Towards a Structural Mechanism for Sister Chromatid Cohesion Establishment at the Eukaryotic Replication Fork
Abstract
Simple Summary
Abstract
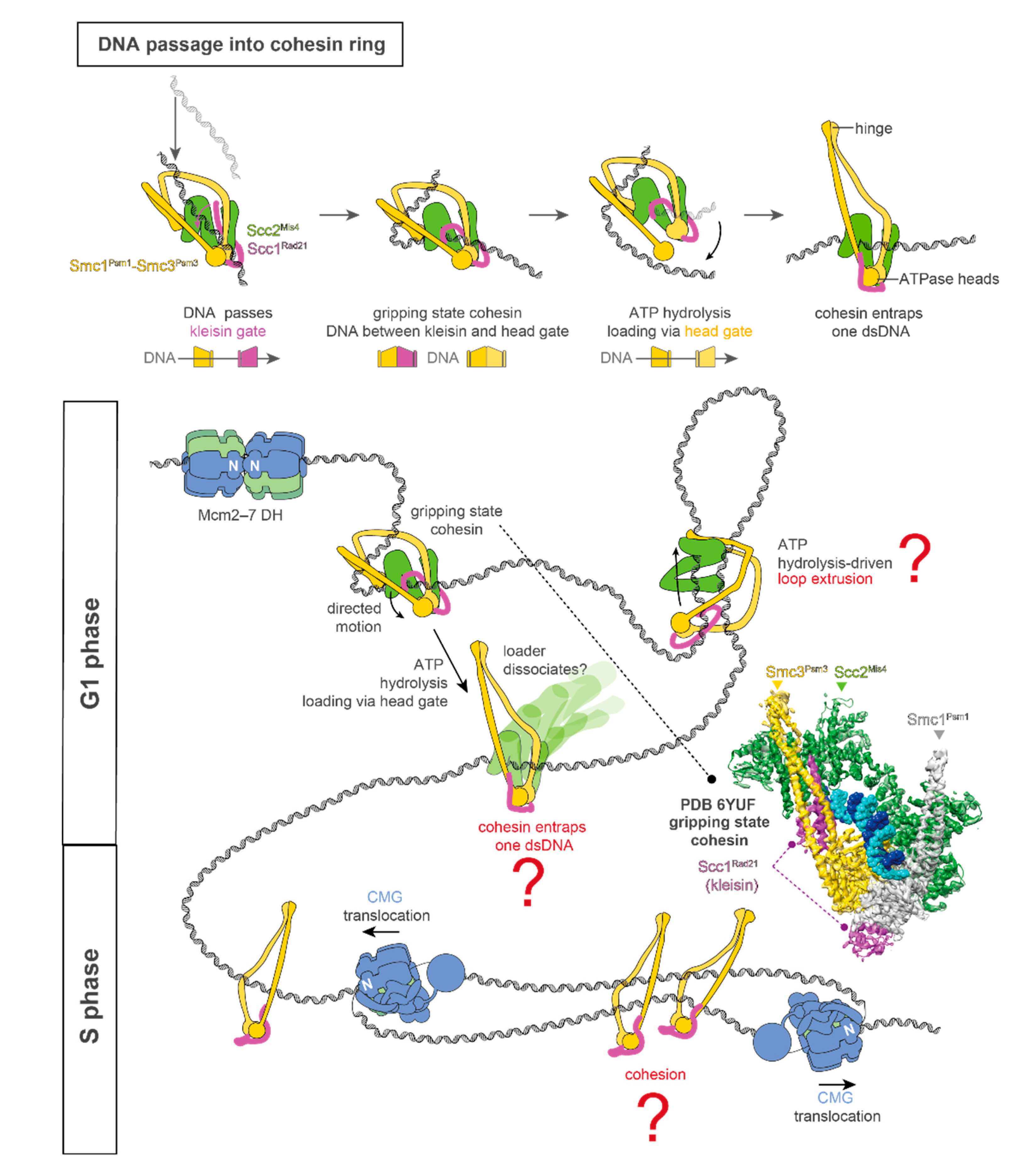
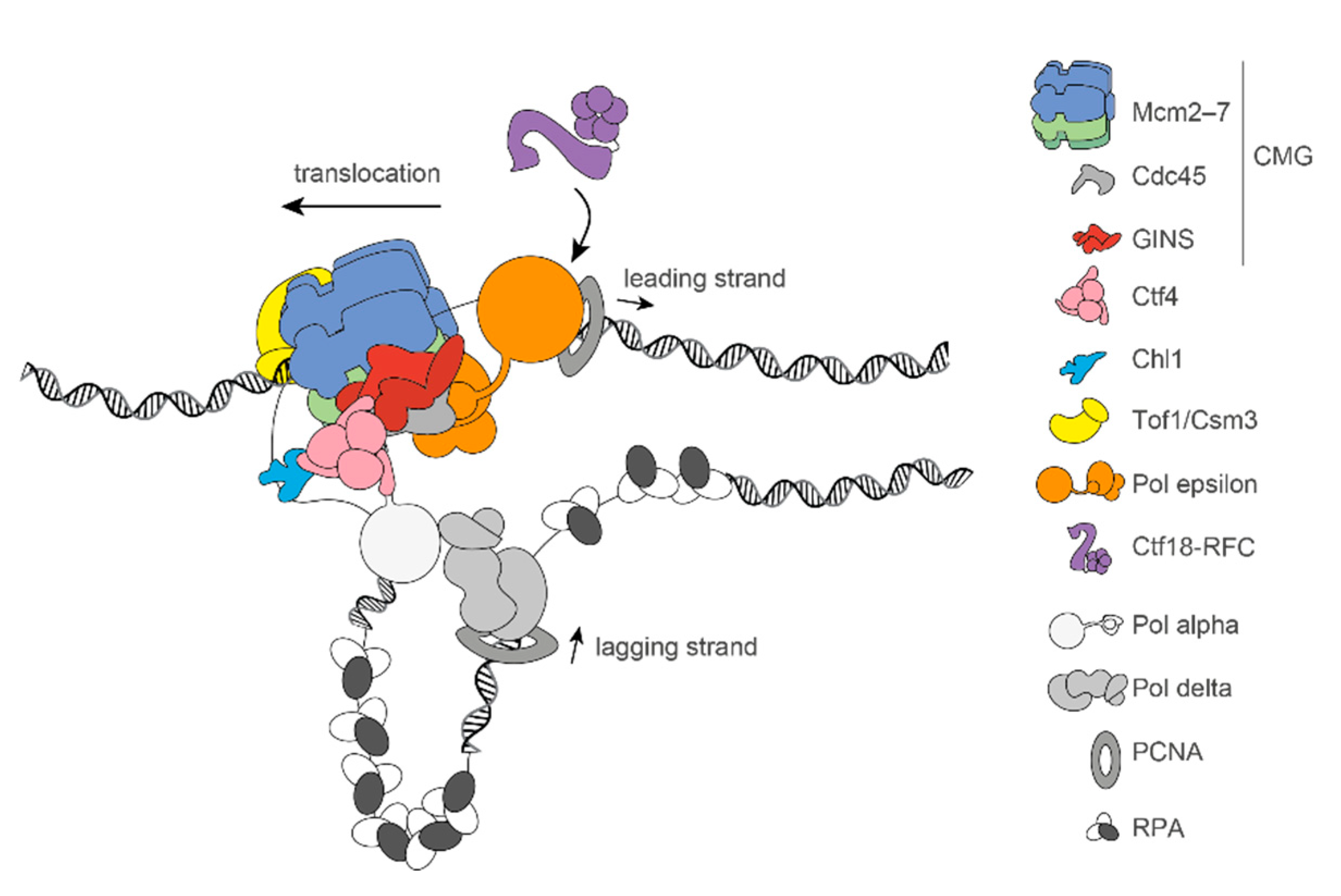
1. DNA Replication and Cohesin Function
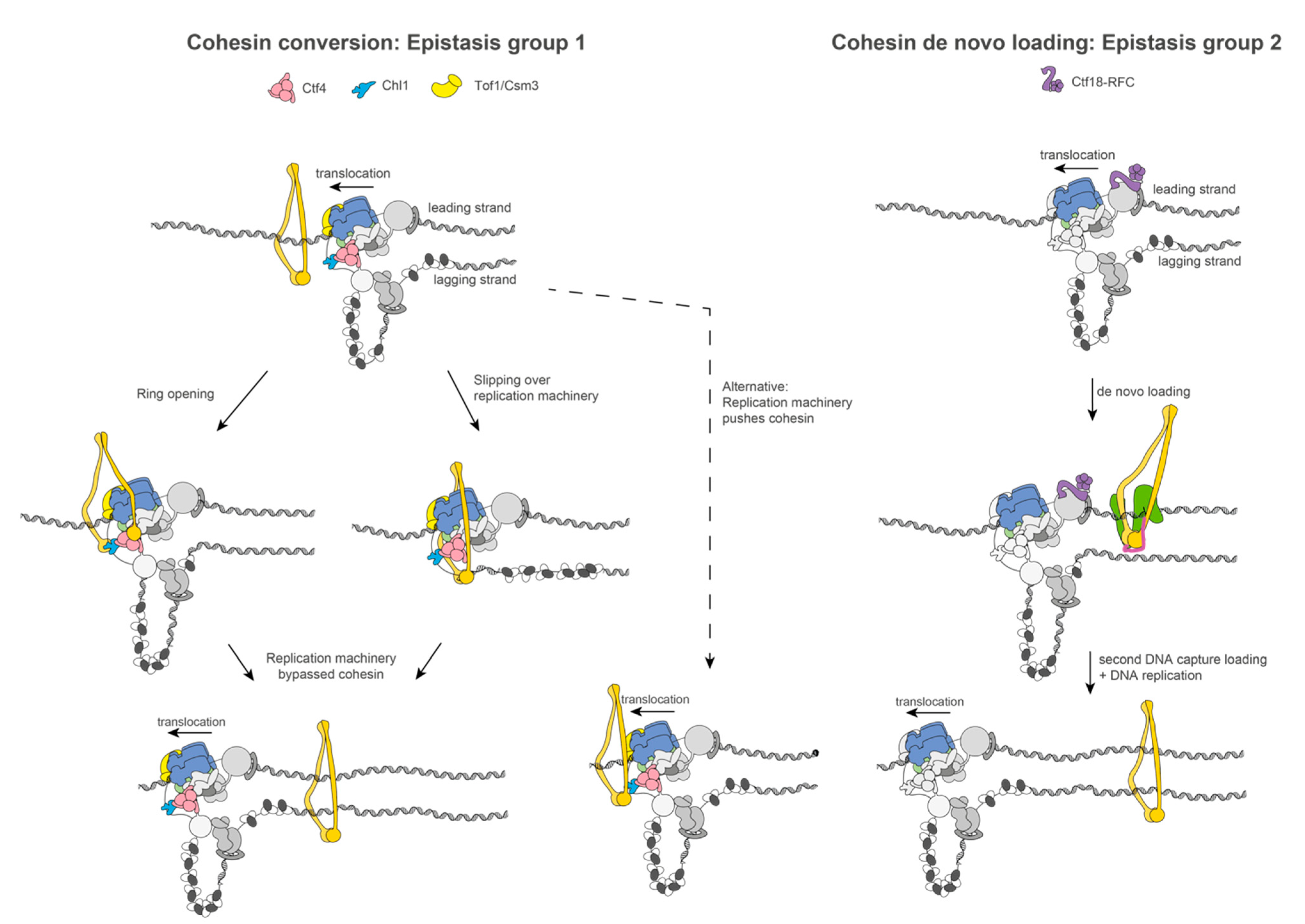
2. Coordination between DNA Replication and Sister Chromatid Cohesion Establishment
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, T.C.R.; Locke, J.; Greiwe, J.F.; Diffley, J.F.X.; Costa, A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 2019, 575, 704–710. [Google Scholar] [CrossRef]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F.X. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Evrin, C.; Clarke, P.; Zech, J.; Lurz, R.; Sun, J.; Uhle, S.; Li, H.; Stillman, B.; Speck, C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 2009, 106, 20240–20245. [Google Scholar] [CrossRef]
- Noguchi, Y.; Yuan, Z.; Bai, L.; Schneider, S.; Zhao, G.; Stillman, B.; Speck, C.; Li, H. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc. Natl. Acad. Sci. USA 2017, 114, E9529–E9538. [Google Scholar] [CrossRef]
- Ali, F.A.; Douglas, M.E.; Locke, J.; Pye, V.E.; Nans, A.; Diffley, J.F.X.; Costa, A. Cryo-EM structure of a licensed DNA replication origin. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Zhai, Y.; Cheng, E.; Wu, H.; Li, N.; Yung, P.Y.K.; Gao, N.; Tye, B.-K. Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat. Struct. Mol. Biol. 2017, 24, 300–308. [Google Scholar] [CrossRef]
- Frigola, J.; He, J.; Kinkelin, K.; Pye, V.E.; Renault, L.; Douglas, M.E.; Remus, D.; Cherepanov, P.; Costa, A.; Diffley, J.F.X. Cdt1 stabilizes an open MCM ring for helicase loading. Nat. Commun. 2017, 8, 15720. [Google Scholar] [CrossRef] [PubMed]
- Coster, G.; Frigola, J.; Beuron, F.; Morris, E.P.; Diffley, J.F.X. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol. Cell 2014, 55, 666–677. [Google Scholar] [CrossRef]
- Kang, S.; Warner, M.D.; Bell, S.P. Multiple functions for Mcm2-7 ATPase motifs during replication initiation. Mol. Cell 2014, 55, 655–665. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Y.; Li, W.; Yang, M.; Lei, J.; Tye, B.-K.; Gao, N. Structure of the eukaryotic MCM complex at 3.8 Å. Nature 2015, 524, 186–191. [Google Scholar] [CrossRef]
- Chong, J.P.J.; Mahbubani, H.M.; Khoo, C.-Y.; Blow, J.J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature 1995, 375, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, F. SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016, 17, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Hagstrom, K.A.; Meyer, B.J. Condensin and cohesin: More than chromosome compactor and glue. Nat. Rev. 2003, 4, 520–534. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006, 7, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Bürmann, F.; Lee, B.; Than, T.; Sinn, L.; O’Reilly, F.; Yatskevich, S.; Rappsilber, J.; Hu, B.; Nasmyth, K.; Löwe, J. A folded conformation of MukBED and cohesin. Nat. Struct. Mol. Biol. 2019, 26, 227–236. [Google Scholar] [CrossRef]
