Bacterial Communities in Lanna Phak-Gard-Dong (Pickled Mustard Green) from Three Different Ethnolinguistic Groups in Northern Thailand
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sample Collection
2.1.1. Sampling Sites
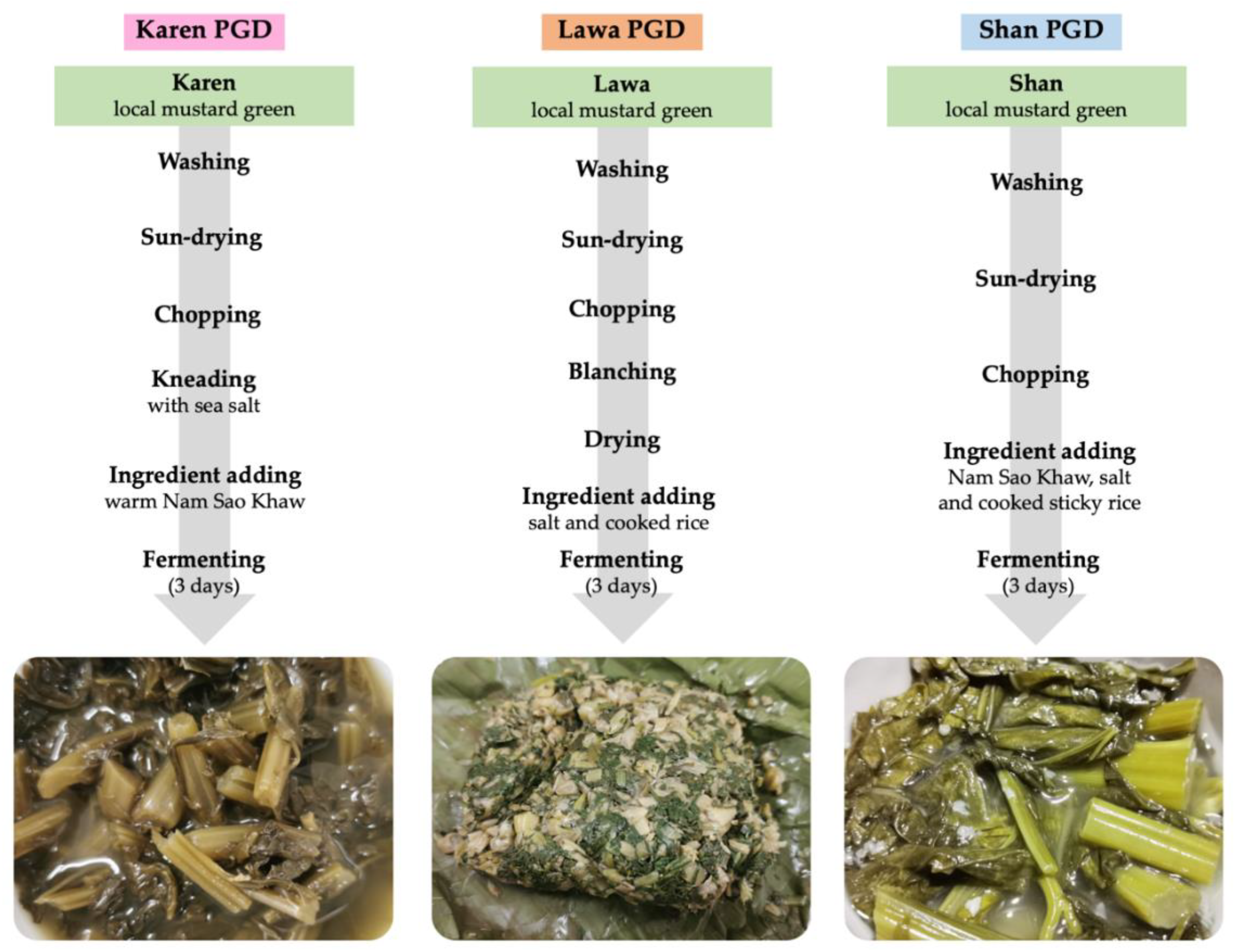
2.1.2. PGD Fermentation Processes
2.1.3. Sample Collection
2.1.4. Determination of pH and Lactic Acid Bacteria Enumeration
2.2. Total DNA Extraction
2.3. DNA Sequencing
2.4. Sequence Processing and Analysis
2.5. Alpha Diversity Analysis
2.6. Beta Diversity Analysis
2.7. Functional Analysis
2.8. Network Analysis
2.9. Sequences Deposit
3. Results
3.1. pH Measuring and Enumeration of Lactic Acid Bacteria
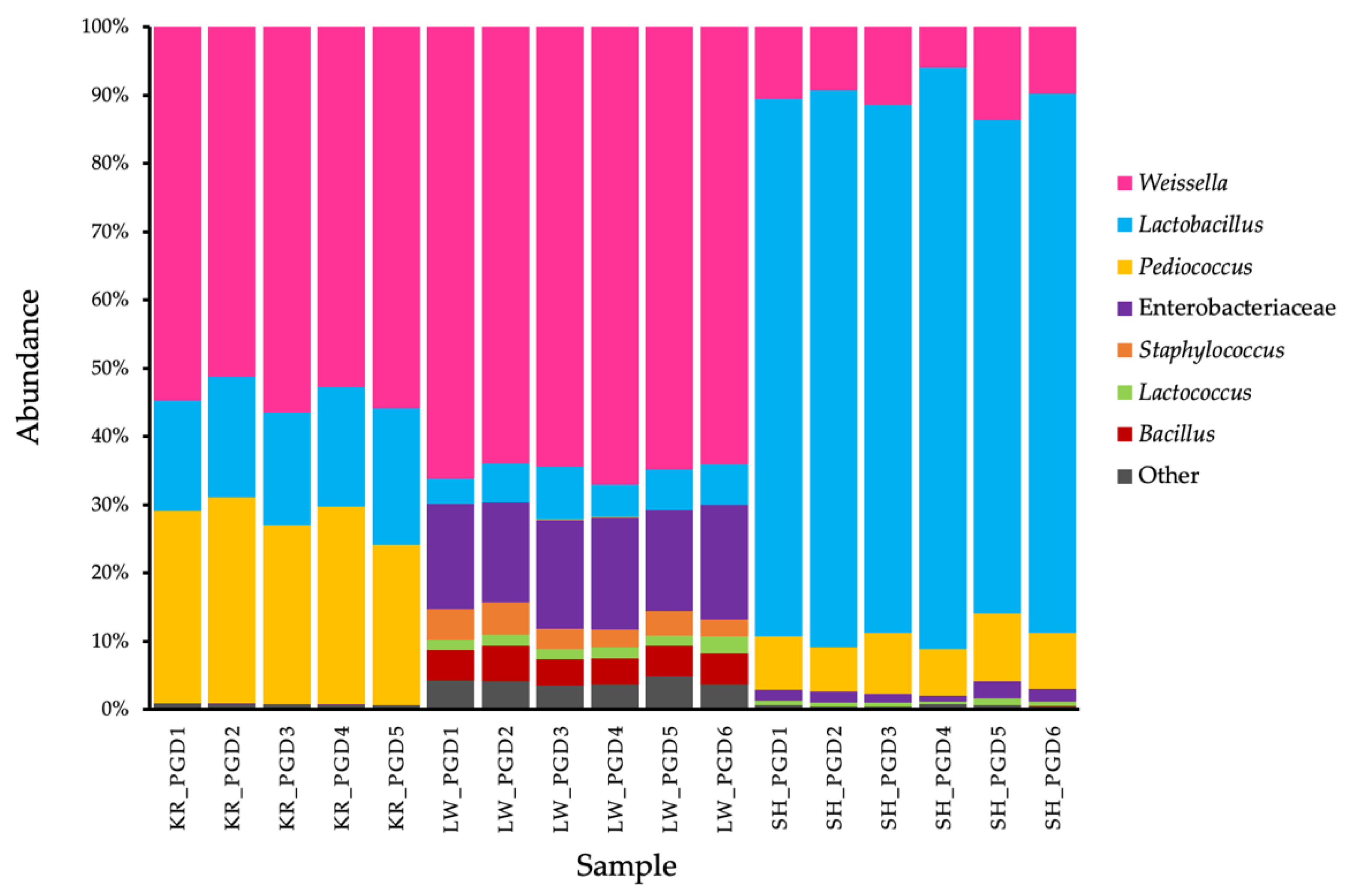
3.2. Bacterial Community Analysis
3.3. Microbiome of Bacteria in Phak-Gard-Dong
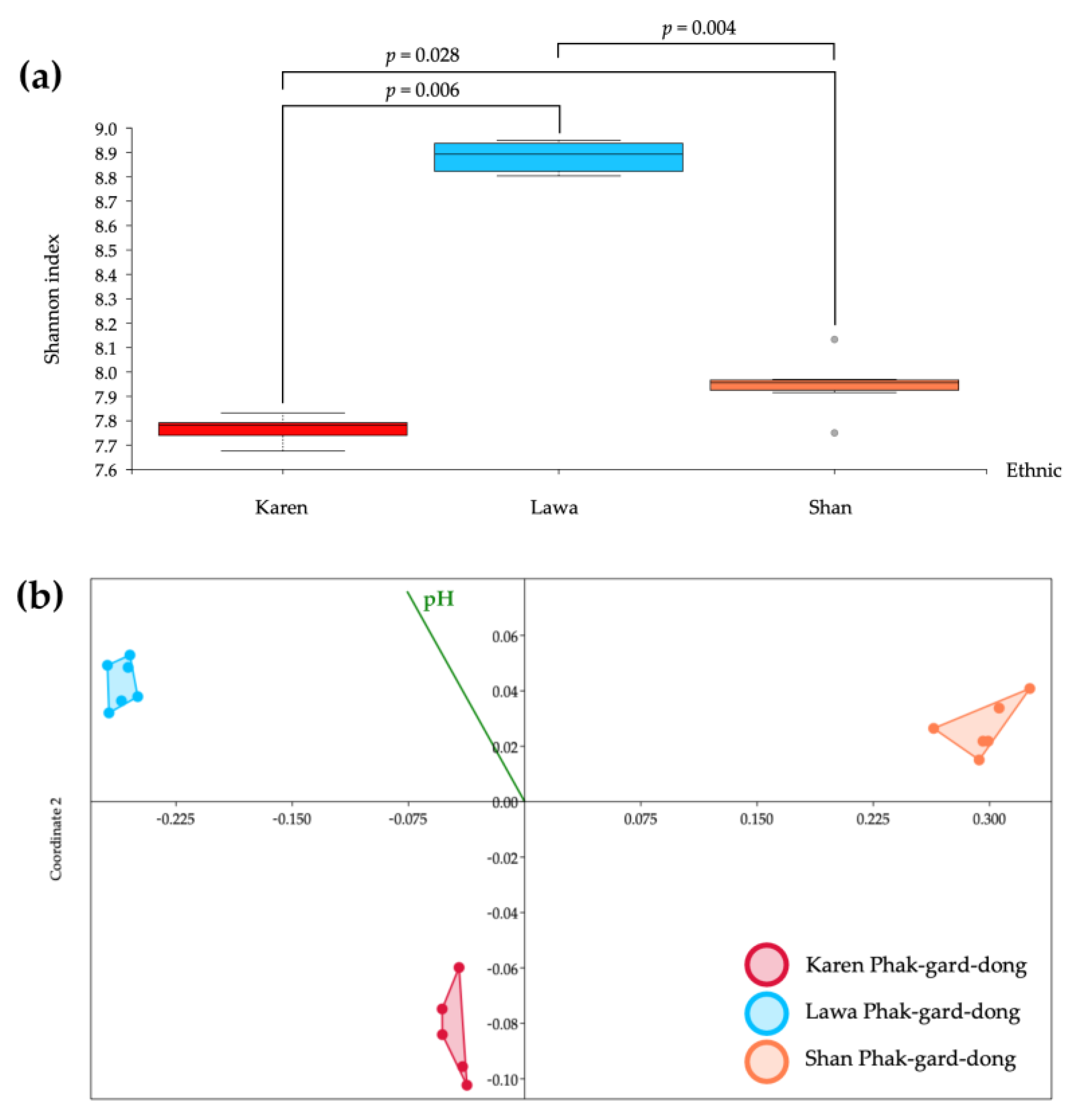
3.4. Alpha and Beta Diversity
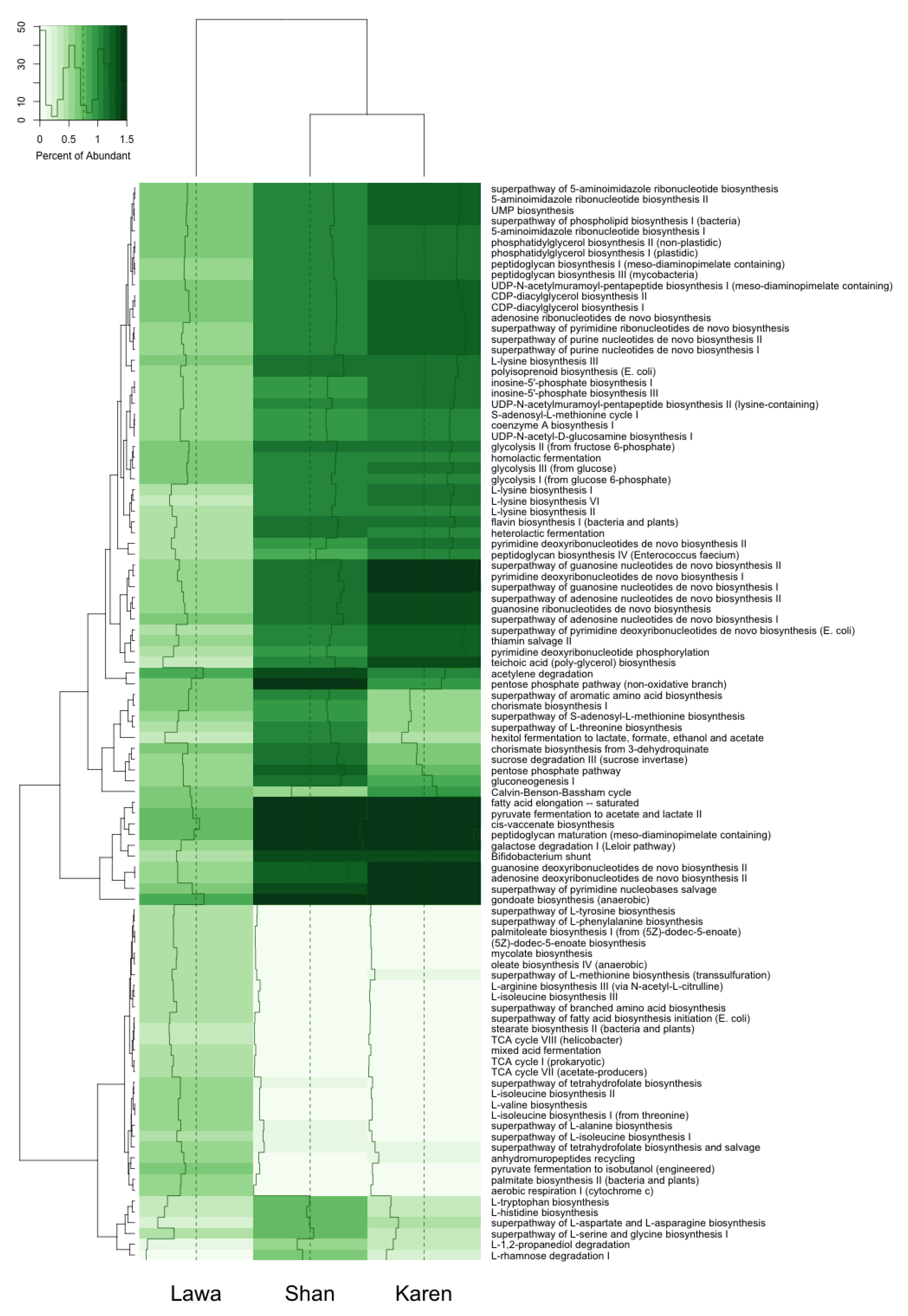
3.5. Functional Gene Prediction
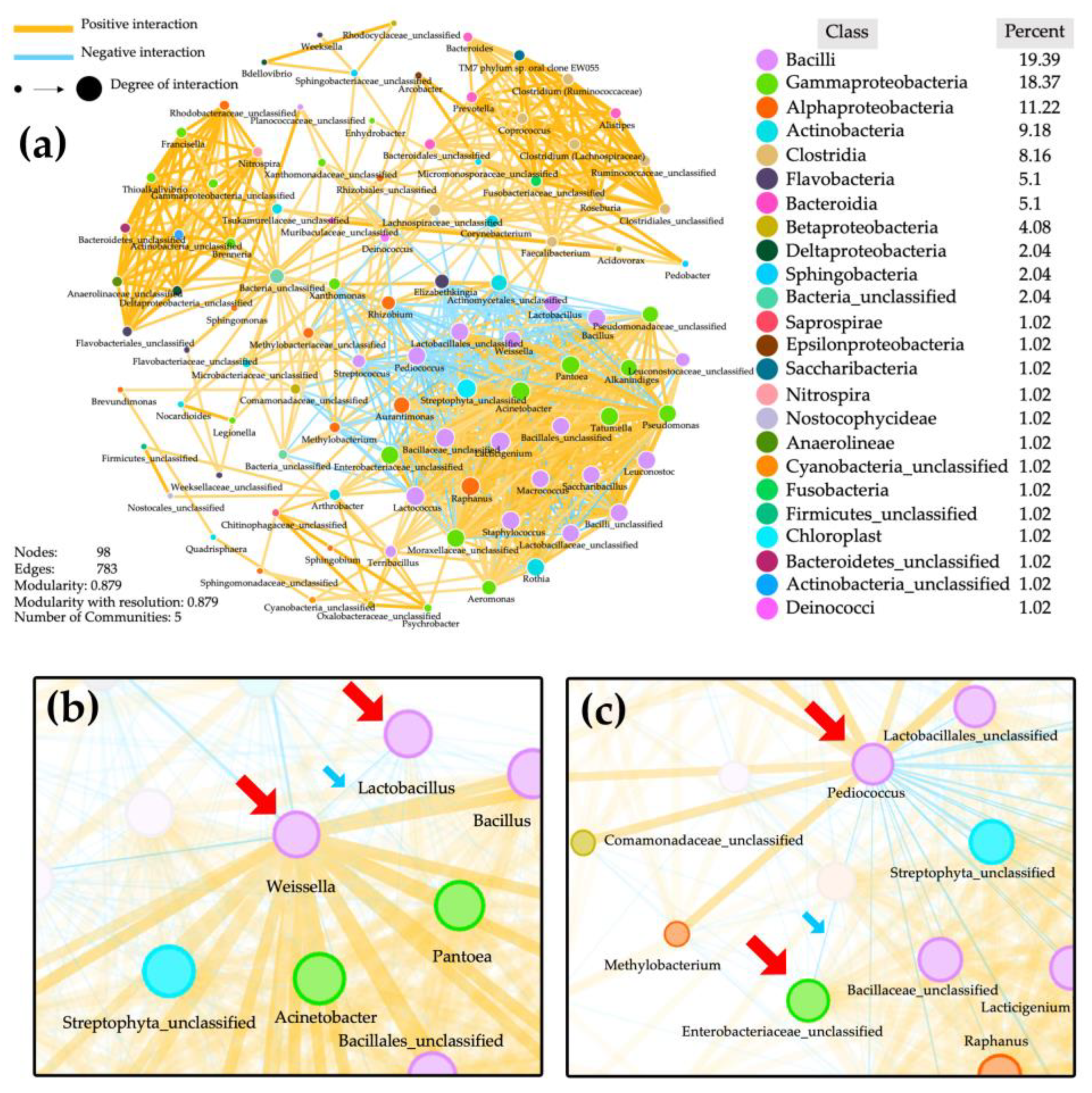
3.6. Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutkins, R.W. Microbiology and Technology of Fermented Foods; John Wiley & Sons: Hoboken, NJ, USA, 2006; ISBN 978-0-8138-0018-9. [Google Scholar]
- Lee, C.-H.; Kim, M.L. History of Fermented Foods in Northeast Asia. In Ethnic Fermented Foods and Alcoholic Beverages of Asia; Tamang, J.P., Ed.; Springer: New Delhi, India, 2016; pp. 1–16. ISBN 978-81-322-2800-4. [Google Scholar]
- Pakwan, C.; Chitov, T.; Chantawannakul, P.; Manasam, M.; Bovonsombut, S.; Disayathanoowat, T. Bacterial Compositions of Indigenous Lanna (Northern Thai) Fermented Foods and Their Potential Functional Properties. PLoS ONE 2020, 15, e0242560. [Google Scholar] [CrossRef]
- Boon-Long, N. Traditional technologies of Thailand: Traditional fermented food products. In Traditional Foods: Some Products and Technologies; Central Food Technological Research Institute: Mysore, India, 1986; pp. 114–133. [Google Scholar]
- Yoon, J.H.; Kang, S.S.; Mheen, T.I.; Ahn, J.S.; Lee, H.J.; Kim, T.K.; Park, C.S.; Kho, Y.H.; Kang, K.H.; Park, Y.H. Lactobacillus kimchii sp. Nov., a New Species from Kimchi. Int. J. Syst. Evol. Microbiol. 2000, 50, 1789–1795. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.-J.; Cheigh, C.-I.; Kim, S.-B.; Lee, J.-C.; Lee, D.-W.; Choi, S.-W.; Park, J.