell, D.M.; Marsh, T.; Garrity, G.M. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef] [Green Version]
- France, J.; Dijkstra, J.; Dhanoa, M.; Lopez, S.; Bannink, A. Estimating the extent of degradation of ruminant feeds from a description of their gas production profiles observed in vitro: Derivation of models and other mathematical considerations. Brit. J. Nutr. 2000, 83, 143–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Martínez, R.; Ranilla, M.; Tejido, M.; Carro, M. Effects of disodium fumarate on in vitro rumen microbial growth, methane production and fermentation of diets differing in their forage: Concentrate ratio. Brit. J. Nutr. 2005, 94, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Kong, F.; Gao, Y.; Tang, M.; Fu, T.; Diao, Q.; Bi, Y.; Tu, Y. Effects of dietary rumen-protected Lys levels on rumen fermentation and bacterial community composition in Holstein heifers. Appl. Microbiol. Biotechnol. 2020, 104, 6623–6634. [Google Scholar] [CrossRef]
- Bach, A.; Calsamiglia, S.; Stern, M. Nitrogen metabolism in the rumen. J. Dairy Sci. 2005, 88, E9–E21. [Google Scholar] [CrossRef] [Green Version]
- Satter, L.D.; Slyter, L.L. Effect of ammonia concentration of rumen microbial protein production in vitro. Br. J. Nutr. 1974, 32, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.K.; Petersen, M.K. Influence of DL-methionine supplementation on growth, ruminal fermentation and dilution rates in heifers. J. Anim. Sci. 1988, 66, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Baghbanzadeh-Nobari, B.; Taghizadeh, A.; Khorvash, M.; Parnian-Khajehdizaj, F.; Maloney, S.K.; Hashemzadeh-Cigari, F.; Ghaffari, A.H. Digestibility, ruminal fermentation, blood metabolites and antioxidant status in ewes supplemented with DL-methionine or hydroxy-4 (methylthio) butanoic acid isopropyl ester. J. Anim. Physiol. Anim. Nutr. 2017, 101, e266–e277. [Google Scholar] [CrossRef]
- Li, Z.; Shen, J.; Xu, Y.; Zhu, W. Metagenomic analysis reveals significant differences in microbiome and metabolic profiles in the rumen of sheep fed low N diet with increased urea supplementation. FEMS Microbiol. Ecol. 2020, 96, fiaa117. [Google Scholar] [CrossRef]
- Wallace, R.J. Effect of ammonia concentration on the composition, hydrolytic activity and nitrogen metabolism of the microbial flora of the rumen. J. Appl. Bacteriol. 1979, 47, 443–455. [Google Scholar] [CrossRef]
- McCracken, B.A.; Judkins, M.B.; Krysl, L.J.; Holcombe, D.W.; Park, K.K. Supplemental methionine and time of supplementation effects on ruminal fermentation, digesta kinetics, and in situ dry matter and neutral detergent fiber disappearance in cattle. J. Anim. Sci. 1993, 71, 1932–1939. [Google Scholar] [CrossRef]
- Kabel, M.A.; Yeoman, C.J.; Han, Y.; Dodd, D.; Abbas, C.A.; de Bont, J.A.; Morrison, M.; Cann, I.K.; Mackie, R.I. Biochemical characterization and relative expression levels of multiple carbohydrate esterases of the xylanolytic rumen bacterium Prevotella ruminicola 23 grown on an ester-enriched substrate. Appl. Environ. Microbiol. 2011, 77, 5671–5681. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.N.; Méndez-García, C.; Geier, R.R.; Iakiviak, M.; Chang, J.; Cann, I.; Mackie, R.I. Metabolic networks for nitrogen utilization in Prevotella ruminicola 23. Sci. Rep. 2017, 7, 7851. [Google Scholar] [CrossRef]
- Wen, Z.; Morrison, M. Glutamate dehydrogenase activity profiles for type strains of ruminal Prevotella spp. Appl. Environ. Microbiol. 1997, 63, 3314–3317. [Google Scholar] [CrossRef] [Green Version]
- Kraatz, M.; Wallace, R.J.; Svensson, L. Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int. J. Syst. Evol. Microbiol. 2011, 61, 795–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, L., Jr.; Hession, A.O. Preventing in vitro lactate accumulation in ruminal fermentations by inoculation with Megasphaera elsdenii. J. Anim. Sci. 1995, 73, 250–256. [Google Scholar] [CrossRef]
