New Species of Talaromyces (Fungi) Isolated from Soil in Southwestern China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Materials
2.2. Morphological Observations
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogenetic Analyses
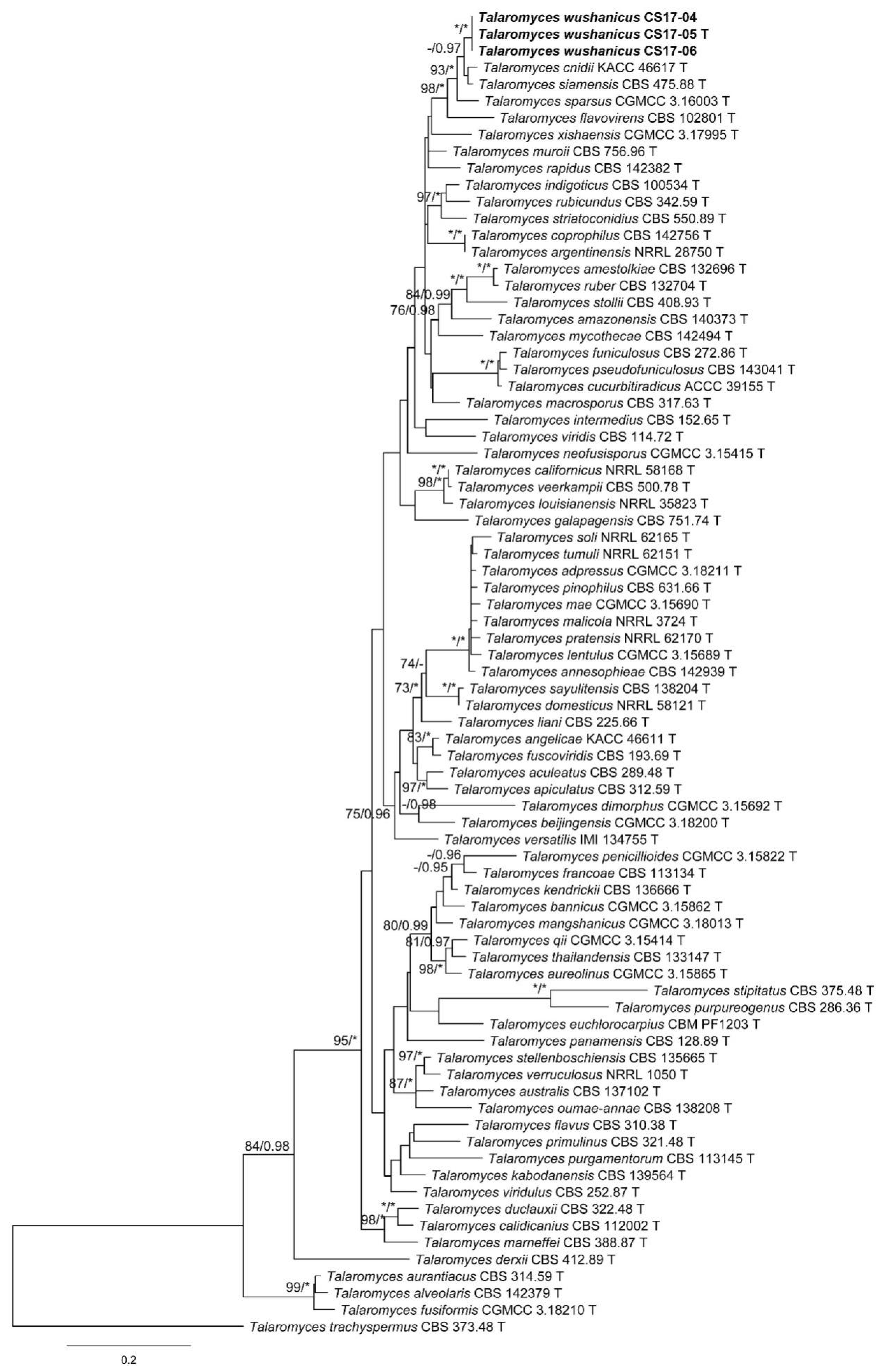
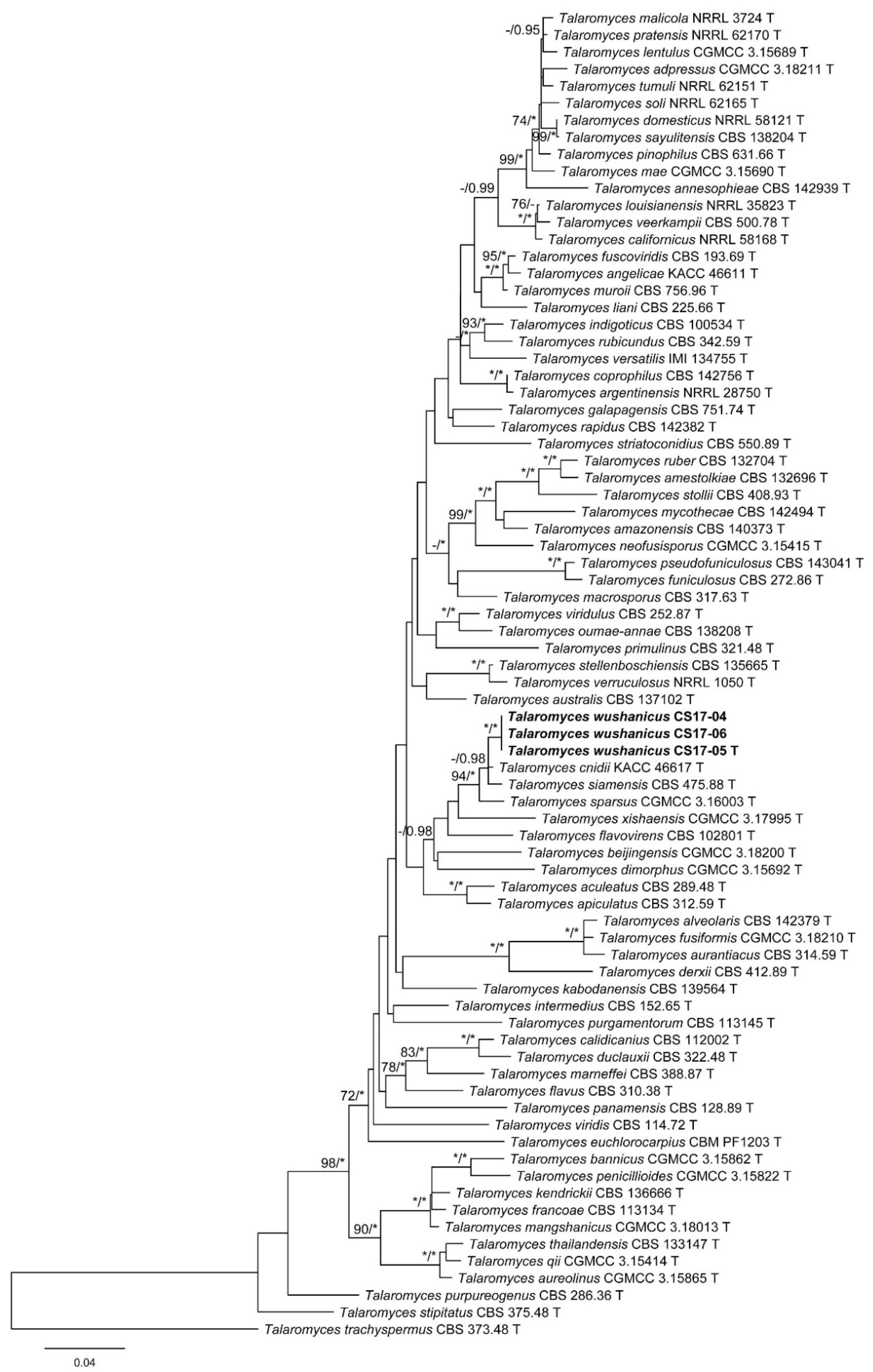
3. Results
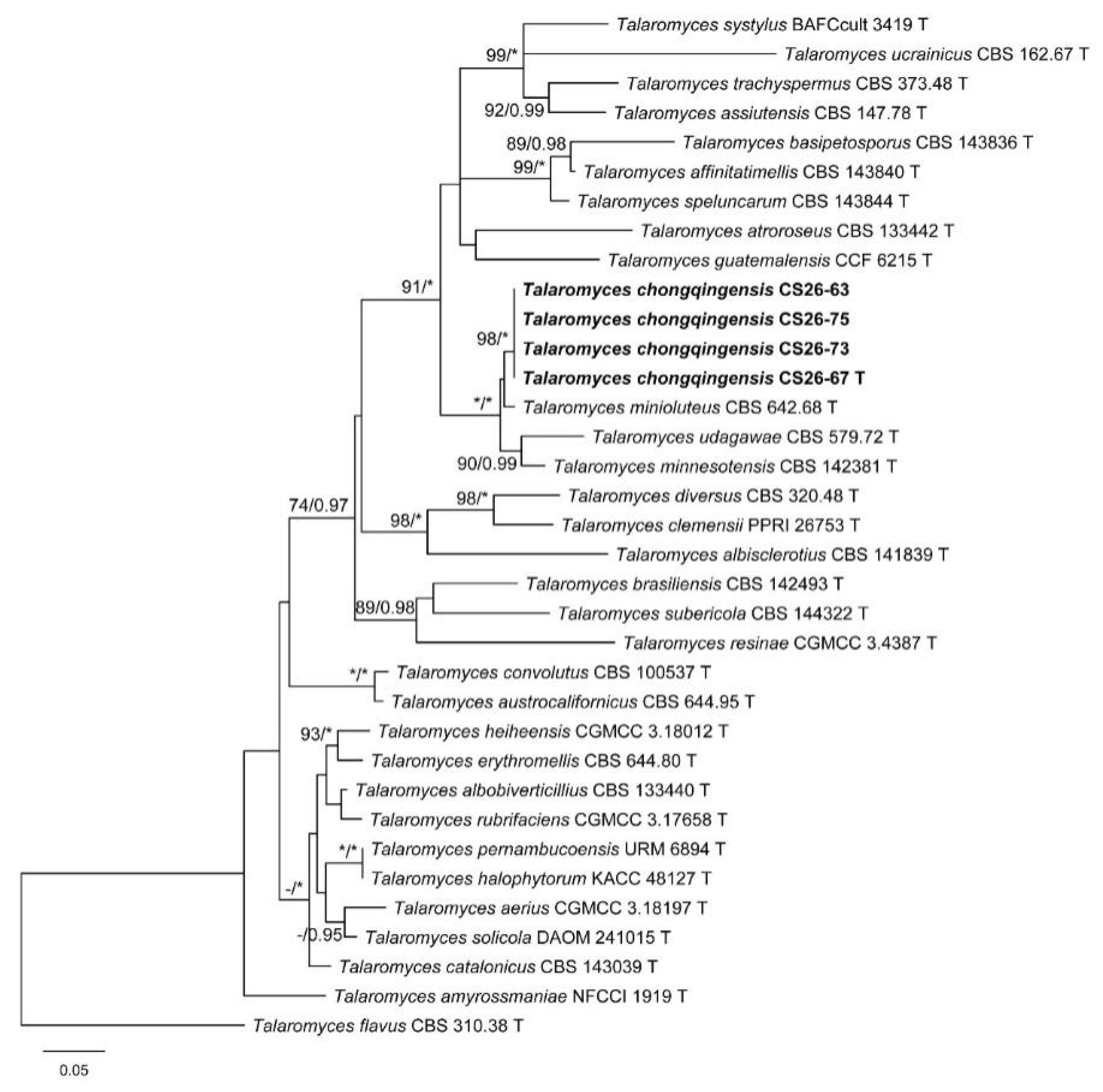
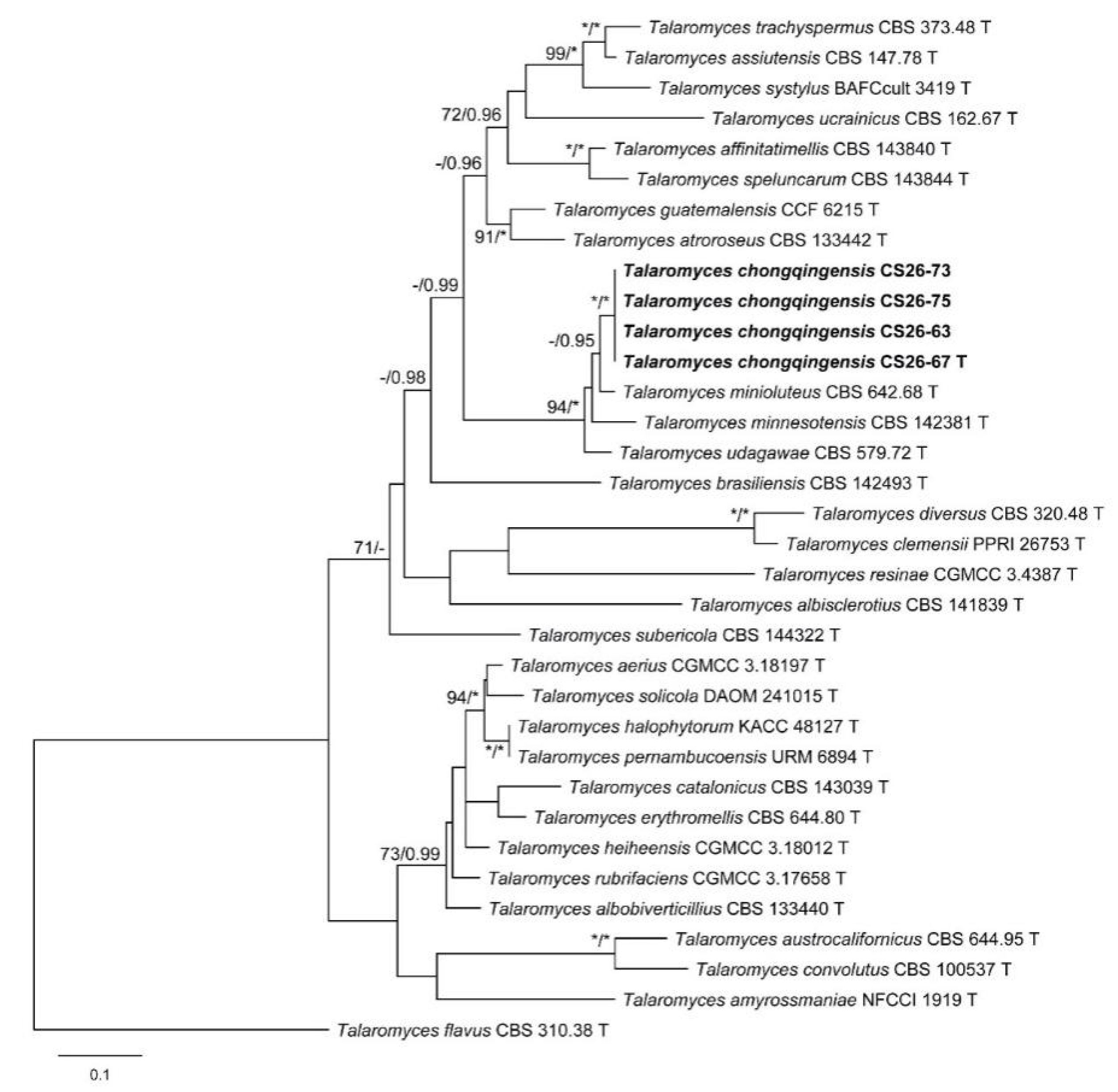
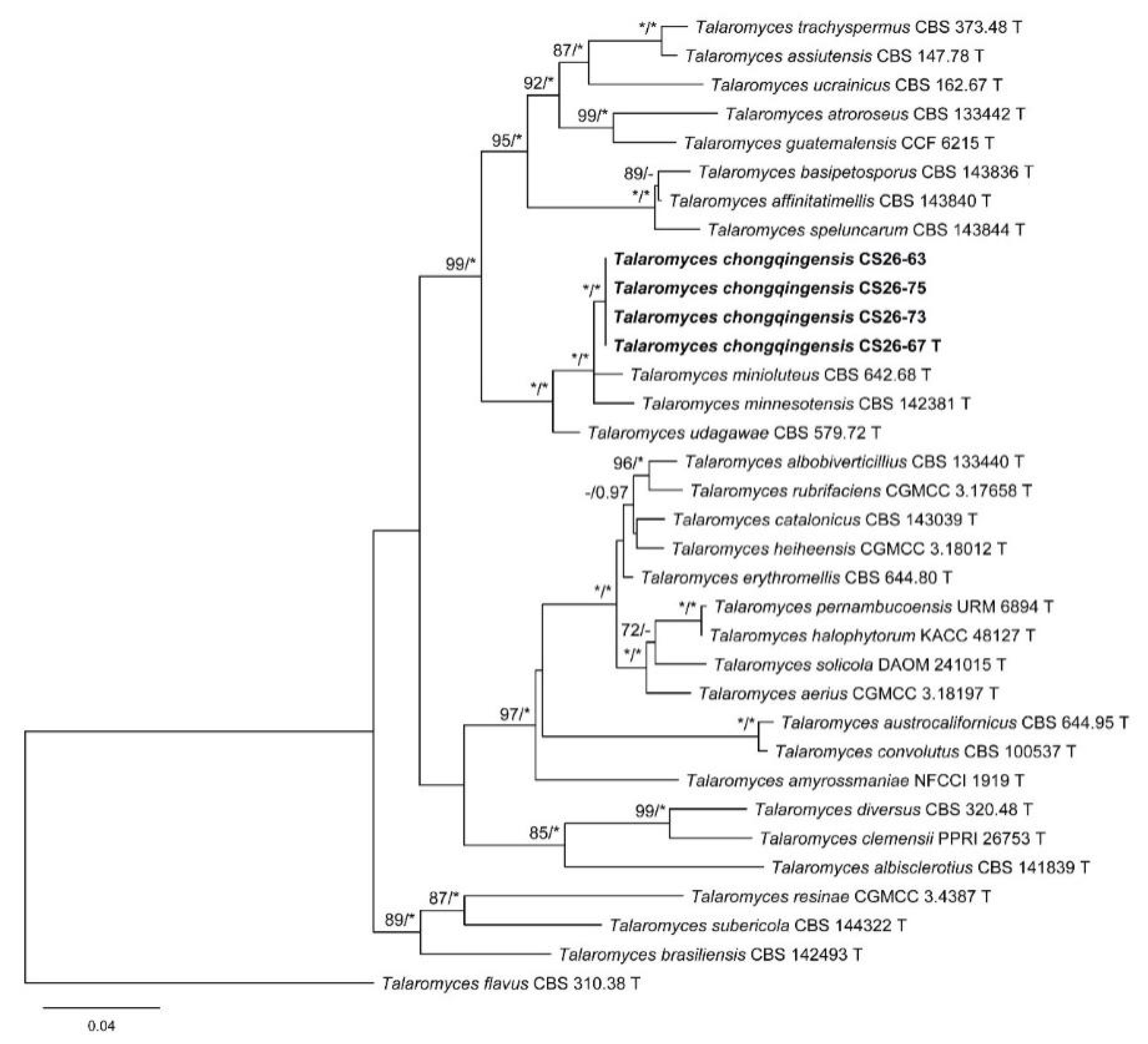
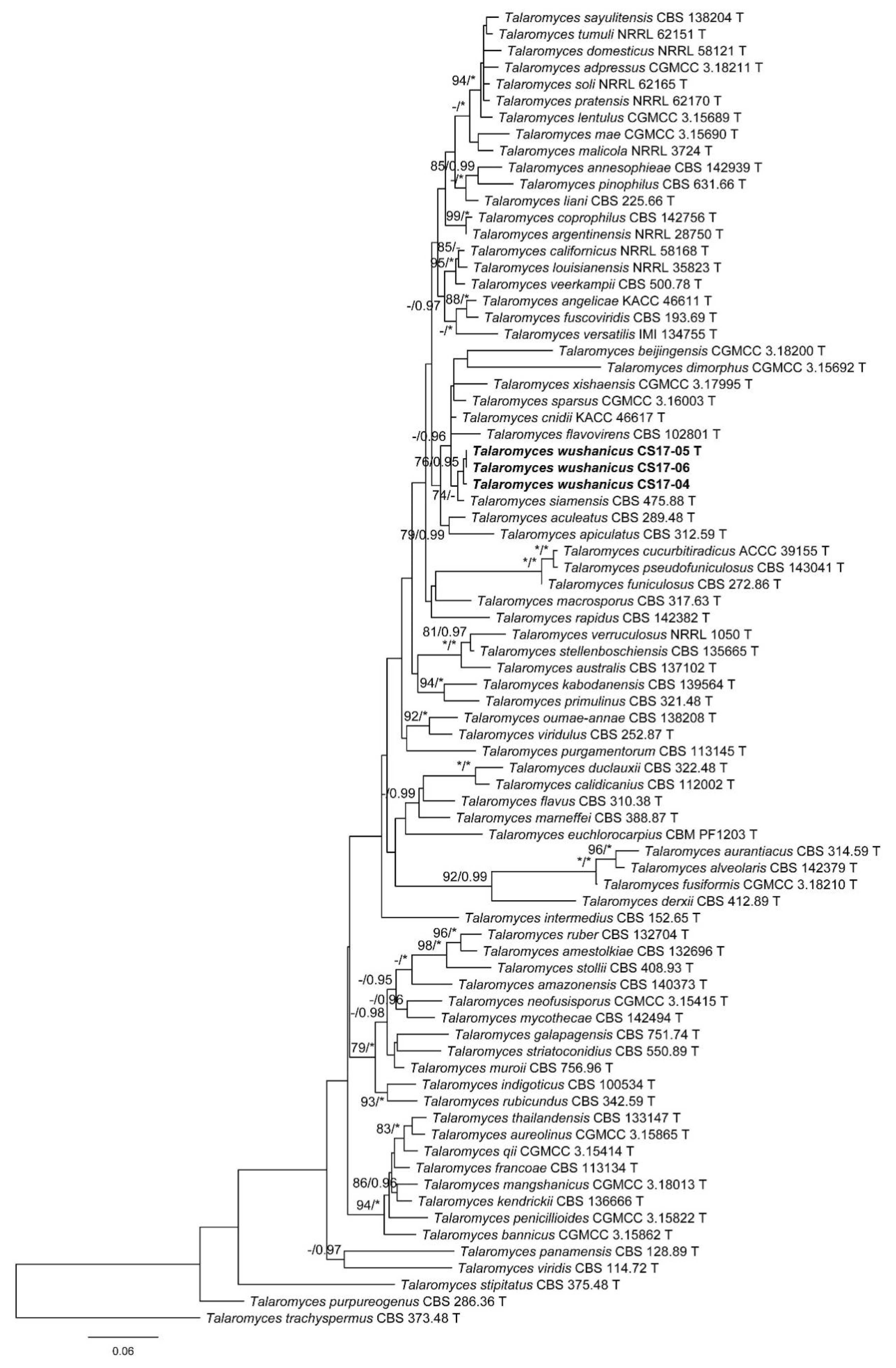
3.1. Phylogenetic Analysis
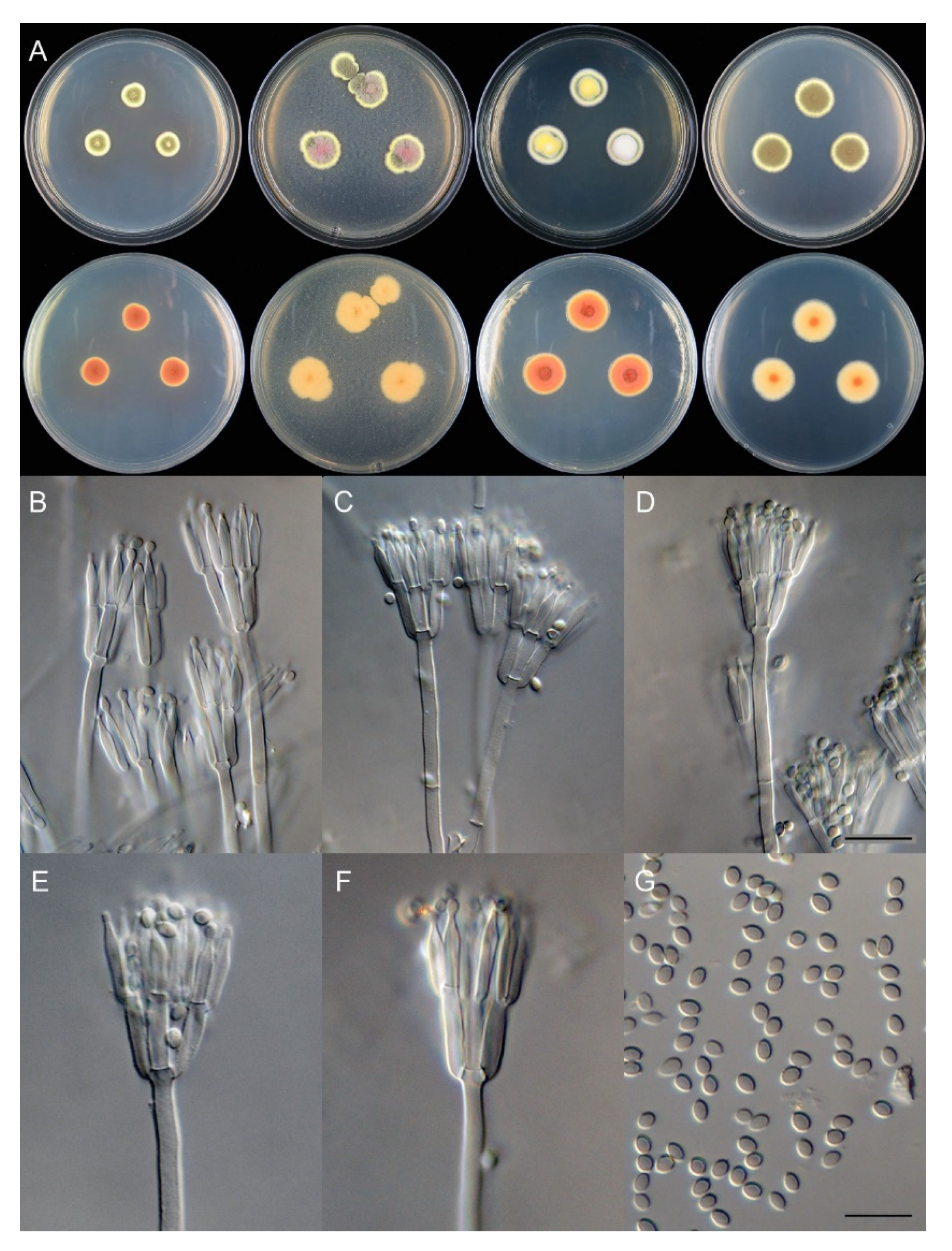
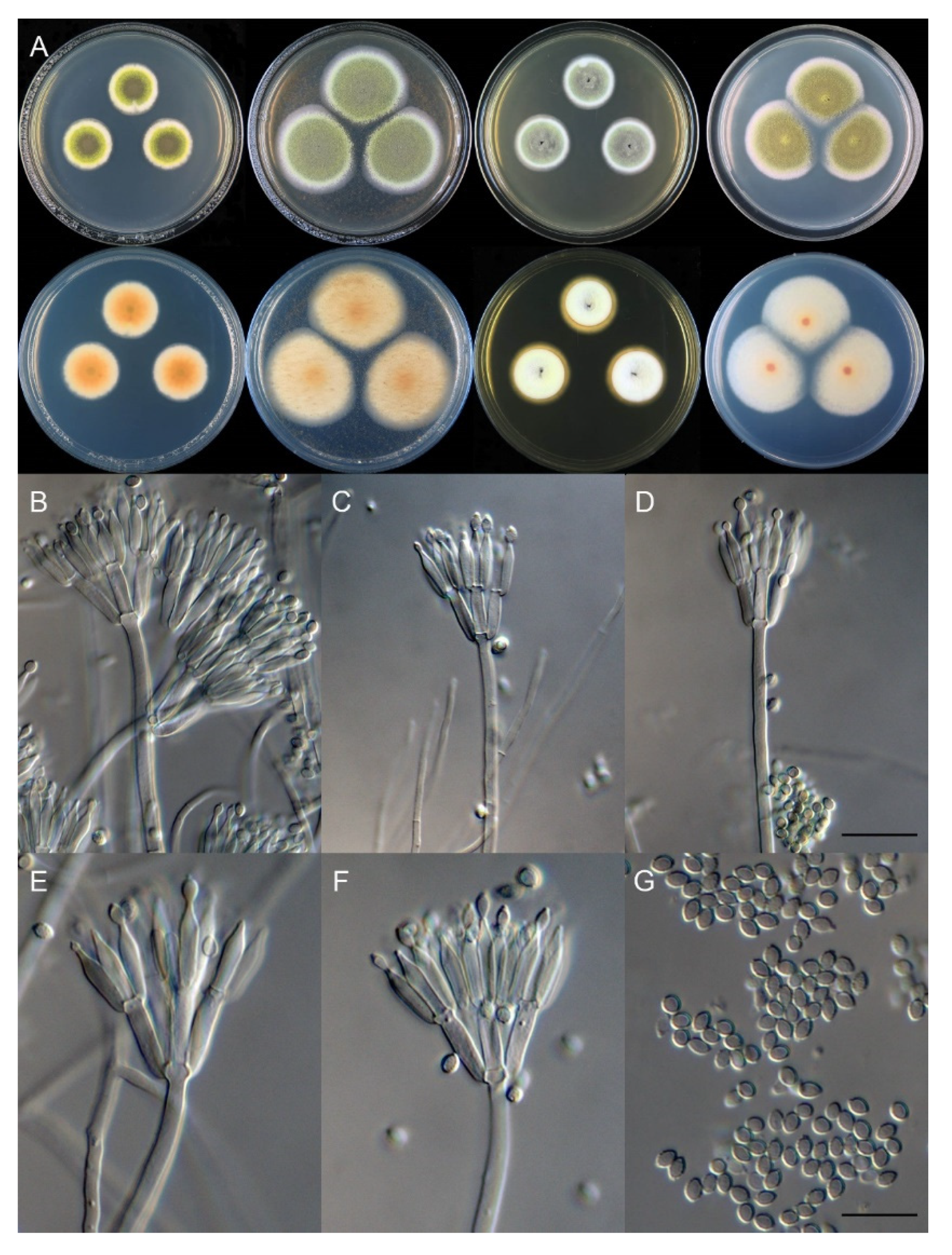
3.2. Taxonomy
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mendez-Liter, J.A.; Nieto-Dominguez, M.; Fernandez de Toro, B.; Gonzalez Santana, A.; Prieto, A.; Asensio, J.L.; Canada, F.J.; de Eugenio, L.I.; Martinez, M.J. A glucotolerant β-glucosidase from the fungus Talaromyces amestolkiae and its conversion into a glycosynthase for glycosylation of phenolic compounds. Microb. Cell Fact. 2020, 19, 127. [Google Scholar] [CrossRef]
- Inoue, H.; Decker, S.R.; Taylor, L.E.; Yano, S.; Sawayama, S. Identification and characterization of core cellulolytic enzymes from Talaromyces cellulolyticus (formerly Acremonium cellulolyticus) critical for hydrolysis of lignocellulosic biomass. Biotechnol. Biofuels 2014, 7, 151. [Google Scholar] [CrossRef]
- Morales-Oyervides, L.; Ruiz-Sanchez, J.P.; Oliveira, J.C.; Sousa-Gallagher, M.J.; Mendez-Zavala, A.; Giuffrida, D.; Dufosse, L.; Montanez, J. Biotechnological approaches for the production of natural colorants by Talaromyces/Penicillium: A review. Biotechnol. Adv. 2020, 43, 107601. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Yilmaz, N.; Thrane, U.; Rasmussen, K.B.; Houbraken, J.; Samson, R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 2013, 8, e84102. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Suarez, M.; Sutton, D.A.; Gene, J.; Garcia, D.; Wiederhold, N.; Guarro, J.; Cano-Lira, J.F. Four new species of Talaromyces from clinical sources. Mycoses 2017, 60, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, K.; Dhungana, N.; Jang, Y.; Chaturvedi, V.; Desmond, E. Characterization of clinical isolates of Talaromyces marneffei and related species, California, USA. Emerg. Infect. Dis. 2019, 25, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Xi, L.; Chaturvedi, V. Talaromycosis (Penicilliosis) due to Talaromyces (Penicillium) marneffei: Insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. Mycopathologia 2019, 184, 709–720. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsube, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Doilom, M.; Guo, J.W.; Phookamsak, R.; Mortimer, P.E.; Karunarathna, S.C.; Dong, W.; Liao, C.F.; Yan, K.; Pem, D.; Suwannarach, N.; et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front. Microbiol. 2020, 11, 585215. [Google Scholar] [CrossRef]