- Higashi, T.L.; Eickhoff, P.; Sousa, J.S.; Locke, J.; Nans, A.; Flynn, H.R.; Snijders, A.P.; Papageorgiou, G.; O’Reilly, N.; Chen, Z.A.; et al. A structure-based mechanism for DNA entry into the cohesin ring. Mol. Cell 2020, 79, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Gao, H.; Bai, X.-C.; Yu, H. Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 2020, 368, 1454–1459. [Google Scholar] [CrossRef]
- Gligoris, T.G.; Scheinost, J.C.; Bürmann, F.; Petela, N.; Chan, K.-L.; Uluocak, P.; Beckouët, F.; Gruber, S.; Nasmyth, K.; Löwe, J. Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science 2014, 346, 963–967. [Google Scholar] [CrossRef]
- Srinivasan, M.; Scheinost, J.C.; Petela, N.J.; Gligoris, T.G.; Wissler, M.; Ogushi, S.; Collier, J.E.; Voulgaris, M.; Kurze, A.; Chan, K.-L.; et al. The cohesin ring uses its hinge to organize DNA using non-topological as well as topological mechanisms. Cell 2018, 173, 1508–1519.e18. [Google Scholar] [CrossRef] [PubMed]
- Murayama, Y.; Uhlmann, F. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 2015, 163, 1628–1640. [Google Scholar] [CrossRef] [PubMed]
- Collier, J.E.; Lee, B.-G.; Roig, M.B.; Yatskevich, S.; Petela, N.J.; Metson, J.; Voulgaris, M.; Gonzalez Llamazares, A.; Löwe, J.; Nasmyth, K.A. Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3. Elife 2020, 9, e59560. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.C.H.; Murayama, Y.; Muñoz, S.; Jones, A.W.; Wade, B.O.; Purkiss, A.G.; Hu, X.-W.; Borg, A.; Snijders, A.P.; Uhlmann, F.; et al. Structure of the cohesin loader Scc2. Nat. Commun. 2017, 8, 13952. [Google Scholar] [CrossRef]
- Kikuchi, S.; Borek, D.M.; Otwinowski, Z.; Tomchick, D.R.; Yu, H. Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc. Natl. Acad. Sci. USA 2016, 113, 12444–12449. [Google Scholar] [CrossRef]
- Murayama, Y.; Uhlmann, F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 2014, 505, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Ciosk, R.; Shirayama, M.; Shevchenko, A.; Tanaka, T.; Toth, A.; Shevchenko, A.; Nasmyth, K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 2000, 5, 243–254. [Google Scholar] [CrossRef]
- Minamino, M.; Higashi, T.L.; Bouchoux, C.; Uhlmann, F. Topological in vitro loading of the budding yeast cohesin ring onto DNA. Life Sci. Alliance 2018, 1, e201800143. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.-M. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef]
- Kim, Y.; Shi, Z.; Zhang, H.; Finkelstein, I.J.; Yu, H. Human cohesin compacts DNA by loop extrusion. Science 2019, 366, 1345–1349. [Google Scholar] [CrossRef]
- Shintomi, K.; Hirano, T. Releasing cohesin from chromosome armsin early mitosis: Opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009, 23, 2224–2236. [Google Scholar] [CrossRef]