-M.; Pyun, Y.-R. Weissella kimchii sp. Nov., a Novel Lactic Acid Bacterium from Kimchi. Int. J. Syst. Evol. Microbiol. 2002, 52, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Lee, J.; Jang, J.; Kim, J.; Han, H. Leuconostocinhae sp. Nov., a Lactic Acid Bacterium Isolated from Kimchi. Int. J. Syst. Evol. Microbiol. 2003, 53, 1123–1126. [Google Scholar] [CrossRef]
- Kim, M.; Chun, J. Bacterial Community Structure in Kimchi, a Korean Fermented Vegetable Food, as Revealed by 16S rRNA Gene Analysis. Int. J. Food Microbiol. 2005, 103, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Tamang, J.P.; Sarkar, P.K. Sinki: A Traditional Lactic Acid Fermented Radish Tap Root Product. J. Gen. Appl. Microbiol. 1993, 39, 395–408. [Google Scholar] [CrossRef] [Green Version]
- Tamang, J.P.; Tamang, B.; Schillinger, U.; Franz, C.M.A.P.; Gores, M.; Holzapfel, W.H. Identification of Predominant Lactic Acid Bacteria Isolated from Traditionally Fermented Vegetable Products of the Eastern Himalayas. Int. J. Food Microbiol. 2005, 105, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Itabashi, M. Sunki-Pickles Prepared by Single Species of Lactic Acid Bacteria. Nippon. Shokuhin Kogyo Gakkaishi 1987, 34, 356–361. [Google Scholar] [CrossRef]
- Watanabe, K.; Fujimoto, J.; Tomii, Y.; Sasamoto, M.; Makino, H.; Kudo, Y.; Okada, S. Lactobacillus kisonensis sp. Nov., Lactobacillus otakiensis sp. Nov., Lactobacillus rapi sp. Nov. and Lactobacillus sunkii sp. Nov., Heterofermentative Species Isolated from Sunki, a Traditional Japanese Pickle. Int. J. Syst. Evol. Microbiol. 2009, 59, 754–760. [Google Scholar] [CrossRef]
- Yan, P.-M.; Xue, W.-T.; Tan, S.-S.; Zhang, H.; Chang, X.-H. Effect of Inoculating Lactic Acid Bacteria Starter Cultures on the Nitrite Concentration of Fermenting Chinese Paocai. Food Control 2008, 19, 50–55. [Google Scholar] [CrossRef]
- Huang, W.-F.; Jiang, J.-H.; Chen, Y.-W.; Wang, C.-H. A Nosema ceranae Isolate from the Honeybee Apis mellifera. Apidologie 2007, 38, 30–37. [Google Scholar] [CrossRef]
- Mingmuang, M. Microbial Study during the Pickling of Leaves of Mustard (Brassica juncea L.) by Fermentation Process. Ph.D. Thesis, Kasetsart University, Bangkok, Thailand, 1974. [Google Scholar]