- Sun, B.D.; Chen, A.J.; Houbraken, J.; Frisvad, J.C.; Wu, W.P.; Wei, H.L.; Zhou, Y.G.; Jiang, X.Z.; Samson, R.A. New section and species in Talaromyces. MycoKeys 2020, 68, 75–113. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs-Kolling, G.; Yilmaz, N.; Larsson, E.; Angelini, C.; Brandrud, T.E.; Dearnaley, J.D.W.; Dima, B.; Dovana, F.; et al. Fungal Planet description sheets: 1112–1181. Persoonia 2020, 45, 251–409. [Google Scholar] [CrossRef]
- Wei, S.; Xu, X.; Wang, L. Four new species of Talaromyces section Talaromyces discovered in China. Mycologia 2021, 113, 492–508. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Xia, Y.W.; Wang, L.; Li, T.H.; Zhuang, W.Y. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa 2016, 267, 187–200. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Qin, W.T.; Zhuang, W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017, 16, 73–81. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Zeng, Z.Q.; Zhuang, W.Y. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci. Rep. 2017, 7, 8233. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef]
- Sang, H.; An, T.J.; Kim, C.S.; Shin, G.S.; Sung, G.H.; Yu, S.H. Two novel Talaromyces species isolated from medicinal crops in Korea. J. Microbiol. 2013, 51, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Marchese, C. Biodiversity hotspots: A shortcut for a more complicated concept. Glob. Ecol. Conserv. 2015, 3, 297–309. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Wang, G.Y.; Zhuang, H.F.; Wang, L.H.; Innes, J.L.; Ma, K.P. Integrating hotspots for endemic, threatened and rare species supports the identification of priority areas for vascular plants in SW China. Forest Ecol. Manag. 2021, 484. [Google Scholar] [CrossRef]
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. aerius A.J. Chen et al. 2016 | CGMCC 3.18197 T | China: Beijing | indoor air | KU866647 | KU866835 | KU866731 | KU866991 |
| T. affinitatimellis Rodr.-Andr. et al. 2019 | CBS 143840 T | Spain | honey | LT906543 | LT906552 | LT906549 | LT906546 |
| T. albisclerotius B.D. Sun et al. 2020 | CBS 141839 T | China: Guizhou | soil | MN864276 | MN863345 | MN863322 | MN863334 |
| T. albobiverticillius (H.M. Hsieh et al.) Samson et al. 2011 | CBS 133440 T | China: Taiwan | decaying leaves | HQ605705 | KF114778 | KJ885258 | KM023310 |
| T. amyrossmaniae Rajeshkumar et al. 2019 | NFCCI 1919 T | India | decaying fruits of Terminalia bellerica | MH909062 | MH909064 | MH909068 | MH909066 |