- Srinivasan, M.; Petela, N.J.; Scheinost, J.C.; Collier, J.; Voulgaris, M.; Roig, M.B.; Beckouët, F.; Hu, B.; Nasmyth, K.A. Scc2 counteracts a Wapl-independent mechanism that releases cohesin from chromosomes during G1. Elife 2019, 8, e44736. [Google Scholar] [CrossRef]
- Sheu, Y.J.; Stillman, B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 2006, 24, 101–113. [Google Scholar] [CrossRef]
- Labib, K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010, 24, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.S.; Basu, A.; Bermudez, V.; Hurwitz, J.; Walter, J.C. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008, 22, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Kanchwala, M.; Xing, C.; Yu, H. MCM2–7-dependent cohesin loading during S phase promotes sister-chromatid cohesion. Elife 2018, 7, e33920. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, S.M.; Makrantoni, V.; Harrison, S.C.; Marston, A.L. The kinetochore receptor for the cohesin loading complex. Cell 2017, 171, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef]
- Lõoke, M.; Maloney, M.F.; Bell, S.P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017, 31, 291–305. [Google Scholar] [CrossRef]
- Langston, L.D.; Mayle, R.; Schauer, G.D.; Yurieva, O.; Zhang, D.; Yao, N.Y.; Georgescu, R.E.; O’Donnell, M.E. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. Elife 2017, e29118. [Google Scholar] [CrossRef]
- Wasserman, M.R.; Schauer, G.D.; O’Donnell, M.E.; Lui, S. Replication fork activation is enabled by a single-stranded DNA gate in CMG helicase. Cell 2019, 178, 600–611e16. [Google Scholar] [CrossRef]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, R.; Yuan, Z.; Bai, L.; de Luna Almeida Santos, R.; Sun, J.; Zhang, D.; Yurieva, O.; Li, H.; O’Donnell, M.E. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. USA 2017, 114, E697–E706. [Google Scholar] [CrossRef] [PubMed]
- Baretić, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Georgescu, R.; Bai, L.; Zhang, D.; Li, H.; O’Donnell, M.E. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun. 2020, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Kose, H.B.; Larsen, N.B.; Duxin, J.P.; Yardimci, H. Dynamics of the eukaryotic replicative helicase at lagging-strand protein barriers support the steric exclusion model. Cell Rep. 2019, 26, 2113–2125. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.V.; Yardimci, H.; Long, D.T.; Ho, T.V.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Schärer, O.D.; Walter, J.C. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef]
- Ali, F.A.; Renault, L.; Gannon, J.; Gahlon, H.L.; Kotecha, A.; Zhou, J.C.; Rueda, D.; Costa, A. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat. Commun. 2016, 10708. [Google Scholar] [CrossRef]