- Palevich, N.; Kelly, W.J.; Leahy, S.C.; Altermann, E.; Rakonjac, J.; Attwood, G.T. The complete genome sequence of the rumen bacterium Butyrivibrio hungatei MB2003. Stand. Genom. Sci. 2017, 12, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawanon, S.; Kobayashi, Y. Synergistic fibrolysis in the rumen by cellulolytic Ruminococcus flavefaciens and non-cellulolytic Selenomonas ruminantium: Evidence in defined cultures. Anim. Sci. J. 2006, 77, 208–214. [Google Scholar] [CrossRef]
- Wood, T.M.; Wilson, C.A.; Stewart, C.S. Preparation of the cellulase from the cellulolytic anaerobic rumen bacterium Ruminococcus albus and its release from the bacterial cell wall. Biochem. J. 1982, 205, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Zhu, W.; Zhu, W.; Liu, J.; Mao, S. Effect of dietary forage sources on rumen microbiota, rumen fermentation and biogenic amines in dairy cows. J. Sci. Food Agric. 2014, 94, 1886–1895. [Google Scholar] [CrossRef]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Phylogenetic diversity and dietary association of rumen Treponema revealed using group-specific 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 2011, 316, 51–60. [Google Scholar] [CrossRef]
- Stanton, T.B.; Canale-Parola, E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 1980, 127, 145–156. [Google Scholar] [CrossRef]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S.; Guan, L.L. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef] [Green Version]
- Sofyan, A.; Uyeno, Y.; Shinkai, T.; Hirako, M.; Kushibiki, S.; Kanamori, H.; Mori, S.; Katayose, Y.; Mitsumori, M. Metagenomic profiles of the rumen microbiota during the transition period in low-yield and high-yield dairy cows. Anim. Sci. J. 2019, 90, 1362–1376. [Google Scholar] [CrossRef]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Guan, L.L.; Liu, J.X. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 2019, 102, 5031–5041. [Google Scholar] [CrossRef] [PubMed]

| Items | Contents |
|---|---|
| Ingredients | |
| Alfalfa | 9.34 |
| Alfalfa silage | 2.12 |
| Corn silage | 25.21 |
| Steam-flaked corn | 17.82 |
| Corn | 12.73 |
| Fatty powder | 1.27 |
| Soybean meal | 15.28 |
| Soybean hull | 4.24 |
| Corn gluten meal | 2.55 |
| Cottonseed meal | 6.37 |
| NaHCO3 | 0.64 |
| Molasses | 1.05 |
| 3% premix 1 | 1.38 |
| Total | 100 |
| Nutrient level 2 | |
| DM, % of air-dried weight | 94.55 ± 0.37 |
| CP | 16.00 ± 0.35 |
| NDF | 38.18 ±0.91 |
| ADF | 25.32 ± 0.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, F.; Liu, Y.; Wang, S.; Zhang, Y.; Wang, W.; Yang, H.; Lu, N.; Li, S. Nutrient Digestibility, Microbial Fermentation, and Response in Bacterial Composition to Methionine Dipeptide: An In Vitro Study. Biology 2022, 11, 93. https://doi.org/10.3390/biology11010093
Kong F, Liu Y, Wang S, Zhang Y, Wang W, Yang H, Lu N, Li S. Nutrient Digestibility, Microbial Fermentation, and Response in Bacterial Composition to Methionine Dipeptide: An In Vitro Study. Biology. 2022; 11(1):93. https://doi.org/10.3390/biology11010093
Chicago/Turabian StyleKong, Fanlin, Yanfang Liu, Shuo Wang, Yijia Zhang, Wei Wang, Hongjian Yang, Na Lu, and Shengli Li. 2022. "Nutrient Digestibility, Microbial Fermentation, and Response in Bacterial Composition to Methionine Dipeptide: An In Vitro Study" Biology 11, no. 1: 93. https://doi.org/10.3390/biology11010093
APA StyleKong, F., Liu, Y., Wang, S., Zhang, Y., Wang, W., Yang, H., Lu, N., & Li, S. (2022). Nutrient Digestibility, Microbial Fermentation, and Response in Bacterial Composition to Methionine Dipeptide: An In Vitro Study. Biology, 11(1), 93. https://doi.org/10.3390/biology11010093