| T. assiutensis Samson and Abdel-Fattah 1978 | CBS 147.78 T | Egypt | soil | JN899323 | KJ865720 | KJ885260 | KM023305 |
| T. atroroseus N. Yilmaz et al. 2013 | CBS 133442 T | South Africa | house dust | KF114747 | KF114789 | KJ775418 | KM023288 |
| T. austrocalifornicus Yaguchi and Udagawa 1993 | CBS 644.95 T | USA | soil | JN899357 | KJ865732 | KJ885261 | MN969147 |
| T. basipetosporus Stchigel et al. 2019 | CBS 143836 T | Argentina | honey | LT906542 | LT906563 | n.a. | LT906545 |
| T. brasiliensis R.N. Barbosa et al. 2018 | CBS 142493 T | Brazil | honey | MF278323 | LT855560 | LT855563 | MN969198 |
| T. catalonicus Guevara-Suarez et al. 2020 | CBS 143039 T | Spain | herbivore dung | LT899793 | LT898318 | LT899775 | LT899811 |
| T. clemensii Visagie and N. Yilmaz 2019 | PPRI 26753 T | South Africa | wood in mine | MK951940 | MK951833 | MK951906 | MN418451 |
| T. convolutus Udagawa 1993 | CBS 100537 T | Nepal | soil | JN899330 | KF114773 | MN969316 | JN121414 |
| T. diversus (Raper and Fennell) Samson et al. 2011 | CBS 320.48 T | USA | mouldy leather | KJ865740 | KJ865723 | KJ885268 | KM023285 |
| T. erythromellis (A.D. Hocking) Samson et al. 2011 | CBS 644.80 T | Australia | soil | JN899383 | HQ156945 | KJ885270 | KM023290 |
| T. guatemalensis A. Nováková et al. 2019 | CCF 6215 T | Guatemala | soil | MN322789 | MN329687 | MN329688 | MN329689 |
| T. halophytorum Y.H. You and S.B. Hong 2020 | KACC 48127 T | South Korea | roots of Limonium tetragonum | MH725786 | MH729367 | MK111426 | MK111427 |
| T. heiheensis X.C. Wang and W.Y. Zhuang 2017 | CGMCC 3.18012 T | China: Heilongjiang | rotten wood | KX447526 | KX447525 | KX447532 | KX447529 |
| T. minioluteus (Dierckx) Samson et al. 2011 | CBS 642.68 T | unknown | unknown | JN899346 | MN969409 | KJ885273 | JF417443 |
| T. minnesotensis Guevara-Suarez et al. 2017 | CBS 142381 T | USA | human ear | LT558966 | LT559083 | LT795604 | LT795605 |
| T. pernambucoensis R. Cruz et al. 2019 | URM 6894 T | Brazil | soil | LR535947 | LR535945 | LR535946 | LR535948 |
| T. resinae (Z.T. Qi and H.Z. Kong) Houbraken and X.C. Wang 2020 | CGMCC 3.4387 T | China: Guizhou | resin of Eucalyptus tereticornis | MT079858 | MN969442 | MT066184 | MN969221 |
| T. rubrifaciens W.W. Gao 2016 | CGMCC 3.17658 T | China: Beijing | hospital air | KR855658 | KR855648 | KR855653 | KR855663 |
| T. solicola Visagie and K. Jacobs 2012 | DAOM 241015 T | South Africa | soil | FJ160264 | GU385731 | KJ885279 | KM023295 |
| T. speluncarum Rodr.-Andr. et al. 2020 | CBS 143844 T | Spain | sparkling wine | LT985890 | LT985901 | LT985906 | LT985911 |
| T. subericola Rodr.-Andr. et al. 2020 | CBS 144322 T | Spain | sparkling wine | LT985888 | LT985899 | LT985904 | LT985909 |