- Eickhoff, P.; Kose, H.B.; Martino, F.; Petojevic, T.; Abid Ali, F.; Locke, J.; Tamberg, N.; Nans, A.; Berger, J.M.; Botchan, M.R.; et al. Molecular basis for ATP-hydrolysis-driven DNA translocation by the CMG helicase of the eukaryotic replisome. Cell Rep. 2019, 28, 2673–2688. [Google Scholar] [CrossRef]
- Goswami, P.; Ali, F.A.; Douglas, M.E.; Locke, J.; Purkiss, A.; Janska, A.; Eickhoff, P.; Early, A.; Nans, A.; Cheung, A.M.; et al. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat. Commun. 2018, 9, 5061. [Google Scholar] [CrossRef]
- Zhou, J.C. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc. Natl. Acad. Sci. USA 2017, 114, 4141–4146. [Google Scholar] [CrossRef]
- Sun, J.; Shi, Y.; Georgescu, R.E.; Yuan, Z.; Chait, B.T.; Li, H.; O’Donnell, M.E. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015, 22, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Langston, L.D.; Al, E. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl. Acad. Sci. USA 2014, 111, 15390–15395. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; van Deursen, F.; Polychronopoulos, D.; Foltman, M.; Jones, R.C.; Edmondson, R.D.; Calzada, A.; Labib, K. A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009, 28, 2992–3004. [Google Scholar] [CrossRef]
- Yuan, Z.; Georgescu, R.; Santos, R.L.A.; Zhang, D.; Bai, L.; Yao, N.Y.; Al, E. Ctf4 organizes sister replisomes and Pol alpha into a replication factory. Elife 2019, 8, e47405. [Google Scholar] [CrossRef]
- Rzechorzek, N.J.; Hardwick, S.W.; Jatikusumo, V.A.; Chirgadze, D.Y.; Pellegrini, L. CryoEM structures of human CMG–ATPγS–DNA and CMG–AND-1 complexes. Nucleic Acids Res. 2020, 48, 6980–6995. [Google Scholar] [CrossRef]
- Simon, A.C.; Zhou, J.C.; Perera, R.; van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinkovic, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 2014, 510, 7504. [Google Scholar] [CrossRef]
- Villa, F.; Simon, A.C.; Bazan, M.A.O.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovic, D.; Pellegrini, L.; Labib, K. Ctf4 is a hub in the eukaryotic replisome that links multiple CIP-box proteins to the CMG helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Serra-Cardona, A.; Hua, X.; Zhou, H.; Labib, K.; Yu, C.; Zhang, Z. The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell 2018, 72, 140–151. [Google Scholar] [CrossRef]
- Haering, C.H.; Löwe, J.; Hochwagen, A.; Nasmyth, K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 2002, 9, 773–788. [Google Scholar] [CrossRef]
- Haering, C.H.; Farcas, A.-M.; Arumugam, P.; Metson, J.; Nasmyth, K. The cohesin ring concatenates sister DNA molecules. Nature 2008, 454, 297–301. [Google Scholar] [CrossRef]
- Gutierrez-Escribano, P.; Newton, M.D.; Llauró, A.; Huber, J.; Tanasie, L.; Davy, J.; Aly, I.; Aramayo, R.; Montoya, A.; Kramer, H.; et al. A conserved ATP- and Scc2/4-dependent activity for cohesin in tethering DNA molecules. Sci. Adv. 2019, 5, eaay6804. [Google Scholar] [CrossRef]