- Dhavises, G. Microbial Studies during the Pickling of the Shoot of Bamboo, Bambusa arundinacea, Wild., and of Pak Sian, Gynandropsis pentaphylla. Ph.D. Thesis, Kasetsart University Bangkok, Bangkok, Thailand, 1972. [Google Scholar]
- Tanasupawat, S.; Komagata, K. Lactic Acid Bacteria in Fermented Foods in Thailand. World J. Microbiol. Biotechnol. 1995, 11, 253–256. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Ezaki, T.; Suzuki, K.-I.; Okada, S.; Komagata, K.; Kozaki, M. Characterization and Identification of Lactobacillus pentosus and Lactobacillus plantarum Strains from Fermented Foods in Thailand. J. Gen. Appl. Microbiol. 1992, 38, 121–134. [Google Scholar] [CrossRef] [Green Version]
- Acedo, A.L., Jr.; Weinberger, K. Best Practices in Postharvest Management of Leafy Vegetables in Greater Mekong Subregion Countries; AVRDC-World Vegetable Center: Tainan, Taiwan, 2009; ISBN 978-92-9058-178-9. [Google Scholar]
- Lesme, H.; Rannou, C.; Famelart, M.-H.; Bouhallab, S.; Prost, C. Yogurts Enriched with Milk Proteins: Texture Properties, Aroma Release and Sensory Perception. Trends Food Sci. Technol. 2020, 98, 140–149. [Google Scholar] [CrossRef]
- Lee, W.J.; Lucey, J.A. Formation and Physical Properties of Yogurt. Asian-Australas. J. Anim. Sci. 2010, 23, 1127–1136. [Google Scholar] [CrossRef]
- Kim, B.; Mun, E.-G.; Kim, D.; Kim, Y.; Park, Y.; Lee, H.-J.; Cha, Y.-S. A Survey of Research Papers on the Health Benefits of Kimchi and Kimchi Lactic Acid Bacteria. J. Nutr. Health 2018, 51, 1–13. [Google Scholar] [CrossRef]
- Kim, M.-S.; Yang, H.J.; Kim, S.-H.; Lee, H.W.; Lee, M.S. Effects of Kimchi on Human Health. Medicine (Baltimore) 2018, 97, e0163. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-Y.; Jeong, J.-K.; Lee, Y.-E.; Daily, J.W. Health Benefits of Kimchi (Korean Fermented Vegetables) as a Probiotic Food. J. Med. Food 2014, 17, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Kimchi Microflora: History, Current Status, and Perspectives for Industrial Kimchi Production. Appl. Microbiol. Biotechnol. 2014, 98, 2385–2393. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Paramithiotis, S.; Shin, H.-S. Kimchi and Other Widely Consumed Traditional Fermented Foods of Korea: A Review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef] [Green Version]