| T. systylus S.M. Romero et al. 2015 | BAFCcult 3419 T | Argentina | soil | KP026917 | KR233838 | KR233837 | n.a. |
| T. trachyspermus (Shear) Stolk and Samson 1973 | CBS 373.48 T | USA | unknown | JN899354 | KF114803 | KJ885281 | JF417432 |
| T. ucrainicus (Panas.) Udagawa 1966 | CBS 162.67 T | Ukraine | potato starch | JN899394 | KF114771 | KJ885282 | KM023289 |
| T. udagawae Stolk and Samson 1972 | CBS 579.72 T | Japan | soil | JN899350 | KF114796 | KX961260 | MN969148 |
| T. flavus (Klöcker) Stolk and Samson 1972 | CBS 310.38 T | New Zealand | unknown | JN899360 | JX494302 | KF741949 | JF417426 |
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. aculeatus (Raper and Fennell) Samson et al. 2011 | CBS 289.48 T | USA | textile | KF741995 | KF741929 | KF741975 | MH793099 |
| T. adpressus A.J. Chen et al. 2016 | CGMCC 3.18211 T | China: Beijing | indoor air | KU866657 | KU866844 | KU866741 | KU867001 |
| T. alveolaris Guevara-Suarez et al. 2017 | CBS 142379 T | USA | human bronchoalveolar lavage | LT558969 | LT559086 | LT795596 | LT795597 |
| T. amazonensis N. Yilmaz et al. 2016 | CBS 140373 T | Colombia | leaf litter | KX011509 | KX011490 | KX011502 | MN969186 |
| T. amestolkiae N. Yilmaz et al. 2012 | CBS 132696 T | South Africa | house dust | JX315660 | JX315623 | KF741937 | JX315698 |
| T. angelicae S.H. Yu et al. 2013 | KACC 46611 T | South Korea | dried root of Angelica gigas | KF183638 | KF183640 | KJ885259 | KX961275 |
| T. annesophieae Houbraken 2017 | CBS 142939 T | Netherlands | soil | MF574592 | MF590098 | MF590104 | MN969199 |
| T. apiculatus Samson et al. 2011 | CBS 312.59 T | Japan | soil | JN899375 | KF741916 | KF741950 | KM023287 |
| T. argentinensis Jurjević and S.W. Peterson 2019 | NRRL 28750 T | Ghana | soil | MH793045 | MH792917 | MH792981 | MH793108 |
| T. aurantiacus (J.H. Mill. et al.) Samson et al. 2011 | CBS 314.59 T | USA | soil | JN899380 | KF741917 | KF741951 | KX961285 |
| T. aureolinus L. Wang 2021 | CGMCC 3.15865 T | China: Yunnan | soil | MK837953 | MK837937 | MK837945 | MK837961 |
| T. australis Visagie et al. 2015 | CBS 137102 T | Australia | soil under pasture | KF741991 | KF741922 | KF741971 | KX961284 |
| T. bannicus L. Wang 2021 | CGMCC 3.15862 T | China: Yunnan | soil | MK837955 | MK837939 | MK837947 | MK837963 |
| T. beijingensis A.J. Chen et al. 2016 | CGMCC 3.18200 T | China: Beijing | indoor air | KU866649 | KU866837 | KU866733 | KU866993 |
| T. calidicanius (J.L. Chen) Samson et al. 2011 | CBS 112002 T | China: Taiwan | soil | JN899319 | HQ156944 | KF741934 | KM023311 |
| T. californicus Jurjević and S.W. Peterson 2019 | NRRL 58168 T | USA | air | MH793056 | MH792928 | MH792992 | MH793119 |