- Rudra, S.; Skibbens, R.V. Sister chromatid cohesion establishment occurs in concert with lagging strand synthesis. Cell Cycle 2012. [Google Scholar] [CrossRef]
- Murayama, Y.; Samora, C.P.; Kurokawa, Y.; Iwasaki, H.; Uhlmann, F. Establishment of DNA-DNA interactions by the cohesin ring. Cell 2018, 172, 465–477. [Google Scholar] [CrossRef]
- Campbell, J.L.; Cohen-Fix, O. Chromosome cohesion: Ring around the sisters? Trends Biochem. Sci. 2002, 27, 492–495. [Google Scholar] [CrossRef]
- Chan, K.L.; Gligoris, T.; Upcher, W.; Kato, Y.; Shirahige, K.; Nasmyth, K.; Beckouët, F. Pds5 promotes and protects cohesin acetylation. Proc. Natl. Acad. Sci. USA 2013, 13020–13025. [Google Scholar] [CrossRef] [PubMed]
- Guillou, E.; Ibarra, A.; Coulon, V.; Casado-Vela, J.; Rico, D.; Casal, I.; Schwob, E.; Losada, A.; Méndez, J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010, 24, 2812–2822. [Google Scholar] [CrossRef]
- Hanna, J.S.; Kroll, E.S.; Lundblad, V.; Spencer, F.A. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell Biol. 2001, 21, 3144–3158. [Google Scholar] [CrossRef]
- Mayer, M.L.; Pot, I.; Chang, M.; Xu, H.; Aneliunas, V.; Kwok, T.; Newitt, R.; Aebersold, R.; Boone, C.; Brown, G.W.; et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Cell Biol. 2004, 15, 1736–1745. [Google Scholar] [CrossRef]
- Petronczki, M.; Chwalla, B.; Siomos, M.F.; Yokobayashi, S.; Helmhart, W.; Deutschbauer, A.M.; Davis, R.W.; Watanabe, Y.; Nasmyth, K. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 2004, 117, 3547–3559. [Google Scholar] [CrossRef]
- Samora, C.P.; Saksouk, J.; Goswami, P.; Wade, B.O.; Singleton, M.R.; Bates, P.A.; Lengronne, A.; Costa, A.; Uhlmann, F. Ctf4 links DNA replication with sister chromatid cohesion establishment by recruiting the Chl1 helicase to the replisome. Mol. Cell 2016, 63, 371–384. [Google Scholar] [CrossRef]
- Rudra, S.; Skibbens, R.V. Chl1 DNA helicase regulates Scc2 deposition specifically during DNA-Replication in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e75435. [Google Scholar] [CrossRef]
- Stokes, K.; Winczura, A.; Song, B.; De Piccoli, G.; Grabarczyk, D.B. Ctf18-RFC and DNA Pol ϵ form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication. Nucleic Acids Res. 2020, 48, 8128–8145. [Google Scholar] [CrossRef]
- Grabarczyk, D.B.; Silkenat, S.; Kisker, C. Structural basis for the recruitment of Ctf18-RFC to the replisome. Cell 2018, 26, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, V.P.; Maniwa, Y.; Tappin, I.; Ozato, K.; Yokomori, K.; Hurwitz, J. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 10237–10242. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shahar, T.R.; Heeger, S.; Lebane, C.; East, P.; Flynn, H.; Skehel, M.; Uhlmann, F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 2008, 321, 563–566. [Google Scholar] [CrossRef]
- Tóth, A.; Ciosk, R.; Uhlmann, F.; Galova, M.; Schleiffer, A.; Nasmyth, K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999, 13, 320–333. [Google Scholar] [CrossRef]