- Chaikaew, S.; Baipong, S.; Sone, T.; Kanpiengjai, A.; Chui-chai, N.; Asano, K.; Khanongnuch, C. Diversity of Lactic Acid Bacteria from Miang, a Traditional Fermented Tea Leaf in Northern Thailand and Their Tannin-Tolerant Ability in Tea Extract. J. Microbiol. 2017, 55, 720–729. [Google Scholar] [CrossRef]
- Khanongnuch, C.; Unban, K.; Kanpiengjai, A.; Saenjum, C. Recent Research Advances and Ethno-Botanical History of Miang, a Traditional Fermented Tea (Camellia sinensis var. assamica) of Northern Thailand. J. Ethn. Foods 2017, 4, 135–144. [Google Scholar] [CrossRef]
- Unban, K.; Khatthongngam, N.; Pattananandecha, T.; Saenjum, C.; Shetty, K.; Khanongnuch, C. Microbial Community Dynamics During the Non-Filamentous Fungi Growth-Based Fermentation Process of Miang, a Traditional Fermented Tea of North Thailand and Their Product Characterizations. Front. Microbiol. 2020, 11, 1515. [Google Scholar] [CrossRef]
- Rungsirivanich, P.; Supandee, W.; Futui, W.; Chumsai-Na-Ayudhya, V.; Yodsombat, C.; Thongwai, N. Culturable Bacterial Community on Leaves of Assam Tea (Camellia sinensis var. assamica) in Thailand and Human Probiotic Potential of Isolated Bacillus spp. Microorganisms 2020, 8, 1585. [Google Scholar] [CrossRef]
- Unban, K.; Chaichana, W.; Baipong, S.; Abdullahi, A.D.; Kanpiengjai, A.; Shetty, K.; Khanongnuch, C. Probiotic and Antioxidant Properties of Lactic Acid Bacteria Isolated from Indigenous Fermented Tea Leaves (Miang) of North Thailand and Promising Application in Synbiotic Formulation. Fermentation 2021, 7, 195. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Bairoch, A. The Enzyme Database in 2000. Nucleic Acids Res. 2000, 28, 304–305. [Google Scholar] [CrossRef] [Green Version]
- Allaire, J. RStudio: Integrated Development Environment for R. Boston MA 2012, 770, 165–171. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Segner, W.P.; Schmidt, C.F.; Boltz, J.K. Effect of Sodium Chloride and pH on the Outgrowth of Spores of Type E Clostridium botulinum at Optimal and Suboptimal Temperatures. Appl. Microbiol. 1966, 14, 49–54. [Google Scholar] [CrossRef]
- Gibson, A.M.; Bratchell, N.; Roberts, T.A. The Effect of Sodium Chloride and Temperature on the Rate and Extent of Growth of Clostridium botulinum Type A in Pasteurized Pork Slurry. J. Appl. Bacteriol. 1987, 62, 479–490. [Google Scholar] [CrossRef]