| T. cnidii S.H. Yu et al. 2013 | KACC 46617 T | South Korea | dried roots of Cnidium sp. | KF183639 | KF183641 | KJ885266 | KM023299 |
| T. coprophilus Guevara-Suarez et al. 2020 | CBS 142756 T | Spain | herbivore dung | LT899794 | LT898319 | LT899776 | LT899812 |
| T. cucurbitiradicus L. Su and Y.C. Niu 2018 | ACCC 39155 T | China: Beijing | endophyte from root of pumpkin (Cucurbita moschata) | KY053254 | KY053228 | KY053246 | n.a. |
| T. derxii Takada and Udagawa 1988 | CBS 412.89 T | Japan | cultivated soil | JN899327 | JX494306 | KF741959 | KM023282 |
| T. dimorphus X.Z. Jiang and L. Wang 2018 | CGMCC 3.15692 T | China: Hainan | forest soil | KY007095 | KY007111 | KY007103 | KY112593 |
| T. domesticus Jurjević and S.W. Peterson 2019 | NRRL 58121 T | USA | floor swab | MH793055 | MH792927 | MH792991 | MH793118 |
| T. duclauxii (Delacr.) Samson et al. 2011 | CBS 322.48 T | France | canvas | JN899342 | JX091384 | KF741955 | JN121491 |
| T. euchlorocarpius Yaguchi et al. 1999 | CBM PF1203 T | Japan | soil | AB176617 | KJ865733 | KJ885271 | KM023303 |
| T. flavovirens (Durieu and Mont.) Visagie et al. 2012 | CBS 102801 T | Spain | unknown | JN899392 | JX091376 | KF741933 | KX961283 |
| T. flavus (Klöcker) Stolk and Samson 1972 | CBS 310.38 T | New Zealand | unknown | JN899360 | JX494302 | KF741949 | JF417426 |
| T. francoae N. Yilmaz et al. 2016 | CBS 113134 T | Colombia | leaf litter | KX011510 | KX011489 | KX011501 | MN969188 |
| T. funiculosus (Thom) Samson et al. 2011 | CBS 272.86 T | India | Lagenaria vulgaris | JN899377 | MN969408 | KF741945 | KM023293 |
| T. fuscoviridis Visagie et al. 2015 | CBS 193.69 T | Netherlands | soil | KF741979 | KF741912 | KF741942 | MN969156 |
| T. fusiformis A.J. Chen et al. 2016 | CGMCC 3.18210 T | China: Beijing | indoor air | KU866656 | KU866843 | KU866740 | KU867000 |
| T. galapagensis Samson and Mahoney 1977 | CBS 751.74 T | Ecuador | soil under Maytenus obovata | JN899358 | JX091388 | KF741966 | KX961280 |
| T. indigoticus Takada and Udagawa 1993 | CBS 100534 T | Japan | soil | JN899331 | JX494308 | KF741931 | KX961278 |
| T. intermedius (Apinis) Stolk and Samson 1972 | CBS 152.65 T | UK | swamp soil | JN899332 | JX091387 | KJ885290 | KX961282 |
| T. kabodanensis Houbraken et al. 2016 | CBS 139564 T | Iran | hypersaline soil | KP851981 | KP851986 | KP851995 | MN969190 |
| T. kendrickii Visagie et al. 2015 | CBS 136666 T | Canada | forest soil | KF741987 | KF741921 | KF741967 | MN969158 |
| T. lentulus X.Z. Jiang and L. Wang 2018 | CGMCC 3.15689 T | China: Shandong | soil | KY007088 | KY007104 | KY007096 | KY112586 |