- Lengronne, A.; Al, E. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell 2006, 787–799. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 2006, 723–732. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, X.; Li, Y.; Kim, B.J.; Jia, J.; Huang, Z.; Yang, T.; Fu, X.; Jung, S.Y.; Wang, Y.; et al. Acetylation of Smc3 by Eco1 Is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 2008, 31, 143–151. [Google Scholar] [CrossRef]
- Ivanov, D.; Schleiffer, A.; Eisenhaber, F.; Mechtler, K.; Haering, C.H.; Nasmyth, K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 2002, 323–328. [Google Scholar] [CrossRef]
- Nasmyth, K.; Haering, C.H. Cohesin: Its role and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef]
- Chao, W.C.H.; Wade, B.O.; Bouchoux, C.; Jones, A.W.; Purkiss, A.G.; Federico, S.; O’Reilly, N.; Snijders, A.P.; Uhlmann, F.; Singleton, M.R. Structural basis of Eco1-mediated cohesin acetylation. Sci. Rep. 2017, 7, 44313. [Google Scholar] [CrossRef]
- Chan, K.-L.; Roig, M.B.; Hu, B.; Beckouët, F.; Metson, J.; Nasmyth, K. Cohesin’s DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 2012, 150, 961–974. [Google Scholar] [CrossRef]
- De, K.; Sterle, L.; Krueger, L.; Yang, X.; Makaroff, C.A. Arabidopsis thaliana WAPL is essential for the prophase removal of cohesin during meiosis. PLoS Genet. 2014. [Google Scholar] [CrossRef]
- Feytout, A.; Vaur, S.; Genier, S.; Vazquez, S.; Javerzat, J.-P. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol. Cell. Biol. 2011, 31, 1771–1786. [Google Scholar] [CrossRef] [PubMed]
- Rowland, B.D.; Roig, M.B.; Nishino, T.; Kurze, A.; Uluocak, P.; Mishra, A.; Beckouët, F.; Underwood, P.; Metson, J.; Imre, R.; et al. Building sister chromatid cohesion: Smc3 acetylation counteracts an antiestablishment activity. Mol. Cell 2009, 33, 763–774. [Google Scholar] [CrossRef]
- Sutani, T.; Kawaguchi, T.; Kanno, R.; Itoh, T.; Shirahige, K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 2009, 19, 492–497. [Google Scholar] [CrossRef]
- Pollard, T.D.; Earnshaw, W.C.; Lippincott-Schwartz, J.; Johnson, G.T. Introduction to the cell cycle. In Cell Biology; Elsevier: Philadelphia, PA, USA, 2017. [Google Scholar]
- Flemming, W. Zellsubstanz, Kern und Zellheilung; F.C.W. Vogel: Leipzig, Germany, 1882. [Google Scholar]
- Borde, V.; de Massy, B. Meiosis: Early DNA double-strand breaks pave the way for inter-homolog repair. Dev. Cell 2015, 32, 663–664. [Google Scholar] [CrossRef]
- Lao, J.P.; Hunter, N. Trying to avoid your sister. PLoS Biol. 2010, 8, e1000519. [Google Scholar] [CrossRef]
- Kim, K.P.; Weiner, B.M.; Zhang, L.; Jordan, A.; Dekker, J.; Kleckner, N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 2010, 143, 924–937. [Google Scholar] [CrossRef]
- Uhlmann, F.; Wernic, D.; Poupart, M.-A.; Koonin, E.V.; Nasmyth, K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 2000, 103, 375–386. [Google Scholar] [CrossRef]