- Brewer, M.S.; Rostogi, B.K.; Argoudelis, L.; Sprouls, G.K. Sodium Lactate/Sodium Chloride Effects on Aerobic Plate Counts and Color of Aerobically Packaged Ground Pork. J. Food Sci. 1995, 60, 58–62. [Google Scholar] [CrossRef]
- Kamdee, S.; Plengvidhya, V.; Chokesajjawatee, N. Changes in Lactic Acid Bacteria Diversity during Fermentation of Sour Pickled Mustard Green. Asia-Pac. J. Sci. Technol. 2014, 19, 26–33. [Google Scholar]
- Jeong, S.H.; Lee, H.J.; Jung, J.Y.; Lee, S.H.; Seo, H.-Y.; Park, W.-S.; Jeon, C.O. Effects of Red Pepper Powder on Microbial Communities and Metabolites during Kimchi Fermentation. Int. J. Food Microbiol. 2013, 160, 252–259. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Sun, H.; Wu, J.; Zhao, C.; Li, T.; Huang, H.; Fang, Q.; Mu, E.; Wu, R. Investigating the Core Microbiota and Its Influencing Factors in Traditional Chinese Pickles. Food Res. Int. 2021, 147, 110543. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Wu, J.; An, F.; Yue, X.; Tao, D.; Wu, R.; Lee, Y. An Integrated Metagenomic/Metaproteomic Investigation of Microbiota in Dajiang-Meju, a Traditional Fermented Soybean Product in Northeast China. Food Res. Int. 2019, 115, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Song, J.H.; Jung, M.Y.; Lee, S.H.; Chang, J.Y. Large-Scale Targeted Metagenomics Analysis of Bacterial Ecological Changes in 88 Kimchi Samples during Fermentation. Food Microbiol. 2017, 66, 173–183. [Google Scholar] [CrossRef]
- Yang, H.; Zou, H.; Qu, C.; Zhang, L.; Liu, T.; Wu, H.; Li, Y. Dominant Microorganisms during the Spontaneous Fermentation of Suan Cai, a Chinese Fermented Vegetable. Food Sci. Technol. Res. 2014, 20, 915–926. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Yu, M.; Liu, X.; Meng, L.; Wang, Q.; Xue, Y.; Wu, J.; Yue, X. Changes in Flavour and Microbial Diversity during Natural Fermentation of Suan-Cai, a Traditional Food Made in Northeast China. Int. J. Food Microbiol. 2015, 211, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Luo, Y.; Zhai, Z.; Zhang, H.; Yang, C.; Tian, H.; Li, Z.; Feng, J.; Liu, H.; Hao, Y. Characterization and Application of an Anti-Listeria Bacteriocin Produced by Pediococcus pentosaceus 05–10 Isolated from Sichuan Pickle, a Traditionally Fermented Vegetable Product from China. Food Control 2009, 20, 1030–1035. [Google Scholar] [CrossRef]