| T. liani (Kamyschko) N. Yilmaz et al. 2014 | CBS 225.66 T | China | soil | JN899395 | JX091380 | KJ885257 | KX961277 |
| T. louisianensis Jurjević and S.W. Peterson 2019 | NRRL 35823 T | USA | air | MH793052 | MH792924 | MH792988 | MH793115 |
| T. macrosporus (Stolk and Samson) Frisvad et al. 1990 | CBS 317.63 T | South Africa | apple juice | JN899333 | JX091382 | KF741952 | KM023292 |
| T. mae X.Z. Jiang and L. Wang 2018 | CGMCC 3.15690 T | China: Shanghai | forest soil | KY007090 | KY007106 | KY007098 | KY112588 |
| T. malicola Jurjević and S.W. Peterson 2019 | NRRL 3724 T | Italy | rhizosphere of an apple tree | MH909513 | MH909406 | MH909459 | MH909567 |
| T. mangshanicus X.C. Wang and W.Y. Zhuang 2016 | CGMCC 3.18013 T | China: Hunan | soil | KX447531 | KX447530 | KX447528 | KX447527 |
| T. marneffei (Segretain et al.) Samson et al. 2011 | CBS 388.87 T | Vietnam | bamboo rat (Rhizomys sinensis) | JN899344 | JX091389 | KF741958 | KM023283 |
| T. muroii Yaguchi et al. 1994 | CBS 756.96 T | China: Taiwan | soil | MN431394 | KJ865727 | KJ885274 | KX961276 |
| T. mycothecae R.N. Barbosa et al. 2018 | CBS 142494 T | Brazil | nest of stingless bee (Melipona scutellaris) | MF278326 | LT855561 | LT855564 | LT855567 |
| T. neofusisporus L. Wang 2016 | CGMCC 3.15415 T | China: Tibet | leaf sample | KP765385 | KP765381 | KP765383 | MN969165 |
| T. oumae-annae Visagie et al. 2014 | CBS 138208 T | South Africa | house dust | KJ775720 | KJ775213 | KJ775425 | KX961281 |
| T. panamensis (Samson et al.) Samson et al. 2011 | CBS 128.89 T | Panama | soil | JN899362 | HQ156948 | KF741936 | KM023284 |
| T. penicillioides L. Wang 2021 | CGMCC 3.15822 T | China: Guizhou | soil | MK837956 | MK837940 | MK837948 | MK837964 |
| T. pinophilus (Hedgc.) Samson et al. 2011 | CBS 631.66 T | France | PVC | JN899382 | JX091381 | KF741964 | KM023291 |
| T. pratensis Jurjević and S.W. Peterson 2019 | NRRL 62170 T | USA | effluent of water treatment plant | MH793075 | MH792948 | MH793012 | MH793139 |
| T. primulinus (Pitt) Samson et al. 2011 | CBS 321.48 T | USA | unknown | JN899317 | JX494305 | KF741954 | KM023294 |
| T. pseudofuniculosus Guevara-Suarez et al. 2020 | CBS 143041 T | Spain | herbivore dung | LT899796 | LT898323 | LT899778 | LT899814 |
| T. purgamentorum N. Yilmaz et al. 2016 | CBS 113145 T | Colombia | leaf litter | KX011504 | KX011487 | KX011500 | MN969189 |
| T. purpureogenus (Stoll) Samson et al. 2011 | CBS 286.36 T | unknown | unknown | JN899372 | JX315639 | KF741947 | JX315709 |
| T. qii L. Wang 2016 | CGMCC 3.15414 T | China: Tibet | leaf sample | KP765384 | KP765380 | KP765382 | MN969164 |
| T. rapidus Guevara-Suarez et al. 2017 | CBS 142382 T | USA | human bronchoalveolar lavage | LT558970 | LT559087 | LT795600 | LT795601 |