- Nieduszynski, C.A.; Knox, Y.; Donaldson, A.D. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006, 20, 1874–1879. [Google Scholar] [CrossRef]
- Jackson, D.A.; Pombo, A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998, 140, 1285–1295. [Google Scholar] [CrossRef]
- Berezney, R.; Dubey, D.D.; Huberman, J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 2000, 108, 471–484. [Google Scholar] [CrossRef]
- Terret, M.-E.; Sherwood, R.; Rahman, S.; Qin, J.; Jallepalli, P.V. Cohesin acetylation speeds the replication fork. Nature 2009, 462, 231–234. [Google Scholar] [CrossRef]
- Liu, J.; Krantz, I.D. Cohesin and human disease. Annu. Rev. Genom. Hum. Genet. 2008, 9, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Gerlich, D.; Koch, B.; Dupeux, F.; Peters, J.-M.; Ellenberg, J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 2006, 16, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Boone, C.; Brown, G.W. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 2007, 176, 1417–1429. [Google Scholar] [CrossRef]
- Srinivasan, M.; Fumasoni, M.; Petela, N.J.; Murray, A.; Nasmyth, K.A. Cohesion is established during DNA replication utilising chromosome associated cohesin rings as well as those loaded de novo onto nascent DNAs. Elife 2020, 9, e56611. [Google Scholar] [CrossRef] [PubMed]
- Akai, Y.; Kurokawa, Y.; Nakazawa, N.; Tonami-Murakami, Y.; Suzuki, Y.; Yoshimura, S.H.; Iwasaki, H.; Shiroiwa, Y.; Nakamura, T.; Shibata, E.; et al. Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol. 2011, 1, 110023. [Google Scholar] [CrossRef]
- P.Rhodes, J.D.; Haarhuis, J.H.I.; Grimm, J.B.; Rowland, B.D.; Lavis, L.D.; Nasmyth, K. Cohesin can remain associated with chromosomes during DNA replication. Cell Rep. 2017, 20, 2749–2755. [Google Scholar] [CrossRef]
- Bernard, P.; Schmidt, C.K.; Vaur, S.; Dheur, S.; Drogat, J.; Genier, S.; Ekwall, K.; Uhlmann, F.; Javerzat, J. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 2007, 27, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Stigler, J.; Çamdere, G.Ö.; Koshland, D.E.; Greene, E.C. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 2016, 15, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Käshammer, L.; Saathoff, J.-H.; Lammens, K.; Gut, F.; Barth, J.; Alt, A.; Kessler, B.; Hopfner, K.-P. Mechanism of DNA end sensing and processing by the Mre11-Rad50 complex. Mol. Cell 2019, 76, 382–394.e6. [Google Scholar] [CrossRef] [PubMed]
- Dequeker, B.J.H.; Brandão, H.B.; Scherr, M.J.; Gassler, J.; Powell, S.; Gaspar, I.; Flyamer, I.M.; Tang, W.; Stocsits, R.; Davidson, I.F.; et al. MCM complexes are barriers that restrict cohesin-mediated loop extrusion. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ryu, J.-K.; Bouchoux, C.; Liu, H.W.; Kim, E.; Minamino, M.; de Groot, R.; Katan, A.J.; Bonato, A.; Marenduzzo, D.; Michieletto, D.; et al. Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci. Adv. 2021, 7, eabe5905. [Google Scholar] [CrossRef]