- Tomita, S.; Watanabe, J.; Nakamura, T.; Endo, A.; Okada, S. Characterisation of the Bacterial Community Structures of Sunki, a Traditional Unsalted Pickle of Fermented Turnip Leaves. J. Biosci. Bioeng. 2020, 129, 541–551. [Google Scholar] [CrossRef]
- Endo, A.; Mizuno, H.; Okada, S. Monitoring the Bacterial Community during Fermentation of Sunki, an Unsalted, Fermented Vegetable Traditional to the Kiso Area of Japan. Lett. Appl. Microbiol. 2008, 47, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Park, E.-J.; Chun, J.; Cha, C.-J.; Park, W.-S.; Jeon, C.O.; Bae, J.-W. Bacterial Community Analysis during Fermentation of Ten Representative Kinds of Kimchi with Barcoded Pyrosequencing. Food Microbiol. 2012, 30, 197–204. [Google Scholar] [CrossRef]
- Battcock, M.; Azam-Ali, S. Fermented Fruits and Vegetables. A Global Perspective; FAO Agricultural Services Bulletin; FAO: Rome, Italy, 1998. [Google Scholar]
- Ward, B. Bacterial Energy Metabolism. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.-W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 201–233. [Google Scholar]
- Ciani, M.; Comitini, F.; Mannazzu, I. Fermentation. In FAO Agricultural Services Bulletin Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Oxford, UK, 2008; pp. 1548–1557. [Google Scholar]
- Anandharaj, M.; Sivasankari, B.; Santhanakaruppu, R.; Manimaran, M.; Rani, R.P.; Sivakumar, S. Determining the Probiotic Potential of Cholesterol-Reducing Lactobacillus and Weissella Strains Isolated from Gherkins (Fermented Cucumber) and South Indian Fermented Koozh. Res. Microbiol. 2015, 166, 428–439. [Google Scholar] [CrossRef]
- Patel, A.; Prajapati, J.B.; Holst, O.; Ljungh, A. Determining Probiotic Potential of Exopolysaccharide Producing Lactic Acid Bacteria Isolated from Vegetables and Traditional Indian Fermented Food Products. Food Biosci. 2014, 5, 27–33. [Google Scholar] [CrossRef]
- Leong, K.-H.; Chen, Y.-S.; Lin, Y.-H.; Pan, S.-F.; Yu, B.; Wu, H.-C.; Yanagida, F. Weissellicin L, a Novel Bacteriocin from Sian-Sianzih-Isolated Weissella hellenica 4–7. J. Appl. Microbiol. 2013, 115, 70–76. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Jiang, M.; Rui, X.; Li, W.; Dong, M. A Newly Discovered Bacteriocin from Weissella hellenica D1501 Associated with Chinese Dong Fermented Meat (Nanx Wudl). Food Control 2014, 42, 116–124. [Google Scholar] [CrossRef]