| T. ruber (Stoll) N. Yilmaz et al. 2012 | CBS 132704 T | UK | aircraft fuel tank | JX315662 | JX315629 | KF741938 | JX315700 |
| T. rubicundus (J.H. Mill. et al.) Samson et al. 2011 | CBS 342.59 T | USA | soil | JN899384 | JX494309 | KF741956 | KM023296 |
| T. sayulitensis Visagie et al. 2014 | CBS 138204 T | Mexico | house dust | KJ775713 | KJ775206 | KJ775422 | MN969146 |
| T. siamensis (Manoch and C. Ramírez) Samson et al. 2011 | CBS 475.88 T | Thailand | forest soil | JN899385 | JX091379 | KF741960 | KM023279 |
| T. soli Jurjević and S.W. Peterson 2019 | NRRL 62165 T | USA | soil | MH793074 | MH792947 | MH793011 | MH793138 |
| T. sparsus L. Wang 2021 | CGMCC 3.16003 T | China: Beijing | soil | MT077182 | MT083924 | MT083925 | MT083926 |
| T. stellenboschiensis Visagie and K. Jacobs 2015 | CBS 135665 T | South Africa | soil | JX091471 | JX091605 | JX140683 | MN969157 |
| T. stipitatus (Thom) C.R. Benj. 1955 | CBS 375.48 T | USA | rotting wood | JN899348 | KM111288 | KF741957 | KM023280 |
| T. stollii N. Yilmaz et al. 2012 | CBS 408.93 T | Netherlands | AIDS patient | JX315674 | JX315633 | JX315646 | JX315712 |
| T. striatoconidium (R.F. Castañeda and W. Gams) Houbraken et al. 2020 | CBS 550.89 T | Cuba | leaf litter of Pachyanthus poirettii | MN431418 | MN969441 | MN969360 | MT156347 |
| T. thailandensis Manoch et al. 2013 | CBS 133147 T | Thailand | forest soil | JX898041 | JX494294 | KF741940 | KM023307 |
| T. tumuli Jurjević and S.W. Peterson 2019 | NRRL 62151 T | USA | soil from prairie | MH793071 | MH792944 | MH793008 | MH793135 |
| T. veerkampii Visagie et al. 2015 | CBS 500.78 T | Columbia | soil | KF741984 | KF741918 | KF741961 | KX961279 |
| T. verruculosus (Peyronel) Samson et al. 2011 | NRRL 1050 T | USA | soil | KF741994 | KF741928 | KF741944 | KM023306 |
| T. versatilis Bridge and Buddie 2013 | IMI 134755 T | UK | unknown | MN431395 | MN969412 | MN969319 | MN969161 |
| T. viridis (Stolk and G.F. Orr) Arx 1987 | CBS 114.72 T | Australia | soil | AF285782 | JX494310 | KF741935 | JN121430 |
| T. viridulus Samson et al. 2011 | CBS 252.87 T | Australia | soil | JN899314 | JX091385 | KF741943 | JF417422 |
| T. xishaensis X.C. Wang et al. 2016 | CGMCC 3.17995 T | China: Hainan | soil | KU644580 | KU644581 | KU644582 | MZ361364 |
| T. trachyspermus (Shear) Stolk and Samson 1973 | CBS 373.48 T | USA | unknown | JN899354 | KF114803 | KJ885281 | JF417432 |
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. chongqingensis X.C. Wang and W.Y. Zhuang sp. nov. | CS26-67 T | China: Chongqing | soil | MZ358001 | MZ361343 | MZ361350 | MZ361357 |
| CS26-63 | China: Chongqing | soil | MZ358002 | MZ361344 | MZ361351 | MZ361358 | |