- Weitzer, S.; Lehane, C.; Uhlmann, F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003, 13, 1930–1940. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Comerci, C.J.; McCarthy, D.G.; Saurabh, S.; Moerner, W.E.; Wysocka, J. Opposing effects of cohesin and transcription on CTCF organization revealed by super-resolution imaging. Mol. Cell 2020, 80, 699–711.e7. [Google Scholar] [CrossRef]
- Hansen, A.S.; Pustova, I.; Cattoglio, C.; Tjian, R.; Darzacq, X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 2017, 6, e25776. [Google Scholar] [CrossRef]
- Huis, P.J.; Herzog, F.; Ladurner, R.; Davidson, I.F.; Piric, S.; Kreidl, E.; Bhaskara, V.; Aebersold, R.; Peters, J.M. Characterization of a DNA exit gate in the human cohesin ring. Science 2014, 346, 968–972. [Google Scholar] [CrossRef]
- Mahamid, J.; Pfeffer, S.; Schaffer, M.; Villa, E.; Danev, R.; Cuellar, L.K.; Förster, F.; Hyman, A.A.; Plitzko, J.M.; Baumeister, W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016, 26, 969–972. [Google Scholar] [CrossRef]
- Turk, M.; Baumeister, W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 2020, 594, 3243–3261. [Google Scholar] [CrossRef] [PubMed]
- Mäeots, M.-E.; Lee, B.; Nans, A.; Jeong, S.-G.; Esfahani, M.M.N.; Ding, S.; Smith, D.J.; Lee, C.-S.; Lee, S.S.; Peter, M.; et al. Modular microfluidics enables kinetic insight from time-resolved cryo-EM. Nat. Commun. 2020, 11, 3465. [Google Scholar] [CrossRef] [PubMed]
- Dandey, V.P.; Budell, W.C.; Wei, H.; Bobe, D.; Maruthi, K.; Kopylov, M.; Eng, E.T.; Kahn, P.A.; Hinshaw, J.E.; Kundu, N.; et al. Time-resolved cryo-EM using Spotiton. Nat. Methods 2020, 17, 897–900. [Google Scholar] [CrossRef]
- Kaledhonkar, S.; Fu, Z.; White, H.; Frank, J. Time-resolved cryo-electron microscopy using a microfluidic chip. In Protein Complex Assembly; Springer: Berlin/Heidelberg, Germany, 2018; pp. 59–71. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henrikus, S.S.; Costa, A. Towards a Structural Mechanism for Sister Chromatid Cohesion Establishment at the Eukaryotic Replication Fork. Biology 2021, 10, 466. https://doi.org/10.3390/biology10060466
Henrikus SS, Costa A. Towards a Structural Mechanism for Sister Chromatid Cohesion Establishment at the Eukaryotic Replication Fork. Biology. 2021; 10(6):466. https://doi.org/10.3390/biology10060466
Chicago/Turabian StyleHenrikus, Sarah S., and Alessandro Costa. 2021. "Towards a Structural Mechanism for Sister Chromatid Cohesion Establishment at the Eukaryotic Replication Fork" Biology 10, no. 6: 466. https://doi.org/10.3390/biology10060466
APA StyleHenrikus, S. S., & Costa, A. (2021). Towards a Structural Mechanism for Sister Chromatid Cohesion Establishment at the Eukaryotic Replication Fork. Biology, 10(6), 466. https://doi.org/10.3390/biology10060466