- Masuda, Y.; Zendo, T.; Sawa, N.; Perez, R.; Nakayama, J.; Sonomoto, K. Characterization and Identification of Weissellicin Y and Weissellicin M, Novel Bacteriocins Produced by Weissella hellenica QU 13. J. Appl. Microbiol. 2012, 112, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M.; Papamichael, E.M. Purification, Amino Acid Sequence and Characterization of the Class IIa Bacteriocin Weissellin A, Produced by Weissella paramesenteroides DX. Bioresour. Technol. 2011, 102, 6730–6734. [Google Scholar] [CrossRef]
- Woraprayote, W.; Pumpuang, L.; Tosukhowong, A.; Roytrakul, S.; Perez, R.H.; Zendo, T.; Sonomoto, K.; Benjakul, S.; Visessanguan, W. Two Putatively Novel Bacteriocins Active against Gram-Negative Food Borne Pathogens Produced by Weissella hellenica BCC 7293. Food Control 2015, 55, 176–184. [Google Scholar] [CrossRef]
- Son, S.-H.; Yang, S.-J.; Jeon, H.-L.; Yu, H.-S.; Lee, N.-K.; Park, Y.-S.; Paik, H.-D. Antioxidant and Immunostimulatory Effect of Potential Probiotic Lactobacillus paraplantarum SC61 Isolated from Korean Traditional Fermented Food, Jangajji. Microb. Pathog. 2018, 125, 486–492. [Google Scholar] [CrossRef]
- Nikolic, M.; López, P.; Strahinic, I.; Suárez, A.; Kojic, M.; Fernández-García, M.; Topisirovic, L.; Golic, N.; Ruas-Madiedo, P. Characterisation of the Exopolysaccharide (EPS)-Producing Lactobacillus paraplantarum BGCG11 and Its Non-EPS Producing Derivative Strains as Potential Probiotics. Int. J. Food Microbiol. 2012, 158, 155–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinić, M.; Pecikoza, U.; Djokić, J.; Stepanović-Petrović, R.; Milenković, M.; Stevanović, M.; Filipović, N.; Begović, J.; Golić, N.; Lukić, J. Exopolysaccharide Produced by Probiotic Strain Lactobacillus paraplantarum BGCG11 Reduces Inflammatory Hyperalgesia in Rats. Front. Pharmacol. 2018, 9, 1. [Google Scholar] [CrossRef]