| CS26-73 | China: Chongqing | soil | MZ358003 | MZ361345 | MZ361352 | MZ361359 | |
| CS26-75 | China: Chongqing | soil | MZ358004 | MZ361346 | MZ361353 | MZ361360 | |
| T. wushanicus X.C. Wang and W.Y. Zhuang sp. nov. | CS17-05 T | China: Chongqing | soil | MZ356356 | MZ361347 | MZ361354 | MZ361361 |
| CS17-04 | China: Chongqing | soil | MZ356357 | MZ361348 | MZ361355 | MZ361362 | |
| CS17-06 | China: Chongqing | soil | MZ356358 | MZ361349 | MZ361356 | MZ361363 |
| Section | Loci | No. of Seq. | Length of Alignment | Model for BI |
|---|---|---|---|---|
| Trachyspermi | BenA | 35 | 533 | TVM+I+G |
| CaM | 34 | 656 | SYM+I+G | |
| RPB2 | 34 | 920 | GTR+I+G | |
| Talaromyces | BenA | 79 | 490 | TrN+I+G |
| CaM | 79 | 565 | SYM+I+G | |
| RPB2 | 78 | 978 | TVM+I+G |
| Species | CYA 25 °C (mm) | CYA 37 °C (mm) | MEA (mm) | YES (mm) | Conidia Shape | Conidia Wall | Conidia Size (μm) | Reference |
|---|---|---|---|---|---|---|---|---|
| T. chongqingensis | 12–13 | no growth | 17–18 | 18–19 | ellipsoidal to broad fusiform | smooth | 2.5–3.5 × 2–2.5 | This study |
| T. minioluteus | 17–18 | no growth | 21–22 | 18 | ellipsoidal | smooth | 2.5–4 × 1.5–2.5 | [24] |
| T. minnesotensis | 24–26 | no growth | 13–15 | 21–24 | ellipsoidal | smooth | 2.5–3.5 × 2–3 | [5] |
| T. udagawae | 6–8 | no growth | 10–11 | 8–9 | subglobose to ellipsoidal | smooth | 3–4 × 2–3 | [24] |
| T. cnidii | 30–35 | 17–20 | 38–43 | 40–45 | ellipsodial | smooth to finely rough | 3–4 × 2–2.5 | [25] |
| T. siamensis | 20–22 | 15 | 32–33 | 27–28 | ellipsoidal to fusiform | smooth to finely rough | 3–4 × 2–3 | [24] |
| T. wushanicus | 21–24 | 17–19 | 40–44 | 24–28 | ellipsoidal to broad fusiform | smooth to finely rough | 3–4 × 2.5–3 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.-K.; Wang, X.-C.; Zhuang, W.-Y.; Cheng, X.-H.; Zhao, P. New Species of Talaromyces (Fungi) Isolated from Soil in Southwestern China. Biology 2021, 10, 745. https://doi.org/10.3390/biology10080745
Zhang Z-K, Wang X-C, Zhuang W-Y, Cheng X-H, Zhao P. New Species of Talaromyces (Fungi) Isolated from Soil in Southwestern China. Biology. 2021; 10(8):745. https://doi.org/10.3390/biology10080745
Chicago/Turabian StyleZhang, Zhi-Kang, Xin-Cun Wang, Wen-Ying Zhuang, Xian-Hao Cheng, and Peng Zhao. 2021. "New Species of Talaromyces (Fungi) Isolated from Soil in Southwestern China" Biology 10, no. 8: 745. https://doi.org/10.3390/biology10080745
APA StyleZhang, Z.-K., Wang, X.-C., Zhuang, W.-Y., Cheng, X.-H., & Zhao, P. (2021). New Species of Talaromyces (Fungi) Isolated from Soil in Southwestern China. Biology, 10(8), 745. https://doi.org/10.3390/biology10080745