- Zivkovic, M.; Miljkovic, M.; Ruas-Madiedo, P.; Strahinic, I.; Tolinacki, M.; Golic, N.; Kojic, M. Exopolysaccharide Production and Ropy Phenotype Are Determined by Two Gene Clusters in Putative Probiotic Strain Lactobacillus paraplantarum BGCG11. Appl. Environ. Microbiol. 2015, 81, 1387–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arasu, M.V.; Al-Dhabi, N.A. In Vitro Antifungal, Probiotic, and Antioxidant Functional Properties of a Novel Lactobacillus paraplantarum Isolated from Fermented Dates in Saudi Arabia. J. Sci. Food Agric. 2017, 97, 5287–5295. [Google Scholar] [CrossRef]
- Tulini, F.L.; Winkelströter, L.K.; De Martinis, E.C.P. Identification and Evaluation of the Probiotic Potential of Lactobacillus paraplantarum FT259, a Bacteriocinogenic Strain Isolated from Brazilian Semi-Hard Artisanal Cheese. Anaerobe 2013, 22, 57–63. [Google Scholar] [CrossRef]
- Punchay, K.; Inta, A.; Tiansawat, P.; Balslev, H.; Wangpakapattanawong, P. Traditional Knowledge of Wild Food Plants of Thai Karen and Lawa (Thailand). Genet. Resour. Crop Evol. 2020, 67, 1277–1299. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yongsawas, R.; Inta, A.; Kampuansai, J.; Pandith, H.; Suwannarach, N.; Lamyong, S.; Chantawannakul, P.; Chitov, T.; Disayathanoowat, T. Bacterial Communities in Lanna Phak-Gard-Dong (Pickled Mustard Green) from Three Different Ethnolinguistic Groups in Northern Thailand. Biology 2022, 11, 150. https://doi.org/10.3390/biology11010150
Yongsawas R, Inta A, Kampuansai J, Pandith H, Suwannarach N, Lamyong S, Chantawannakul P, Chitov T, Disayathanoowat T. Bacterial Communities in Lanna Phak-Gard-Dong (Pickled Mustard Green) from Three Different Ethnolinguistic Groups in Northern Thailand. Biology. 2022; 11(1):150. https://doi.org/10.3390/biology11010150
Chicago/Turabian StyleYongsawas, Rujipas, Angkana Inta, Jatupol Kampuansai, Hataichanok Pandith, Nakarin Suwannarach, Saisamorn Lamyong, Panuwan Chantawannakul, Thararat Chitov, and Terd Disayathanoowat. 2022. "Bacterial Communities in Lanna Phak-Gard-Dong (Pickled Mustard Green) from Three Different Ethnolinguistic Groups in Northern Thailand" Biology 11, no. 1: 150. https://doi.org/10.3390/biology11010150
APA StyleYongsawas, R., Inta, A., Kampuansai, J., Pandith, H., Suwannarach, N., Lamyong, S., Chantawannakul, P., Chitov, T., & Disayathanoowat, T. (2022). Bacterial Communities in Lanna Phak-Gard-Dong (Pickled Mustard Green) from Three Different Ethnolinguistic Groups in Northern Thailand. Biology, 11(1), 150. https://doi.org/10.3390/biology11010150







